1. Introduction
The The Pacific oyster (
Crassostrea gigas) is one of the world’s most commonly cultivated shellfish [
1].
Ostreid herpesvirus 1 (OsHV-1) is a viral pathogen of Pacific oysters that has been implicated in numerous mortality outbreaks in this species [
2]. OsHV-1 is a member of the
Herpesvirales order, which contains herpesviruses that infect and cause disease in a wide range of vertebrate and invertebrate animal hosts [
3]. The
Herpesvirales order contains the
Alloherpesviridae,
Herpesviridae, and
Malacoherpesviridae families. Fish and amphibian herpesviruses belong to the
Alloherpesviridae family; avian, mammalian, and reptilian herpesviruses belong to the
Herpesviridae family; and molluscan herpesviruses, including OsHV-1, belong to the
Malacoherpesviridae family [
4].The
Malacoherpesviridae family comprises two genera with one species each (ICTV 2019), OsHV-1 and
Haliotid herpesvirus 1 (HaHV-1). Multiple variants of OsHV-1 exist with different levels of virulence in Pacific oysters [
5,
6,
7]; however, the mutations in the viral genome that lead to differences in virulence have yet to be determined.
One of the unique features of herpesviruses is a latent infection following an initial lytic infection [
8]. Upon initial infection of a susceptible host, the virus enters a lytic state where it replicates exponentially at the entry site and in peripheral tissues. Following the initial infection, the virus can enter a latent state in the surviving hosts in either peripheral nerve ganglia neurons or lymphatic cells [
8]. During latent infections, the viral genome usually is “hiding” in a few cells and remains dormant and therefore causes no harm to the infected host. However, the latent viral genome can be reactivated to produce infectious viral progeny under stress conditions, such as elevated temperature, injury, or immune suppression. Latency and reactivation have been well-studied in herpesviruses in the
Alloherpesviridae and
Herpesviridae [
9,
10,
11], but little is known about these processes in the
Malacoherpesviridae despite their economic importance.
OsHV-1 latency has important implications for Pacific oyster aquaculture. Oysters are often moved around for different purposes, e.g., adults from the field are often moved into hatcheries as broodstock, and these transfers have the potential to unintentionally spread OsHV-1. Detecting latent infections can be difficult due to the small number of cells that are infected [
9]. This can be a problem because a false negative can result in the unintentional introduction of OsHV-1 into a hatchery or a bay. Currently recommended OIE methods for the detection of OsHV-1, specifically qPCR on DNA extracted from oyster tissues [
12], cannot detect latent infection; for example, Pernet et al. (2015) found OsHV-1 lytic infection occurred in oysters that had tested negative when seawater temperature was suddenly raised to 21°C [
13]. One way of increasing the likelihood of detecting latent infections is to test the tissue where OsHV-1 becomes latent and use more sensitive methods to detect low copy numbers of viral DNA. The predominate sites of latency of the
Alloherpesviridae and
Herpesviridae are either neurons or immune cells [
9,
11,
14]; however, the location of latency for OsHV-1 has yet to be determined. In this study, OsHV-1 latency was investigated in both experimentally infected oysters and survivors of an OsHV-1 mortality event in the field.
2. Materials and Methods
2.1. Virus stock
OsHV-1 stocks were prepared using moribund oysters from a mortality event in Tomales Bay, California, in 2019, as previously described by Divilov et al. [
15]. Whole oyster tissues were ground with a mortar and pestle in phosphate-buffered saline (PBS) at a 1:5 ratio (w/v). The homogenized tissues were first centrifuged at 4000 × g for 15 min at 4°C, and then the supernatant was centrifuged at 10,000 × g at 4°C to remove extracellular organelles. The supernatant was filtered through a 0.45 μm filter and saved as viral stocks at -80°C. The viral concentrations of the viral stocks were estimated by the OIE OsHV-1 qPCR assay described below.
2.2. OIE OsHV-1 qPCR assay
The OIE OsHV-1 quantitative PCR (qPCR) with the primers (OsHV1BF: 5’-GTCGCATCTTTGGATTTAACAA-3′; reverse B4: 5’-ACTGGGATCCGACTGACAAC-3′) and Taqman probe (5’-TGCCCCTGTCATCTTGAGGTATAGACAATC-3′) developed by Martenot et al. [
16] was performed according to the manufacturer's instructions for the TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific). Each reaction contained 5 µl of total DNA and was run for 40 cycles of 95°C for 15 s, 48°C for 30 s, and 68°C for 30 s on a StepOne Real-Time PCR System (Thermo Fisher Scientific). To allow for the absolute quantification of OsHV-1 copies in samples, the PCR product was cloned into the pCR2.1-TOPO vector (Thermo Fisher Scientific) and confirmed via Sanger sequencing at Oregon State University’s Center for Quantitative Life Sciences. Plasmids with the OsHV-1 DNA insert were then used to establish a standard curve.
2.3. Nested PCR
The selection of primers for OsHV-1 sequence amplification was based on OsHV-1 genome sequence data available through Genbank (NC_005881). Nested primers specific for ORF100 and ORF88 (Table 1), which code for DNA polymerase and a transmembrane protein, respectively, were used to detect the OsHV-1 genome. PCR amplification was performed using a 25 µl solution consisting of 2.5 µl amplification buffer (10X PCR buffer with 15 mM MgCl2), 0.4 mM dNTP, 0.4 µM each primer, 1 U Taq (Thermo Fisher Scientific), and 500 ng–1 µg DNA sample. The mixture was subjected to 94°C for 2 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by a 5 min elongation reaction at 72 °C after the final cycle. The nested PCR was performed using nested set primers (Table 1). A 2.5 µl aliquot of the first-run PCR product was included as a template in the second (nested) amplification under the same PCR running conditions. The 1 Kb Plus DNA Ladder (Thermo Fisher Scientific) served as size markers in gel electrophoresis.

2.4. Oyster infection and stress
Adult Pacific oysters, one or two years old (~7 cm or 14 cm in shell length, respectively), free of OsHV-1, were obtained from Yaquina Bay, Oregon, and brought to a BSL-2 laboratory in the Carlson College of Veterinary Medicine at Oregon State University Corvallis, Oregon. Oysters in this bay were determined to be free of OsHV-1 through yearly qPCR testing conducted by a USDA-APHIS accredited diagnostic laboratory (AquaTechnics Inc.). Oysters were anesthetized overnight at room temperature (20-22°C) in an aquarium filled with Coral Pro Salt (Red Sea) seawater with MgCl
2 added at 50 g/L. The next day, 100 μl of an OsHV-1 (Tomales Bay strain) stock at 1×10
7 OsHV-1 DNA copies/ml was injected into the adductor muscle of each oyster with a 1 ml syringe with a 27-gauge needle, and the oysters were returned to the aquarium 30 min after injection. For the next 21 days, oysters were kept in seawater at room temperature (20-22°C) and fed ad libitum with LPB Frozen Shellfish Diet (Reed Mariculture). The aquarium water was changed every 7 days. Aged-matched oysters without infection were kept in a separate container as a control. To induce OsHV-1 reactivation from latency, oysters at 21 days post-infection (dpi) were relaxed in 50 g/L MgCl
2 overnight and injected with 100 µl of sodium butyrate (125mg/ml) (Cat No. B5887, Sigma-Aldrich) per oyster. Sodium butyrate is a histone deacetylase inhibitor and can reactivate HSV-1 latency in mouse models [
17,
18]. The injected oysters were kept in seawater containing 1µM TPCA-1 (an inhibitor of the NF-kB innate immune pathway) at 30ºC for 6 h before returning the stressed oysters to seawater at room temperature. Three infected oysters without stress at 21 dpi were kept in a separate container as a control.
