Submitted:
21 June 2023
Posted:
21 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Specimen Examination, and Isolation
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analyses
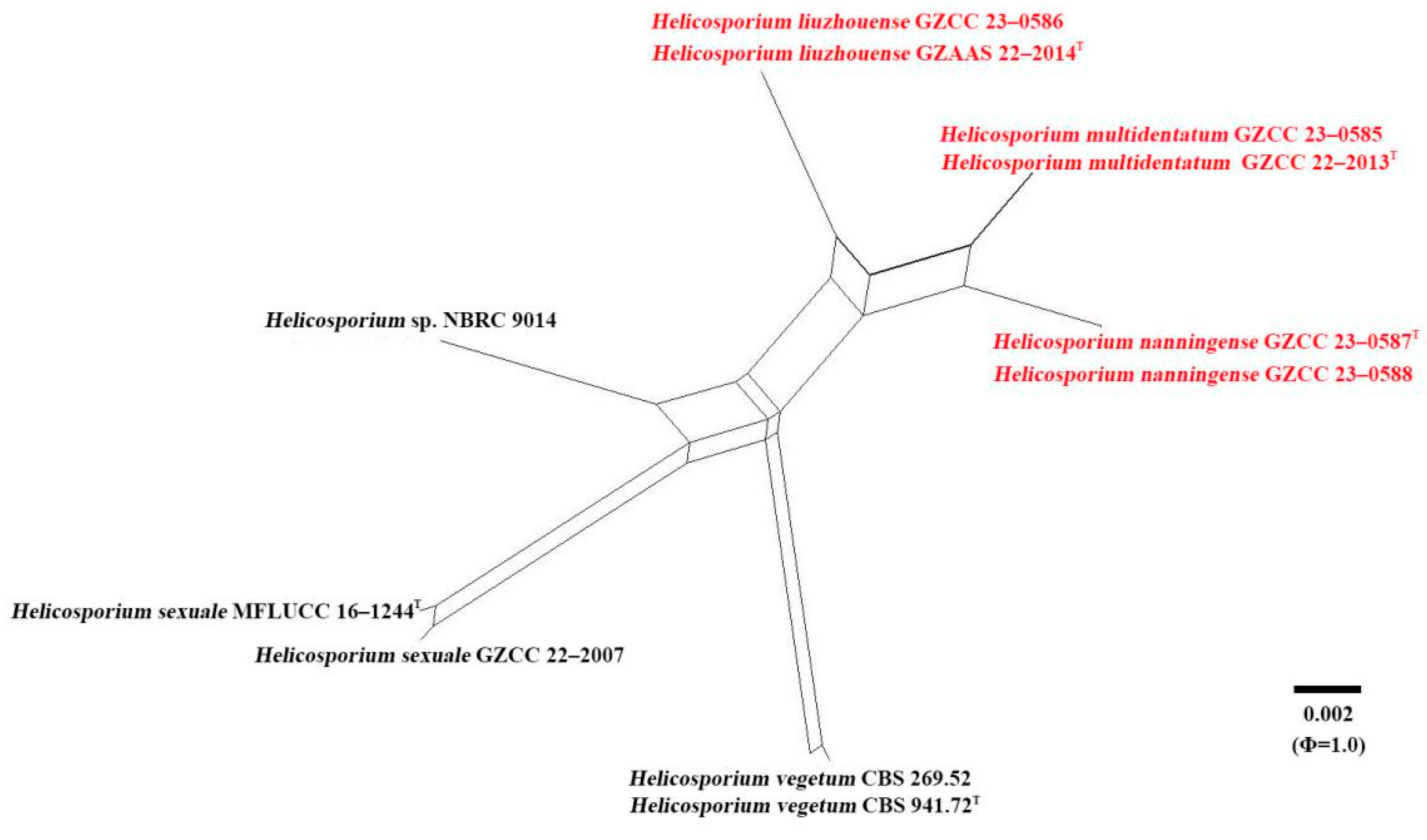
2.4. Genealogical Concordance Phylogenetic Species Recognition (GCPSR) analysis
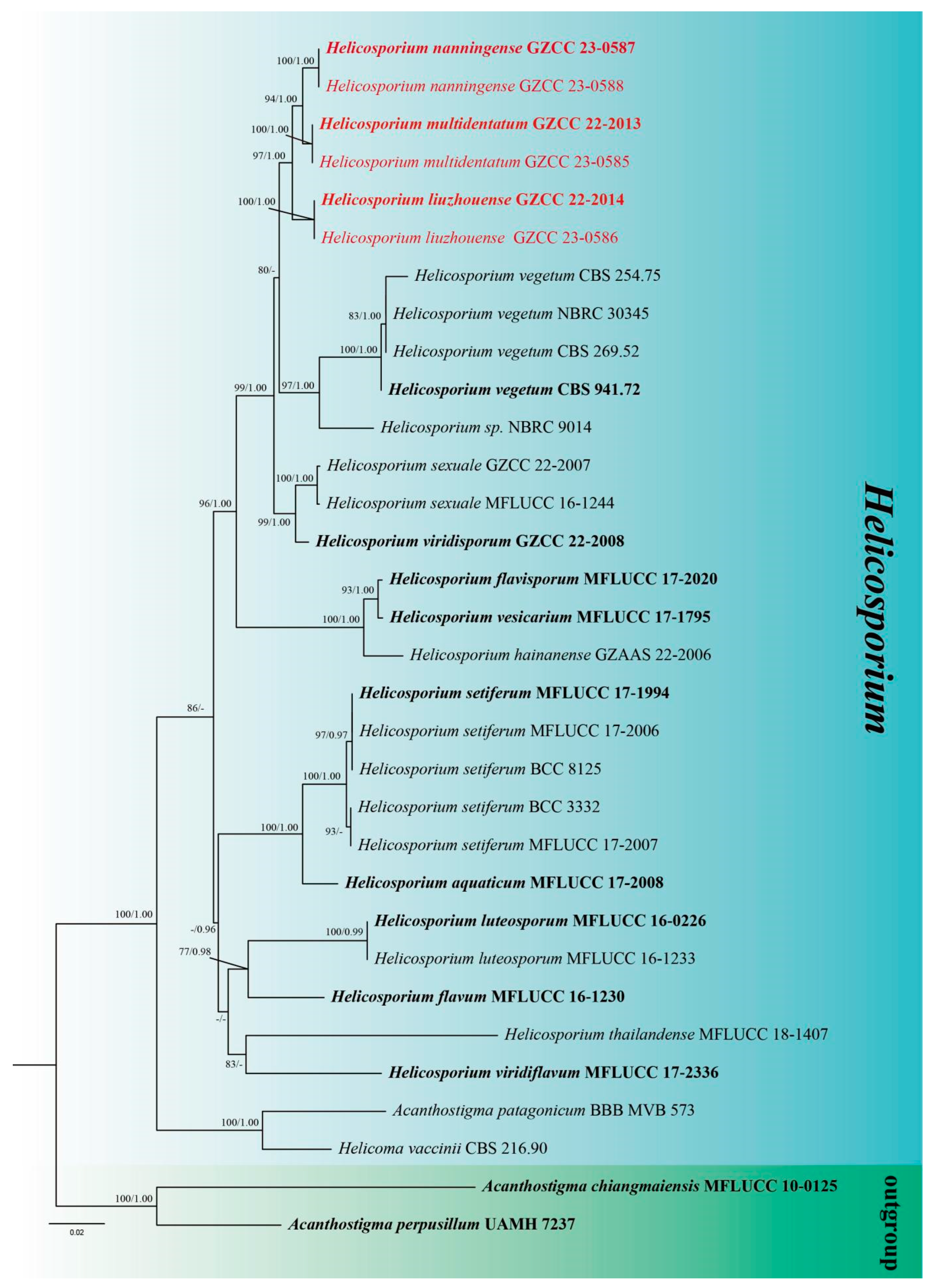
3. Phylogenetic Results
4. Genealogical Concordance Phylogenetic Species Recognition (GCPSR) analysis
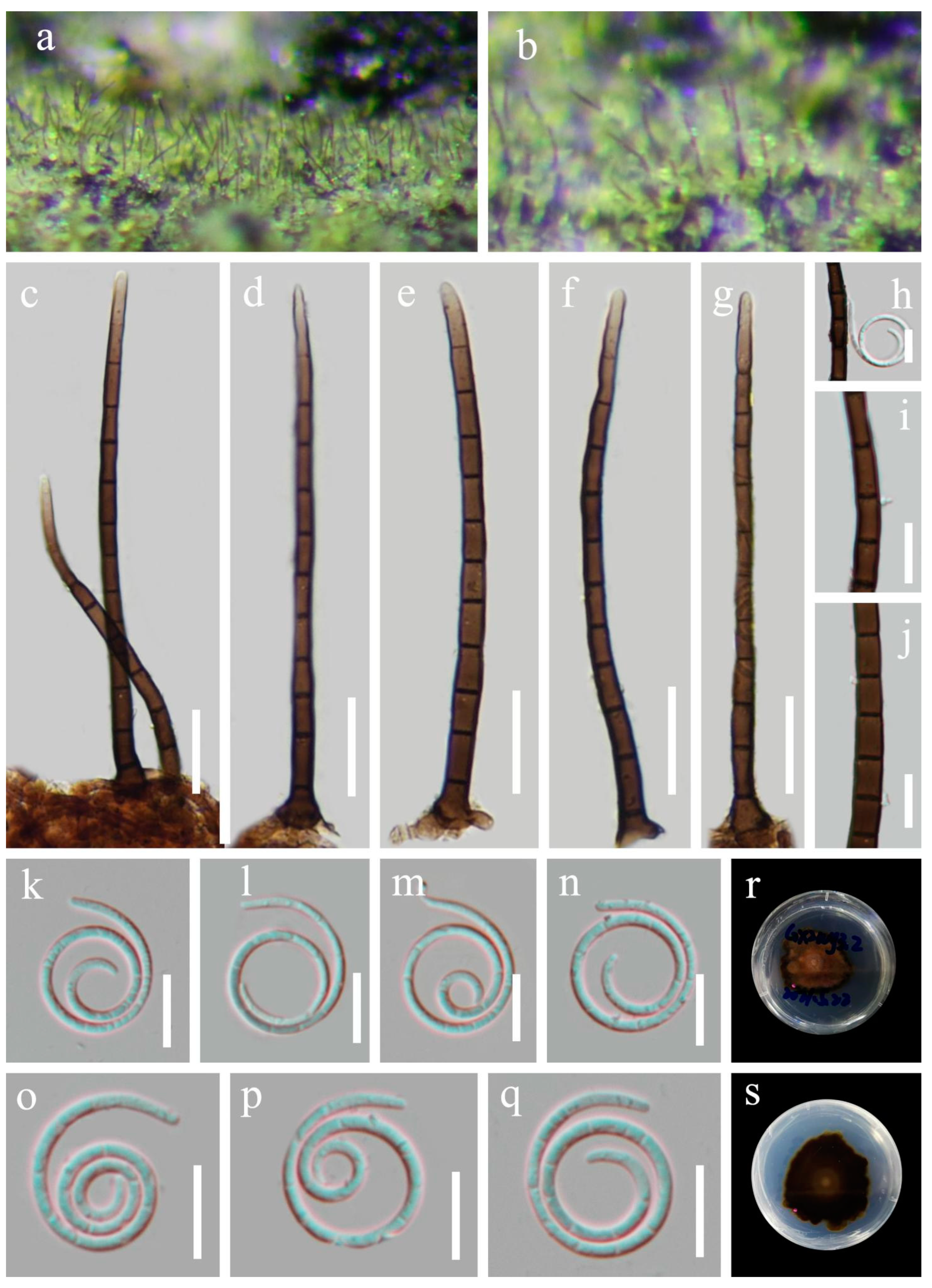
5. Taxonomy
6. Discussion
| 1. Ascomata present | 2 |
| 1. Ascomata lacking | 4 |
| 2. Ascomata bright yellow to brown yellow, without setae | H. vegetum |
| 2. Ascomata bright yellow to brown yellow, with sparsely setae | 3 |
| 3. Ascomata globose to subglobose, 185–235 µm high, 116–214 µm diam. | H. sexuale |
| 3. Ascomata globose to subglobose, 320–350 µm high, 220–260 µm diam. | H. flavum |
| 4. Fresh colonies on decaying woody substrate are bright green | 5 |
| 4. Fresh colonies on decaying woody substrate are hyaline, gray, yellow, yellow green, or dark brown | 7 |
| 5. Conidiophores unbranched, with multi-dentate protrusions conidiogenous cells | H. multidentatum |
| 5. Conidiophores unbranched, with tiny tooth-like protrusions conidiogenous cells | 6 |
| 6. Conidiophores < 120 μm long | H. nanningense |
| 6. Conidiophores > 120 μm long | H. liuzhouense or H. viridisporum |
| 7. Fresh colonies on decaying woody substrate are yellow green | 8 |
| 7. Fresh colonies on decaying woody substrate are hyaline, gray, yellow, or dark brown | 14 |
| 8. Conidiophores with bladder-like protrusions conidiogenous cells | 9 |