2.5. OsHV-1 shedding into seawater
Seawater samples were collected with three replicates per time point. According to the manufacturer's instructions, viral DNA was isolated from 200 µl of each seawater sample using the QIAamp DNA Blood Mini Kit (QIAGEN). The OsHV-1 qPCR assay described above was then used to quantify viral load in the seawater samples.
2.6. OsHV-1 detection and quantification in tissues
The total DNA of tissues was isolated by first homogenizing 0.1-0.2 g tissue in 400 µl of tissue lysis buffer from the EZNA Tissue DNA Kit (Omega Bio-tek), then extracted according to the manufacturer’s instructions for the EZNA Tissue DNA Kit. Total DNA was eluted in 100 µl of TE buffer (10 mM Tris-HCL [pH 8.0]-1 mM EDTA) and then quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Approximately 500 ng of DNA per tissue was used to perform the OsHV-1 qPCR assay.
2.7. DNA sequencing of PCR product
PCR or reverse-transcription-polymerase chain reaction (RT-PCR) products were purified using a ChargeSwitch PCR Clean-Up kit (Invitrogen), and the cleaned PCR product was directly sequenced by the Center for Quantitative Life Sciences at Oregon State University. The nucleotide sequences were analyzed with the Geneious Prime 2023 software.
2.8. OsHV-1 detection and quantification in hemocytes
One to two ml of hemolymph was drawn from each oyster’s adductor muscle with a 1 ml syringe fitted with a 27 gauge needle. The hemolymph was diluted in 3 volumes of 1X PBS and centrifuged at 4000 × g for 10 min to pellet the hemocytes. The cell pellet was washed once in 1X PBS and resuspended in 400 µl of hypotonic buffer (50 mM Tris–HCl pH 8.0, 5 mM EDTA, 50 g/ml RNAse, 2% Triton X-100) at 4°C for 24 h (19). Cell nuclei from the post-hypotonic treatment were pelleted by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was then loaded directly to the HiBind DNA Mini Column (Omega Bio-tek). The DNA binding column was washed according to the manufacturer’s instructions for the EZNA Tissue DNA Kit, and DNA was eluted in 50 µl of TE buffer. Around 10-50ng of DNA isolated from the supernatant was used in the PCR. Total DNA from the pellet of the hypnotically-treated cell nuclei was isolated according to the manufacturer’s instructions for the EZNA Tissue DNA Kit, and the DNA was also eluted in 50 µl of TE buffer. Approximately 100-500 ng of DNA isolated from the cell pellet was used in the PCR.
2.9. OsHV-1 gene expression
Total RNA was extracted from oyster hemocytes using the High Pure RNA Isolation Kit (Roche). cDNA was synthesized with the qScript cDNA Synthesis Kit (Quantabio) and used as a template for the detection of mRNA of ORF100 and ORF88 (Table 1). Three μl of cDNA were used as templates in the first run of the nested PCR assay described above. A 2.5 µl aliquot of the original PCR product was included as a template in the second (nested) amplification. RT-PCR with and without reverse transcription was also performed with primers EF1a_F and EF1a_R specific to the C. gigas housekeeping gene EF1a (elongation factor 1-alpha) (LOC105338957) to ensure that comparable levels of input RNA were used in RT-PCR and to ensure that amplification did not originate from residual DNA. The RT-PCR product of EF1a is expected to be at 143 bp from processed mRNA and 228 bp from genomic DNA.
2.10. OsHV-1 persistence in Pacific oysters surviving a mortality event in the field
The Pacific oyster spat were planted in Tomales Bay, California, in April 2022 for the purpose of commercial production and experienced an OsHV-1 mortality event in July 2022. Survivors of this mortality event were kept in the bay until they were harvested in April 2023 and shipped overnight on ice to a BSL-2 laboratory in the Carlson College of Veterinary Medicine at Oregon State University Corvallis, Oregon. Upon arrival, adductor muscle, gill, mantle tissue, and hemocytes were collected from these oysters. DNA and RNA were then extracted from these tissues to conduct the qPCR and nested PCR assays described above.
3. Results
3.1. OsHV-1 reactivation in experimentally infected one-year-old adult oysters at 21 days post-infection
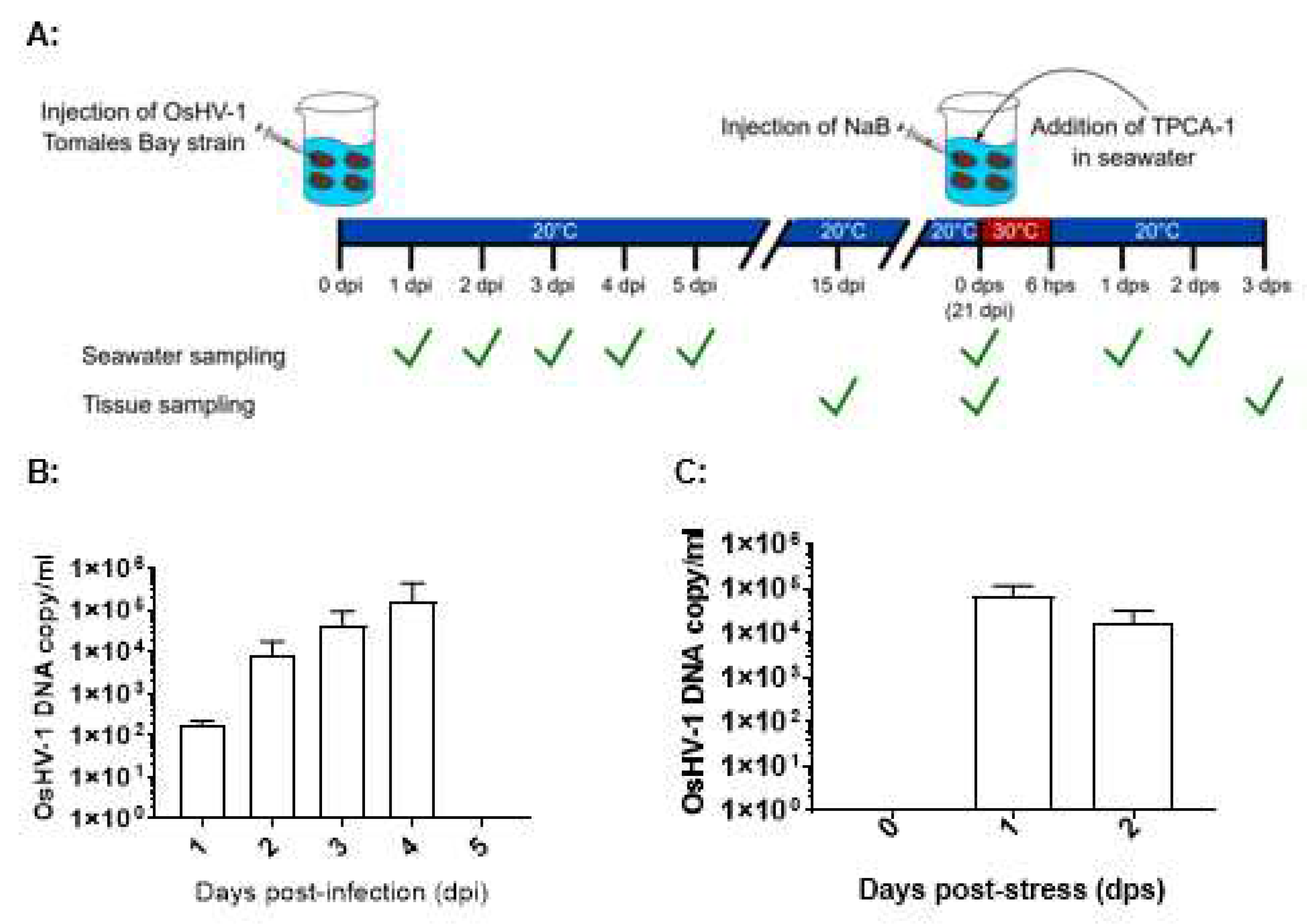
OsHV-1 lytic shedding in seawater immediately after infection was examined from infected oysters kept together in 3.5 L of seawater in 9 L tanks at 20 ℃ (
Figure 1A). OsHV-1 DNA copy number was estimated by the OIE-approved TaqMan qPCR assay in three aliquots of 200 µl of seawater from the tanks. As shown in
Figure 1B, OsHV-1 lytic shedding was detected at 1, 2, 3, and 4 dpi with mean OsHV-1 copies of 1.9×10
2, 1.1×10
4, 6.1×10
4, and 2.2×10
5 OsHV-1 copies/ml, respectively. The total amount of virus shed in the tank ranges between 7×10
5 to 8×10
8 OsHV-1 DNA copies within the first 4 dpi. No OsHV-1 shedding was detected 5 dpi. No OsHV-1 was detected in DNA samples extracted from adductor muscle, gill, intestine, labial palp, or mantle tissues collected from three oysters at 15 and 21 dpi.
If OsHV-1 becomes latent in these surviving oysters, OsHV-1 shedding in seawater after stress will suggest OsHV-1 reactivation from latency. OsHV-1 shedding in seawater was again examined in six stressed oysters at 21 dpi, kept in 1 L seawater. As shown in
Figure 1C, OsHV-1 shedding at 7.0×10
4 and 1.7×10
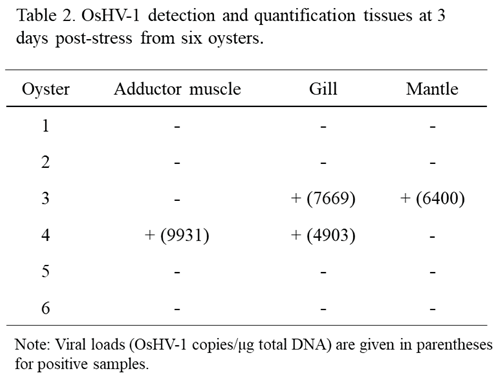
4 OsHV-1 copies/ml were again detected at 1 and 2 days post-stress (dps), respectively. To further confirm OsHV-1 reactivation from latency, all oysters were harvested on 3 dps. OsHV-1 was detected in the adductor muscle, gill, and mantle from two of the six oysters, with OsHV-1 copies/µg ranging from 4903 and 9931 (Table 2). No OsHV-1 was detected in all three infected oysters that were not stressed. These results suggest OsHV-1 can be reactivated from latency under stress conditions.
3.2. OsHV-1 persistence in experimentally infected two-year-old adult Pacific oysters
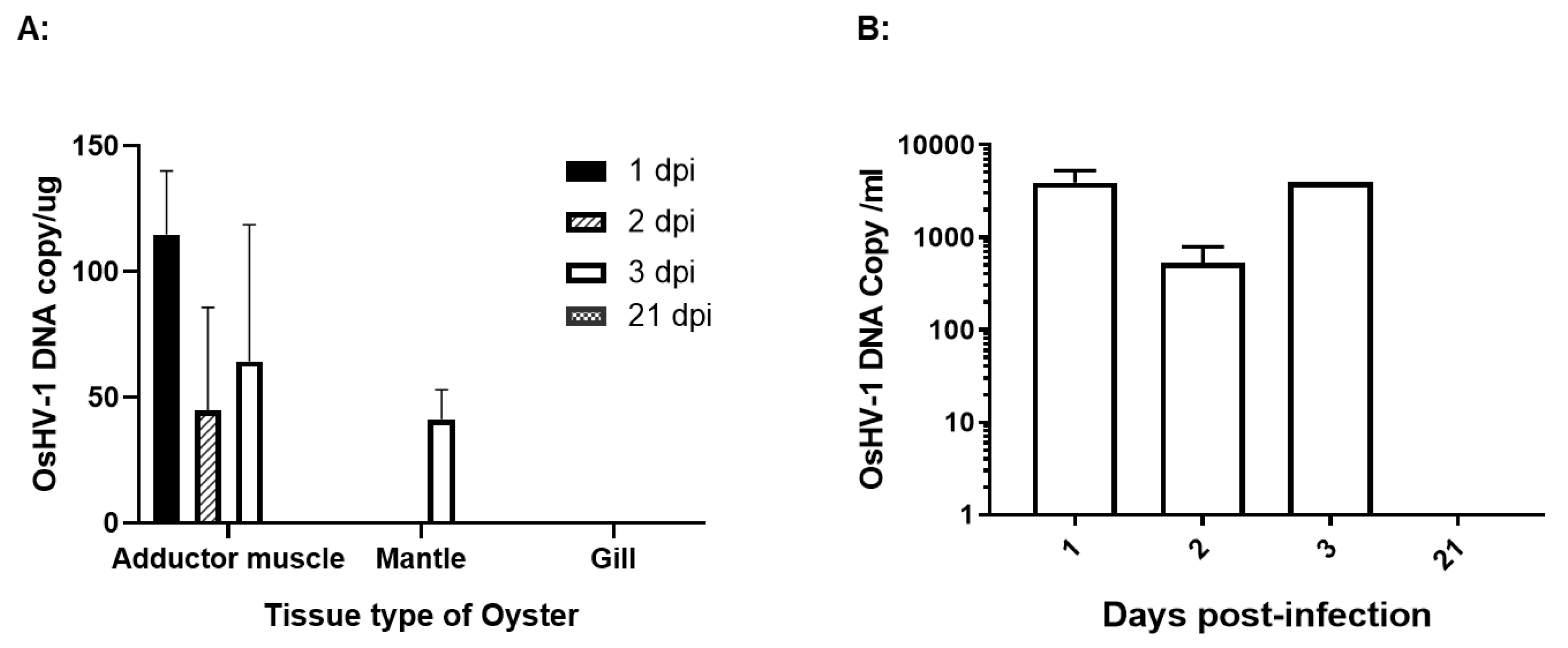
To investigate the latency sites of OsHV-1 infection, 25 two-year-old oysters, ~14 cm in shell length, were infected similarly as above. OsHV-1 shedding in the seawater was monitored daily for 3 dpi, as described above. No OsHV-1 shedding was detectable by qPCR in the seawater at 1, 2, and 3 dpi. To prove acute infection occurred in the infected oysters, adductor muscle, gill, and mantle tissues were collected from three oysters at 1 dpi and three oysters at 3 dpi. OsHV-1 at concentrations of 20 to 133 copies/μg were detected in total DNA samples of adductor muscle and mantle tissues, but no OsHV-1 was detected in the total DNA samples of the gill tissues (
Figure 2A). In addition, viral DNA at 280 to 5450 OsHV-1 copies/ml were also detected in hemolymph samples within the first 3 dpi (
Figure 2B); however, no OsHV-1 DNA was detectable by qPCR in the total DNA of tissues or hemocytes tested on 21 dpi.