| 8. Conidiophores with tiny tooth-like protrusions conidiogenous cells | 10 |
| 9. Conidiophores < 120 μm long | H. vesicarium |
| 9. Conidiophores > 120 μm long | H. hainanense |
| 10. Conidia are acropleurogenous | H. thailandense |
| 10. Conidia are pleurogenous and apical sterile in conidiophores | 11 |
| 11. Conidiophores < 180 μm long | 12 |
| 11. Conidiophores > 180 μm long | 13 |
| 12. Conidia 10–14 µm diam., 70–90 µm long | H. aquaticum |
| 12. Conidia 13–21 µm diam., 100–130 µm long | H. setiferum |
| 13. Conidiophores 184–257 µm long | H. flavidum |
| 13. Conidiophores 250–425 µm long | H. viridiflavum |
| 14. Fresh colonies on decaying woody substrate are yellow | 15 |
| 14. Fresh colonies on decaying woody substrate are hyaline, gray, or dark brown | 16 |
| 15. Conidiophores with bladder-like protrusions conidiogenous cells | H. flavisporum |
| 15. Conidiophores with tiny tooth-like protrusions conidiogenous cells | H. luteosporum |
| 16. Fresh colonies on decaying woody substrate are hyaline, branched conidiophores 200–300 µm long | H. albidum |
| 16. Fresh colonies on decaying woody substrate are gray, or dark brown | 17 |
| 17. Fresh colonies on decaying woody substrate are gray, conidiophores 350–400 µm long, with bladder-like protrusions conidiogenous cells | H. neesii |
| 17. Fresh colonies on decaying woody substrate are dark brown | 18 |
| 18. Conidiophores are unbranched with tooth-like protrusions conidiogenous cells | H. murinum |
| 18. Conidiophores are rarely branched | 19 |
| 19. Tooth- or bladder-like protrusions conidiogenous cells, conidia 6–9 µm diam. | H. decumbens |
| 19. Tooth-like protrusions conidiogenous cells, conidia 11.4–19 µm diam. | H. melghatianum |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nees Von Esenbeck, C.D.G. System der Pilze und Schwämme. Würtzburg, 1817; 1–334. [Google Scholar]
- Linder, D.H. A monograph of the helicosporous fungi imperfecti. Ann. Mo. Bot. Gard. 1929, 16, 227–388. [Google Scholar] [CrossRef]
- Goos, R.D. On the anamorph genera Helicosporium and Drepanospora. Mycologia 1989, 81, 356–374. [Google Scholar] [CrossRef]
- Ho, W.H.; Hyde, K.D.; Hodgkiss, I.J. Seasonality and sequential occurrence of fungi on wood submerged in Tai Po Kau Forest Stream, Hong Kong. Fungal Divers. 2002, 10, 21–43. [Google Scholar]
- Pinruan, U.; Hyde, K.D.; Lumyong, S.; McKenzie, E.H.C.; Jones, E.B.G. Occurrence of fungi on tissues of the peat swamp palm Licuala longicalycata. Fungal Divers. 2007, 25, 157–173. [Google Scholar]
- Zhao, G.Z.; Liu, X.Z.; Wu, W.P. Helicosporous hyphomycetes from China. Fungal Divers. 2007, 26, 313–524. [Google Scholar]
- Boonmee, S.; Rossman, A.Y.; Liu, J.K.; Li, W.J.; Dai, D.Q.; Bhat, J.D.; Jones, E.B.G.; McKenzie, E.H.C.; Xu, J.C.; Hyde, K.D. Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers. 2014, 68, 239–298. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.U.; Jones, G.E.B.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Boonmee, S.; Bhat, D.J.; Hyde, K.D.; Kang, J.C. Helicosporium luteosporum sp. nov. and Acanthohelicospora aurea (Tubeufiaceae, Tubeufiales) from terrestrial habitats. Phytotaxa 2017, 319, 241–253. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Liu, J.K.; Hyde, K.D.; Jeewon, R.; Kang, J.C.; Fan, C.; Boonmee, S.; Bhat, D.J.; Luo, Z.L.; Lin, C.G.; Eungwanichayapant, P.D. A taxonomic reassessment of Tubeufiales based on multi-locus phylogeny and morphology. Fungal Divers. 2018, 92, 131–344. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Ma, J.; Xiao, X.J.; Zhang, L.J.; Xiao, Y.P.; Kang, J.C. Four new species and three new records of helicosporous hyphomycetes from China and their multi-gene phylogenies. Front. Microbiol. 2022, 13, 1053849. [Google Scholar] [CrossRef]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: https://www.indexfungorum.org/Names/Names.asp (accessed on 20 May 2023).
- Brahmanage, R.S.; Lu, Y.Z.; Bhat, D.J.; Wanasinghe, D.N.; Yan, J.Y.; Hyde, K.D.; Boonmee, S. Phylogenetic investigations on freshwater fungi in Tubeufiaceae (Tubeufiales) reveals the new genus Dictyospora and new species Chlamydotubeufia aquatica and Helicosporium flavum. Mycosphere 2017, 8, 917–933. [Google Scholar] [CrossRef]
- Hardy, G.E.S.J.; Sivasithamparam, K. Antagonism of fungi and actinomycetes isolated from composted Eucalyptus bark to Phytophthora drechsleri in a steamed and non-steamed composted Eucalyptus bark-amended container medium. Soil Biol. Biochem. 1995, 27, 243–246. [Google Scholar] [CrossRef]
- Choi, H.J.; Lee, S.M.; Kim, S.H.; Kim, D.W.; Choi, Y.W.; Joo, W.H. A novel Helicosporium isolate and its antimicrobial and cytotoxic pigment. J. Microbiol. and Biotechn. 2012, 22, 1214–1217. [Google Scholar] [CrossRef]