To determine whether hemocytes might be the sites of OsHV-1 latency, viral DNA from hemocytes harvested at 21 dpi was isolated by treating them with a hypotonic buffer to exclude cellular DNA, as reported previously [
19]. The presence of OsHV-1 DNA in the supernatant of hemocytes after this treatment was examined by qPCR and nested PCR with primers specific to ORF100, which encodes the DNA polymerase (Table 1). The qPCR failed to detect OsHV-1 in hemocytes tested on 21 dpi (
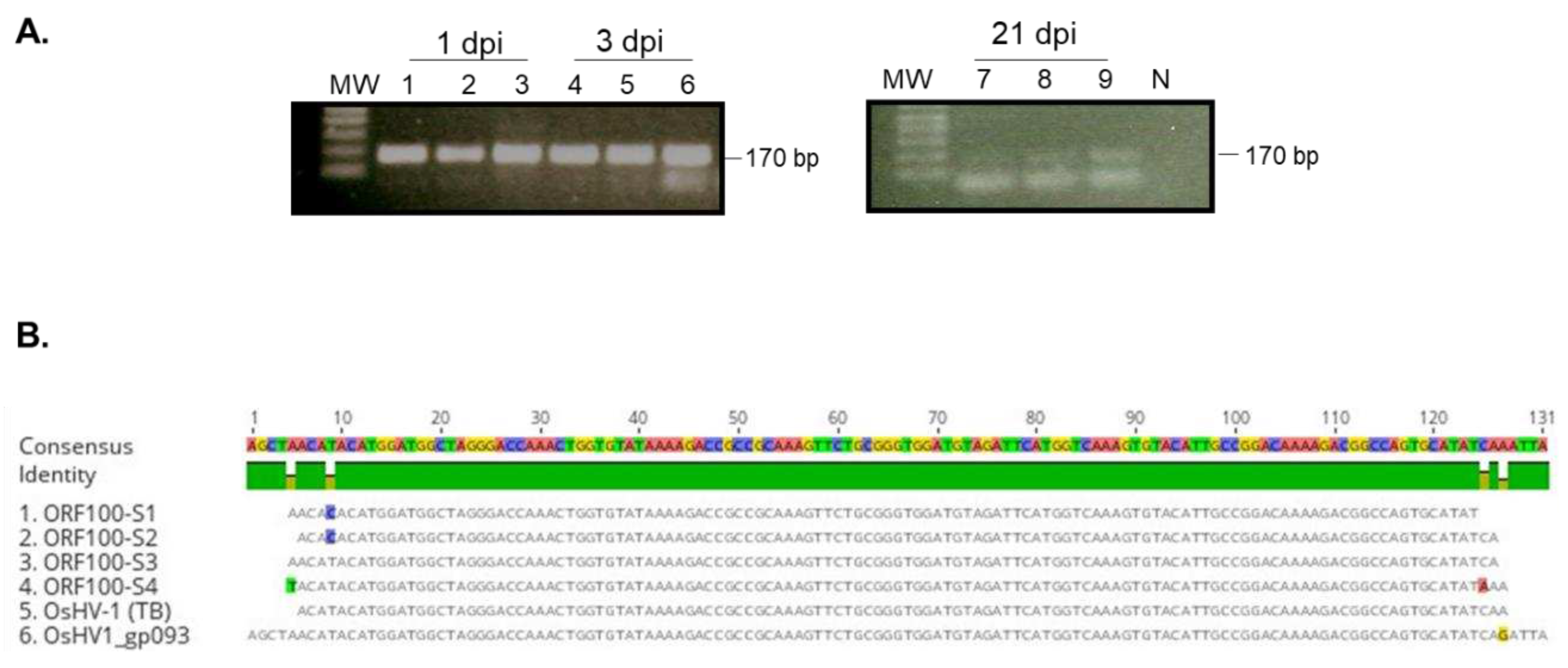
Figure 2B), however, as shown in
Figure 3, a correct-sized fragment, 170bp, was amplified by nested PCR from hemocytes of all three tested oysters on 1, 3 and 21 dpi, although the amplification was weak on 21 dpi (
Figure 3). DNA sequences of the PCR product from oyster one at 1 dpi (
Figure 3 lane 1,
Figure 3B, ORF100-S1) and oyster four at 3 dpi (
Figure 3 lane 4,
Figure 3B, ORF100-S2) are perfectly aligned with the PCR product of the Tomales Bay strain of OsHV-1 (TB) (
Figure 3B). This result suggests that the OsHV-1 viral genome persists in hemocytes of oysters after acute infection.
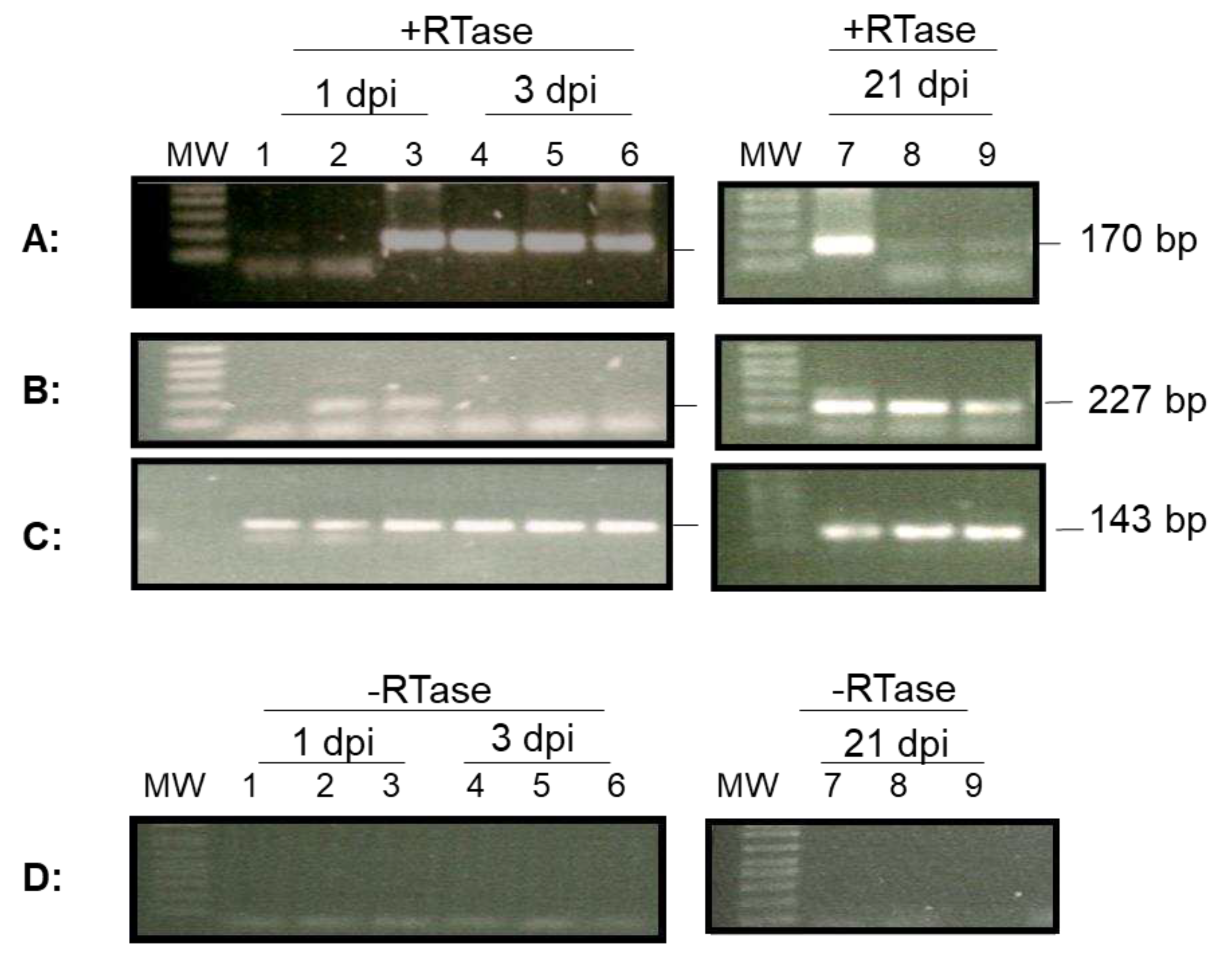
3.3. OsHV-1 lytic gene expression persists in hemocytes of oysters
To determine whether lytic gene transcription occurred in the hemocytes during the acute phase of infection, lytic gene expression was examined in the total RNA isolated from 1-2 ml of hemocytes from three infected two-year-old oysters on 1 and 3 dpi. The cDNA synthesized from the total RNA of hemocytes was analyzed with primers specific for the DNA polymerase gene (ORF100) and a transmembrane gene (ORF88), respectively [
3,
20]. As shown in
Figure 4, ORF100 mRNA was detected by RT-PCR in hemocytes from one out of three oysters on 1 dpi and from all three oysters on 3 dpi. Similarly, mRNA of ORF88 was detected by RT-PCR in hemocytes from two out of three oysters at 1 dpi and from none of the three oysters at 3 dpi (
Figure 4). To determine if lytic viral gene transcription was still present at 21 dpi, total RNA was also isolated from 1-2 ml of hemocytes from three oysters on 21 dpi. As shown in
Figure 4, mRNA from both ORF100 and ORF88 were detectable in all three oysters sampled at 21 dpi. The amplification signal at 21 dpi was similar to that during the acute phase. The amplification of the housekeeping gene EF1a was performed as an internal control to ensure that comparable levels of input RNA were used in RT-PCR. EF1a amplification was similar from all three oyster hemocytes tested on 1, 3, and 21 dpi (
Figure 4C). No ORF100 amplification was detected by RT-PCR without reverse transcriptase in the reaction using total RNA extracted from hemocytes at 1, 3, and 21 dpi (
Figure 4D).