- Chomnunti, P.; Hongsanan, S.; Hudson, B.A.; Tian, Q.; Peršoh, D.; Dhami, M.K.; Alias, A.S.; Xu, J.C.; Liu, X.Z.; Stadler, M.; Hyde, K.D. The sooty moulds. Fungal Divers. 2014, 66, 1–36. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97(1), 84–98. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Boonmee, S.; Dai, D.Q.; Liu, J.K.; Hyde, K.D.; Bhat, D.J.; Ariyawansa, H.; Kang, J.C. Four new species of Tubeufia (Tubeufiaceae, Tubeufiales) from Thailand. Mycol. Prog. 2017, 16, 403–417. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Boonmee, S.; Liu, J.K.; Hyde, K.D.; McKenzie, E.H.C.; Eungwanichayapant, P.D.; Kang, J.C. Multi-gene phylogenetic analyses reveals Neohelicosporium gen. nov. and five new species of helicosporous hyphomycetes from aquatic habitats. Mycol. Prog. 2018, 17, 631–646. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, J.Y.; Xiao, X.J.; Xiao, Y.P.; Tang, X.; Boonmee, S.; Kang, J.C.; Lu, Y.Z. Multi-gene phylogenetic analyses revealed five new species and two new records of Distoseptisporales from China. J. Fungi 2022, 8. [Google Scholar] [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Goos, R.D. Some helicosporous fungi from Hawaii. Mycologia 1980, 72, 595–610. [Google Scholar] [CrossRef]
- Goos, R.D. A review of the anamorph genus Helicomyces. Mycologia 1985, 77, 606–618. [Google Scholar] [CrossRef]
- Goos, R.D. A review of the anamorph genus Helicoma. Mycologia 1986, 78, 744–761. [Google Scholar] [CrossRef]
- Luo, Z.L.; Bhat, D.J.; Jeewon, R.; Boonmee, S.; Bao, D.F.; Zhao, Y.C.; Chai, H.M.; Su, H.Y.; Su, X.J.; Hyde, K.D. Molecular phylogeny and morphological characterization of asexual fungi (Tubeufiaceae) from freshwater habitats in Yunnan, China. Cryptogamie Mycol. 2017, 38, 27–53. [Google Scholar] [CrossRef]
- Liu, J.K.; Lu, Y.Z.; Cheewangkoon, R.; To-Anun, C. Phylogeny and morphology of Helicotubeufia gen. nov, with three new species in Tubeufiaceae from aquatic habitats. Mycosphere 2018, 9, 495–509. [Google Scholar] [CrossRef]
- Tian, X.; Karunarathna, S.C.; Xu, R.; Lu, Y.; Suwannarach, N.; Mapook, A.; Bao, D.; Xu, J.; Tibpromma, S. Three new species, two new records and four new collections of Tubeufiaceae from Thailand and China. J. Fungi 2022, 8. [Google Scholar] [CrossRef]
- Zeng, X.; Qian, S.; Lu, Y.; Li, Y.; Chen, L.; Qian, Y.; He, Z.; Kang, J. A novel Nitrogen-containing Glyceride from fungal saprobe Tubeufia rubra reverses MDR of tumor cell lines to Doxorubicin. Rec. Nat. Prod. 2022, 16, 622–632. [Google Scholar] [CrossRef]
- Zhang, L.J.; Yang, M.F.; Ma, J.; Xiao, X.J.; Ma, X.Y.; Zheng, D.G.; Han, M.Y.; Xia, M.L.; Jayawardena, R.S.; Mapook, A.; et al. Neogrisphenol A, a potential ovarian cancer inhibitor from a new record fungus Neohelicosporium griseum. Metabolites 2023, 13, 435. [Google Scholar] [CrossRef]
- Zheng, W.; Han, L.; He, Z.J.; Kang, J.C. A new alkaloid derivative from the saprophytic fungus Neohelicomyces hyalosporus PF11-1. Nat. Prod. Res. 2023, 1–5. [Google Scholar] [CrossRef]
- Qian, S.Y.; Zeng, X.B.; Qian, Y.X.; Lu, Y.Z.; He, Z.J.; Kang, J.C. A saprophytic fungus Tubeufia rubra Produces novel rubracin D and E reversing multidrug resistance in cancer cells. J. Fungi 2023, 9, 309. [Google Scholar] [CrossRef]
- Link, H.F. Observationes in ordines plantarum naturales. Dissertatio I. Mag Ges Naturf Fr. Berl. 1809, 3, 3–42. [Google Scholar]
- Corda, A.K.J. Icones fungorum hucusque cognitorum, Vol 1. Prague: J.G. Calve, 1837, 1–32.