3.4. OsHV-1 persistence in hemocytes of Pacific oysters surviving a mortality event in the field
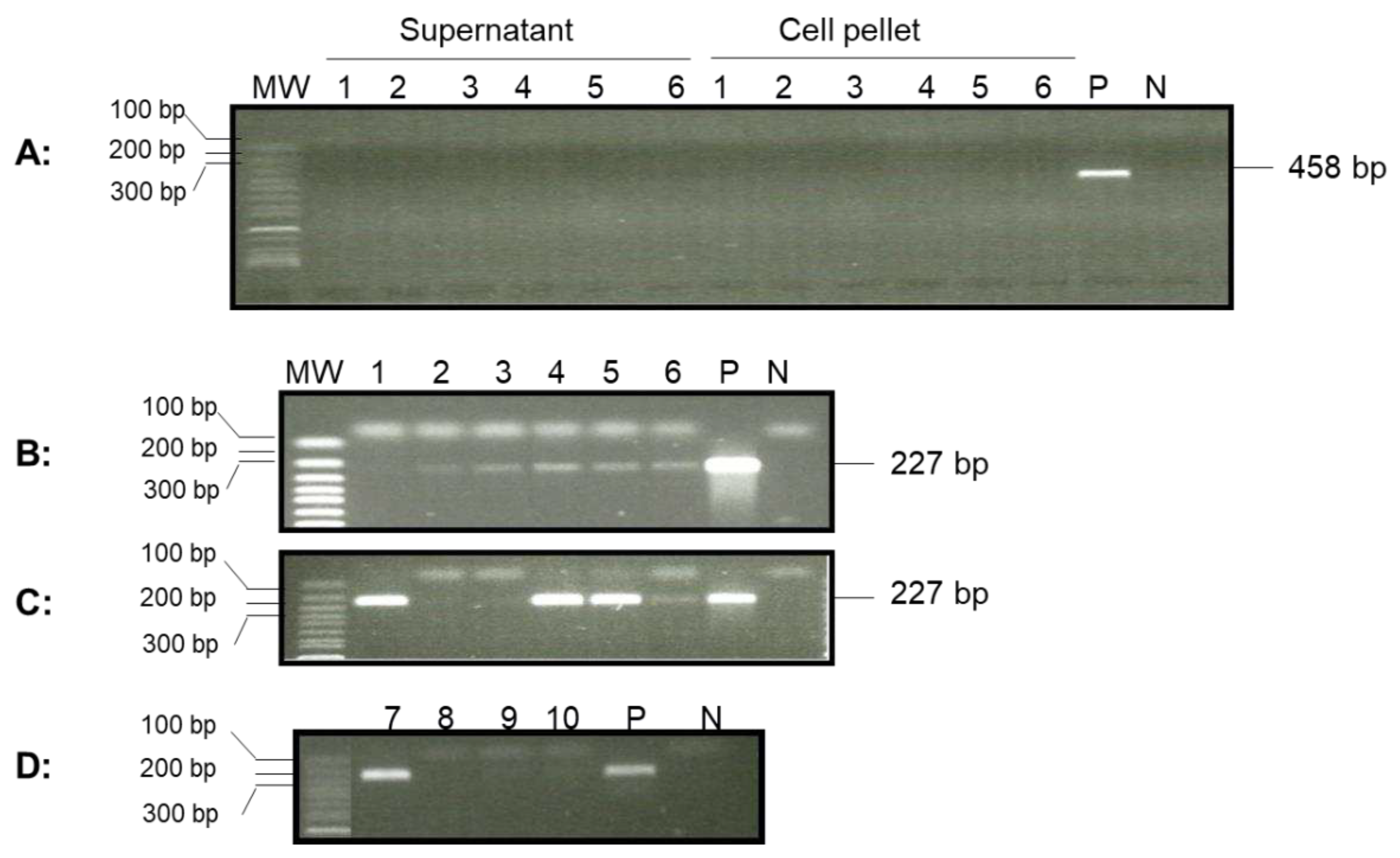
To determine whether OsHV-1 was still present in surviving one-year-old oysters (~7 cm in shell length) from a summer mortality event in Tomales Bay after overwintering in the bay, adductor muscle, gill, and mantle tissues from four randomly selected oysters were screened for the OsHV-1 genome by qPCR. No OsHV-1 DNA was detected by qPCR in the total DNA isolated from these tissues; however, a correct-sized fragment, 227 bp, was amplified by nested PCR, with primer specific to ORF88, from five out of six oyster hemocyte supernatants post-hypotonic buffer treatment (
Figure 5B, lanes 1 to 6). Two of the six hemocyte supernatants also tested positive for OsHV-1 by qPCR. In addition, OsHV-1 was also detected in four out of six hemocyte DNA samples isolated from the cell pellets after post-hypotonic buffer treatment (
Figure 5C). Only one out of four oysters tested positive using the total DNA of hemocytes isolated by the EZNA Tissue DNA Kit (
Figure 5D). To confirm that the amplified products were indeed OsHV-1 DNA, the nested-PCR products from oysters 1 and 4 (
Figure 5C, lanes 1 and 4), labeled as ORF88-S1 and ORF88-S2 in
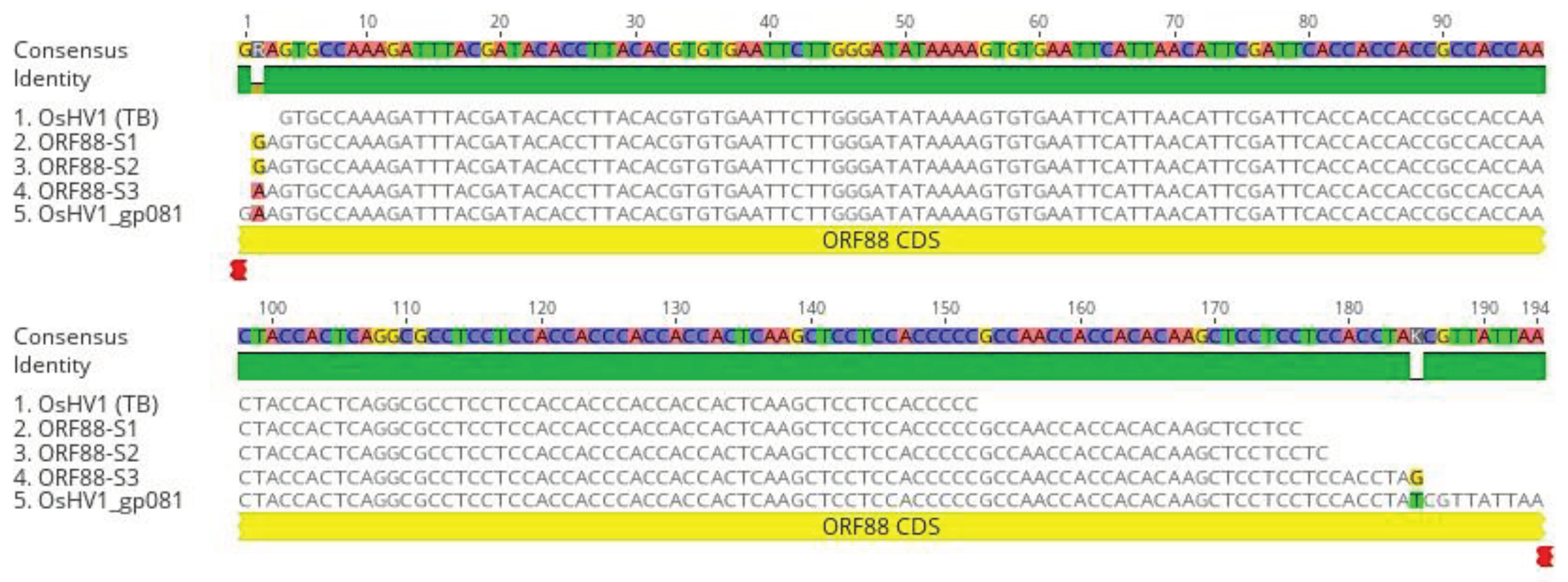
Figure 7A, respectively, were sequenced directly. As shown in Figure7A, ORF88-S1 and ORF88-S2 have 100% homology with the Tomales Bay strain of OsHV-1. The above result confirms that OsHV-1 persists in the hemocytes.