- Moore, R.T. Index to the Helicosporae. Mycologia 1955, 47, 90–103. [Google Scholar] [CrossRef]
- Tsui, C.K.; Sivichai, S.; Berbee, M.L. Molecular systematics of Helicoma, Helicomyces and Helicosporium and their teleomorphs inferred from rDNA sequences. Mycologia 2006, 98, 94–104. [Google Scholar] [CrossRef]
- Kuo, C.H.; Goh, T.K. Two new species of helicosporous hyphomycetes from Taiwan. Mycol. Prog. 2018, 17, 557–569. [Google Scholar] [CrossRef]
- Dharkar, N.; Subhedar, A.; Hande, D.; Shahezad, M.A. Two new fungal species from Vidarbha, India. J. Mycol. Plant Pathol. 2010, 40, 235–237. [Google Scholar]
- Grove, W.B. New or noteworthy fungi. III. J. Bot. Br. Foreign 1886, 24, 197–207. [Google Scholar]
- Hsieh, S.Y.; Goh, T.K.; Kuo, C.H. New species and records of Helicosporium sensu lato from Taiwan, with a reflection on current generic circumscription. Mycol. Prog. 2021, 20, 169–190. [Google Scholar] [CrossRef]
- Moore, R.T. Index to the Helicosporae: addenda. Mycologia 1957, 49, 580–587. [Google Scholar] [CrossRef]


| Taxon | Strain | GenBank Accessions | |||
|---|---|---|---|---|---|
| ITS | LSU | tef1α | rpb2 | ||
| Acanthostigma chiangmaiensis | MFLUCC 10-0125T | JN865209 | JN865197 | KF301560 | - |
| Acanthostigma patagonicum | BBB MVB 573 | JN127358 | JN127359 | - | - |
| Acanthostigma perpusillum | UAMH 7237T | AY916492 | AY856892 | - | - |
| Helicoma vaccinii | CBS 216.90 | AY916486 | AY856879 | - | - |
| Helicosporium aquaticum | MFLUCC 17-2008T | MH558733 | MH558859 | MH550924 | MH551049 |
| Helicosporium flavisporum | MFLUCC 17-2020T | MH558734 | MH558860 | MH550925 | MH551050 |
| Helicosporium flavum | MFLUCC 16-1230T | KY873626 | KY873621 | KY873285 | - |
| Helicosporium hainanense | GZCC 22-2006T | OP508730 | OP508770 | OP698081 | OP698070 |
| Helicosporium liuzhouense | GZCC 22-2014T | OQ981394 | OQ981402 | OQ980476 | OQ980474 |
| Helicosporium liuzhouense | GZCC 23-0586 | OR066416 | OR066423 | OR058862 | OR058855 |
| Helicosporium luteosporum | MFLUCC 16-0226T | KY321324 | KY321327 | KY792601 | - |
| Helicosporium luteosporum | MFLUCC 16-1233 | - | KY873624 | - | - |
| Helicosporium multidentatum | GZCC 22-2013T | OQ981395 | OQ981403 | OQ980477 | OQ980475 |
| Helicosporium multidentatum | GZCC 23-0585 | OR066417 | OR066424 | OR058863 | OR058856 |
| Helicosporium nanningense | GZCC 22-2175T | OR066418 | OR066425 | OR058864 | OR058857 |
| Helicosporium nanningense | GZCC 23-0588 | OR066419 | OR066426 | OR058865 | OR058858 |
| Helicosporium setiferum | BCC 3332 | AY916490 | AY856907 | - | - |
| Helicosporium setiferum | BCC 8125 | AY916491 | - | - | - |
| Helicosporium setiferum | MFLUCC 17-1994T | MH558735 | MH558861 | MH550926 | MH551051 |