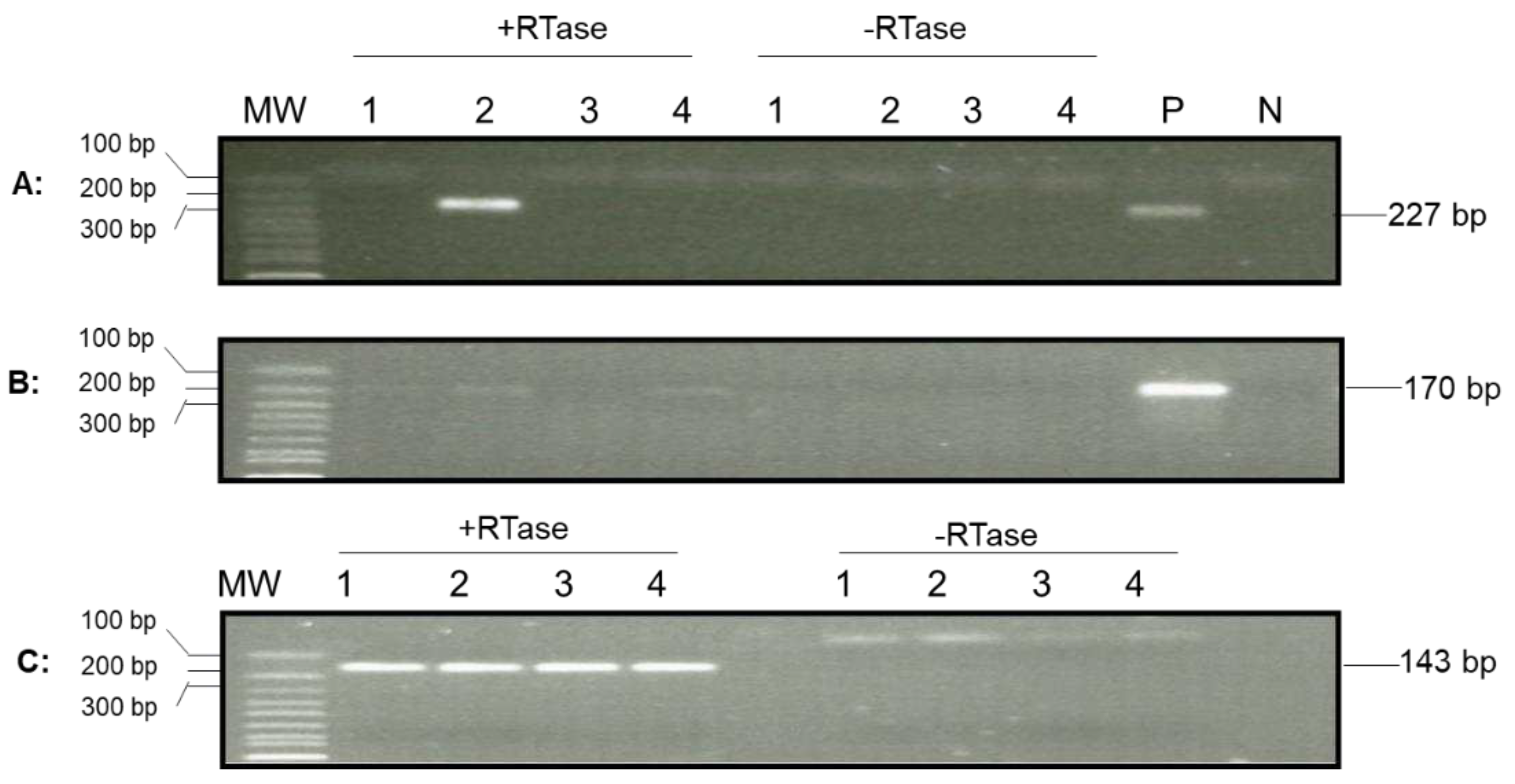
To determine whether lytic gene transcription was present in the hemocytes of these oysters, viral lytic gene expression was examined in the total RNA isolated from 1-2 ml of hemocytes from another four oysters. The synthesized cDNA made from the total RNA of hemocytes was analyzed with primers specific for the DNA polymerase gene (ORF100) and transmembrane gene (ORF88) as above. As shown in
Figure 6, ORF88 mRNA was detected by nested RT-PCR in hemocytes from one of the four oysters (
Figure 6A), and the mRNA of ORF100 was detected in three of the four tested oysters (
Figure 6B). All four samples had similar amplification with primers specific to a host gene, EF1a, used as a control with total cDNA as the template (
Figure 6C). No product was amplified with cDNA from RT without reverse transcriptase (
Figure 6). To confirm that the amplified products were indeed OsHV-1 DNA, the nested-RT-PCR products of ORF88 in
Figure 6A, lane 2 (+RTase) labeled as ORF88-S3 in
Figure 7A, and ORF100 in
Figure 6B, lanes 1 and 2 (+RTase) labeled as ORF100-S3 and ORF100-S4 in
Figure 7B, respectively, were sequenced. As shown in Figure7, the DNA sequence of the nested RT- PCR products of ORF88 and ORF100 are almost identical to the OsHV-1 reference gene sequences OsHV1_gp081 (ORF88) and OsHV1_gp093 (ORF100), respectively. These results indicate that lytic gene transcription exists in some latently infected oysters.
Figure 6.