| Helicosporium setiferum | MFLUCC 17-2006 | MH558736 | MH558862 | MH550927 | MH551052 |
| Helicosporium setiferum | MFLUCC 17-2007 | MH558737 | MH558863 | MH550928 | MH551053 |
| Helicosporium sexuale | GZCC 22-2007 | OP508731 | OP508771 | OP698082 | OP698071 |
| Helicosporium sexuale | MFLUCC 16-1244T | MZ538503 | MZ538537 | MZ567082 | MZ567111 |
| Helicosporium sp. | NBRC 9014 | AY916489 | AY856903 | - | - |
| Helicosporium thailandense | MFLUCC 18-1407T | MT627698 | MN913718 | MT954371 | - |
| Helicosporium vegetum | CBS 254.75 | - | DQ470982 | DQ471105 | - |
| Helicosporium vegetum | CBS 269.52 | AY916487 | AY856893 | - | - |
| Helicosporium vegetum | CBS 941.72T | AY916488 | AY856883 | - | - |
| Helicosporium vegetum | NBRC 30345 | - | AY856896 | - | - |
| Helicosporium vesicarium | MFLUCC 17-1795T | MH558739 | MH558864 | MH550930 | MH551055 |
| Helicosporium viridiflavum | MFLUCC 17-2336T | MH558738 | - | MH550929 | MH551054 |
| Helicosporium viridisporum | GZCC 22-2008T | OP508736 | OP508776 | OP698087 | OP698076 |
| Species | Habitats | Distribution | Molecular data | References |
|---|---|---|---|---|
| H. albidum | Terrestrial | Belgium, Britain (Birminghan) | No | Grove 1886 |
| H. aquaticum | Freshwater | Thailand | Yes | Lu et al. 2018b |
| H. decumbens | Terrestrial | Austria, Brazil | No | Linder 1929 |
| flavidum | Freshwater | China | No | Hsieh 2021 |
| H. flavisporum | Freshwater | Thailand | Yes | Lu et al. 2018b |
| H. flavum | Freshwater | Thailand | Yes | Brahamanage et al. (2017) |
| H. hainanense | Terrestrial | China | Yes | Lu et al. 2022 |
| H. liuzhouense | Freshwater | China | Yes | This study |
| H. luteosporum | Terrestrial | Thailand | Yes | Lu et al. 2017a |
| H. melghatianum | Terrestrial | India | No | Dharkar et al. 2010 |
| H. murinum | Terrestrial | Argentina, Austria, Brazil, Canada, China, Cuba, Malaysia, USA | No | Linder 1929; Goos 1989; Zhao et al. 2007 |
| H. multidentatum | Terrestrial | China | Yes | This study |
| H. nanningense | Terrestrial | China | Yes | This study |
| H. neesii | Terrestrial | USA | No | Moore 1957 |
| H. setiferum | Freshwater | Thailand | Yes | Lu et al. 2018b |
| H. sexuale | Freshwater, Terrestrial | China, Thailand | Yes | Boonmee et al. 2021; Lu et al. 2022 |
| H. thailandense | Freshwater | Thailand | Yes | Dong et al. 2020 |
| H. vegetum | Terrestrial | Worldwide | Yes | Boonmee et al. 2014 |
| H. vesicarium | Freshwater | Thailand | Yes | Lu et al. 2018b |
| H. viridiflavum | Terrestrial | Thailand | Yes | Lu et al. 2018b |
| viridisporum | Freshwater | China | Yes | Lu et al. 2022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