Detection of OsHV-1 lytic gene transcripts in hemocytes by nested PCR. Total RNA was isolated from four oysters from Tomales Bay, California, that survived a summer OsHV-1 mortality event and overwintered in the bay. The cDNA synthesized by random primers from the total RNA in the presence of reverse transcriptase (+RTase) and absence of reverse transcriptase (-RTase) was amplified with primers specific to ORF88 (A), ORF100 (B), and EF1a (C). Lanes 1-4: total RNA of hemocytes from oysters 1-4. P: OsHV-1 Tomales Bay strain DNA. N: negative control. MW: 1 Kb Plus DNA Ladder (Thermo Fisher Scientific).
Figure 6.
Detection of OsHV-1 lytic gene transcripts in hemocytes by nested PCR. Total RNA was isolated from four oysters from Tomales Bay, California, that survived a summer OsHV-1 mortality event and overwintered in the bay. The cDNA synthesized by random primers from the total RNA in the presence of reverse transcriptase (+RTase) and absence of reverse transcriptase (-RTase) was amplified with primers specific to ORF88 (A), ORF100 (B), and EF1a (C). Lanes 1-4: total RNA of hemocytes from oysters 1-4. P: OsHV-1 Tomales Bay strain DNA. N: negative control. MW: 1 Kb Plus DNA Ladder (Thermo Fisher Scientific).
Figure 7.
DNA sequence alignments of nested PCR and RT-PCR products amplified with primers specific to ORF88 (A). ORF88 DNA sequence alignment with the OsHV-1 reference genome’s (AY509253) ORF88 gene (OsHV1_gp081). ORF88-S1 and ORF88-S2: Nested PCR product amplified from DNA isolated from oyster 1 and 4 hemocytes (
Figure 5C, lanes 1 and 4), respectively, ORF88-S3: Nested RT-PCR product amplified from total RNA isolated from oyster 2 hemocytes (
Figure 6A, lane 2). OsHV-1 (TB): OsHV-1 (Tomales Bay strain).
Figure 7.
DNA sequence alignments of nested PCR and RT-PCR products amplified with primers specific to ORF88 (A). ORF88 DNA sequence alignment with the OsHV-1 reference genome’s (AY509253) ORF88 gene (OsHV1_gp081). ORF88-S1 and ORF88-S2: Nested PCR product amplified from DNA isolated from oyster 1 and 4 hemocytes (
Figure 5C, lanes 1 and 4), respectively, ORF88-S3: Nested RT-PCR product amplified from total RNA isolated from oyster 2 hemocytes (
Figure 6A, lane 2). OsHV-1 (TB): OsHV-1 (Tomales Bay strain).
4. Discussion
Shedding of OsHV-1 virions into the seawater is a sign of a lytic OsHV-1 infection in Pacific oysters. Upon infection of one-year-old Pacific oysters with OsHV-1, shedding of viral particles into the seawater was observed for the first four days but was not observed afterward at 21 dpi. After a stress event, the same oysters that had stopped shedding viruses by 21 dpi started to shed viral particles again into the seawater at a similar level as in the initial shedding (
Figure 1). This indicates that OsHV-1 can be reactivated from a latent state. Other researchers have also found evidence of OsHV-1 reactivation. An OsHV-1 mortality event was reported in Pacific oysters in UV-treated, flow-through tanks for up to 64 days at low temperatures after they were briefly exposed to a higher temperature [
13]. In another experiment, Pacific oysters that survived an OsHV-1 field mortality event experienced another OsHV-1 mortality event approximately two months later [
21]. In the second study, it is interesting to note that OsHV-1 was not reactivated when oysters were initially moved from the field, where the temperature was approximately 5°C, into heated tanks at approximately 20°C; rather, OsHV-1 was only reactivated approximately two months later after incoming seawater temperatures reached 16°C [
21]. In contrast to these two experiments, we held oysters in artificial seawater without water exchange, which ensured that no OsHV-1 virion could escape the experimental container. From the study by Lionel et al. (2013), it seems that elevated temperature was necessary but not sufficient to reactivate OsHV-1; however, research is lacking concerning the appropriate conditions necessary to reactivate OsHV-1 in Pacific oysters.
No detectable shedding of viral particles was observed upon experimental infection of two-year-old Pacific oysters with OsHV-1 compared to one-year-old oysters, which might have been due to the increased tolerance of older oysters to OsHV-1 [
22]. Nevertheless, OsHV-1 was detected in tissues and hemocytes of two-year old oysters on 1, 2, and 3 dpi, with higher levels of OsHV-1 in the hemocytes (
Figure 2). During lytic infection in Pacific oysters, OsHV-1 predominately infects hemocytes, which undergo degeneration and death [
23,
24,
25,
26]. OsHV-1 has been detected in epithelial cells of Pacific oysters in some studies [
27,
28,
29] but not in others [
23,
30,
31], suggesting that epithelial cells are not the primary sites of replication during lytic infection by OsHV-1. It is unusual for immune cells to be the primary sites of replication during lytic infection by herpesviruses, since epithelial cells are primary sites of replication during lytic infection for most vertebrate herpesviruses and most fish alloherpesviruses [
9,
10,
32]. The tropism of OsHV-1 to hemocytes suggests their use in OsHV-1 detection assays could provide increased sensitivity.
In the present study, we used a hypotonic buffer to release viral DNA into the solution to obtain DNA free of the chromosomal DNA [
19]. Viral DNA is much smaller than chromosomal DNA and can leak out of cells in a hypotonic solution. Centrifugation can separate soluble DNA from the chromosomal DNA trapped inside the cellular nuclei. This likely significantly decreased the amount of host DNA, which has been shown to act as a PCR inhibitor when present in high amounts [
33]. The DNA isolated from the hypotonic buffer is mostly viral DNA and fragmented cellular DNA; therefore, its concentration was relatively low, often less than 10 ng/µl from 1-2 ml hemocytes. However, 5 µl to 10 µl of the DNA extract was enough to detect OsHV-1 by PCR (Figs. 2B, 3, and 5). Viral DNA was also detected in the total DNA isolated from the cell pellet post-hypotonic treatment; however, the detection rate was lower than the supernatant-derived DNA. Hemocytes have been previously found to contain more OsHV-1 copies than mantle tissue during experimental infections [
34,
35], although conversely, Evans et al. [
36], using commercial kits to co-extract host and viral DNA, did not find that total DNA extracted from hemolymph was more sensitive for OsHV-1 detection than total DNA extracted from gill tissue.
OsHV-1 DNA could only be detected in hemocytes at 21 dpi using nested PCR and not with qPCR, which suggests that only a very small number of hemocytes contained OsHV-1 DNA (Figs. 2 and 3). This finding is similar to other herpesviruses that become latent in immune cells; for example, koi herpesvirus (KHV) is latently present in 0.6% of koi peripheral white blood cells [
37], and cytomegalovirus (CMV) is latently present in 0.004-0.01% of human mononuclear cells [
38]. On the other hand, hemocytes may not be the only place that OcHV-1 becomes latent. Other researchers have found that OsHV-1 DNA and proteins were not restricted to a particular tissue or cell type in asymptomatic juvenile and adult oysters [
27,
28], which suggests that there might be more cell types capable of being latently infected with OsHV-1. There is precedence for a herpesvirus to infect different cell types latently; for example, varicella–zoster virus is latently present in 2-5% of human neurons as well as <0.1% of nonneuronal cells [
39].
The DNA polymerase gene (ORF100), considered an early gene, and transmembrane genes (e.g., ORF88), usually expressed later, are typically expressed during lytic infection. However, the mRNAs of ORF100 and ORF88 were also detectable in hemocytes at 21 dpi and oysters sampled 9 months after surviving an OsHV-1 mortality event in Tomales Bay (Figs.4 and 6). The detection of mRNA from genes expressed during lytic infection at 21 dpi suggests that OsHV-1 latency might be similar to CMV latency, where genes expressed during latency are similar to those expressed late in its lytic state [
40]. Other herpesviruses, such as HSV-1, have usually been thought to transcribe only one or a few genes during latency, but a growing body of literature suggests that lytic gene expression is not uncommon in latent cells [
41]. Depending on how many transcripts OsHV-1 produces for a particular gene per latently infected cell, RT-PCR might be a more sensitive method for detecting latent infections than PCR.
OsHV-1 was found to be present in hemocytes of oysters sampled 9 months after surviving an OsHV-1 mortality event in Tomales Bay (
Figure 5). Reactivation of OsHV-1 in latently infected overwintered oysters may be one source of inoculum for infection during the following season. Persistence of OsHV-1 after mortality events have been observed in other locations; for example, OsHV-1 has been found in survivors of OsHV-1 mortality events in La Cruz lagoon, Mexico; Marennes-Oléron Bay, France; and Woolooware Bay, Australia [
21,
36,
42]. The detection of OsHV-1 in survivors of a mortality event again suggests that OsHV-1 can persist in the population and reactivate when the conditions favor virus replication.
5. Conclusions
In summary, OsHV-1 latent or persistent infection was demonstrated in the experimentally infected oysters using an OsHV-1 isolate from Tomales Bay, California. Like other herpesviruses, OsHV-1 can also reactivate and be shed into the seawater from surviving oysters. OsHV-1 persistency was also shown in Tomales Bay oysters sampled 9 months after surviving a mortality event. OsHV-1 latent or persistent infections are not detectable with the qPCR assay recommended by OIE in tissues; however, OsHV-1 DNA can be detected in hemocytes using a nested PCR. The persistency of OsHV-1 can be detected at a high rate when DNA samples were isolated from hemocytes treated with a hypotonic buffer. In addition to detecting the latent viral genome during latency, OsHV-1 lytic gene transcription can also be detected in hemocytes during latent infection, which could be used to screen for OsHV-1 in Pacific oysters.
Author Contributions
Conceptualization, L.J, and K.D.; methodology, L.J., X.W., B.S., G.F. and K.D.; software, L.J.; validation, X.W.; formal analysis, L.J.; investigation, K.D., X.W., B.S., and G.F.; resources, L.J., C.L; data curation, K.D. and L.J.; writing—original draft preparation, L.J.; writing—review and editing, K.D., C.L., and B.S.; visualization, L.J.; supervision, L.J.; project administration, L.J.; funding acquisition, C.L. and L.J. All authors have read and agreed to the published version of the manuscript.
Funding
Support for KD and partial support for XW was provided by the United States Department of Agriculture - Agricultural Research Service (USDA-ARS) project “Identifying Genetic Factors Associated with the Expression and Regulation of Economically Important Traits in Cultured Pacific Oysters” (Project No. 2076-63000-005-002-S) and the Pacific States Marine Fisheries Commission (PSMFC) project “Preparing for Future Challenges – Threats from Ocean Acidification, Vibrio coralliilyticus and OsHV-1 μvar to West Coast Oyster Farmers” (Award No. NA18NMF4720007).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Dr. George Rohrmann (Oregon State University) for help editing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Botta R: A review of global oyster aquaculture production and consumption. Marine Policy 2020, 117(103952). [CrossRef]
- Fuhrmann M, Pathirana, E., de Kantzow, M. and Hick, P.: Ostreid herpesvirus disease. In Aquaculture Pathophysiology: Academic Press; 2022.
- Davison AJ, Trus BL, Cheng N, Steven AC, Watson MS, Cunningham C, Deuff RL, Renault T: A novel class of herpesvirus with bivalve hosts. The Journal of general virology 2005, 86(Pt 1):41-53. [CrossRef]
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E: The order Herpesvirales. Arch Virol 2009, 154(1):171-177. [CrossRef]
- Agnew MV, Friedman CS, Langdon C, Divilov K, Schoolfield B, Morga B, Degremont L, Dhar AK, Kirkland P, Dumbauld B et al.: Differential Mortality and High Viral Load in Naive Pacific Oyster Families Exposed to OsHV-1 Suggests Tolerance Rather than Resistance to Infection. Pathogens 2020, 9(12). [CrossRef]
- Segarra A, Pepin JF, Arzul I, Morga B, Faury N, Renault T: Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus research 2010, 153(1):92-99. [CrossRef]
- Trancart S, Tweedie A, Liu O, Paul-Pont I, Hick P, Houssin M, Whittington RJ: Diversity and molecular epidemiology of Ostreid herpesvirus 1 in farmed Crassostrea gigas in Australia: Geographic clusters and implications for "microvariants" in global mortality events. Virus research 2022, 323:198994. [CrossRef]
- Knipe DM, Howley PM: Fields virology, 6th edn. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013.
- Cohen JI: Herpesvirus latency. J Clin Invest 2020, 130(7):3361-3369. [CrossRef]
- Hanson L, Dishon A, Kotler M: Herpesviruses that infect fish. Viruses 2011, 3(11):2160-2191. [CrossRef]
- Reed AN, Izume S, Dolan BP, LaPatra S, Kent M, Dong J, Jin L: Identification of B cells as a major site for cyprinid herpesvirus 3 latency. Journal of virology 2014, 88(16):9297-9309. [CrossRef]
- E: Manual of Diagnostic Tests for Aquatic Animals. In Infection with ostreid herpesvirus 1 microvariants. 2019.
- Pernet FT, D.; Petton, B.: Influence of low temperatures on the survival of the Pacific oyster (Crassostrea gigas) infected with ostreid herpes virus type 1. Aquaculture 2015, 445(1):57-62. [CrossRef]
- Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S: Marek's disease virus: from miasma to model. Nat Rev Microbiol 2006, 4(4):283-294. [CrossRef]
- Divilov K, Blaine S, Cortez DM, Wang X, Fleener GB, Jin L, Dumbauld BR, Langdon C: Genetic improvement of survival in Pacific oysters to the Tomales Bay strain of OsHV-1 over two cycles of selection. Aquaculture 2021, 543. [CrossRef]
- Martenot C, Oden E, Travaille E, Malas JP, Houssin M: Comparison of two real-time PCR methods for detection of ostreid herpesvirus 1 in the Pacific oyster Crassostrea gigas. Journal of virological methods 2010, 170(1-2):86-89. [CrossRef]
- Danaher RJ, Jacob RJ, Steiner MR, Allen WR, Hill JM, Miller CS: Histone deacetylase inhibitors induce reactivation of herpes simplex virus type 1 in a latency-associated transcript-independent manner in neuronal cells. J Neurovirol 2005, 11(3):306-317. [CrossRef]
- Neumann DM, Bhattacharjee PS, Hill JM: Sodium butyrate: a chemical inducer of in vivo reactivation of herpes simplex virus type 1 in the ocular mouse model. Journal of virology 2007, 81(11):6106-6110. [CrossRef]
- Jin L, Perng GC, Brick DJ, Naito J, Nesburn AB, Jones C, Wechsler SL: Methods for detecting the HSV-1 LAT anti-apoptosis activity in virus infected tissue culture cells. Journal of virological methods 2004, 118(1):9-13. [CrossRef]
- Burioli EAV, Prearo M, Houssin M: Complete genome sequence of Ostreid herpesvirus type 1 microVar isolated during mortality events in the Pacific oyster Crassostrea gigas in France and Ireland. Virology 2017, 509:239-251. [CrossRef]
- Lionel D, Guyader, T., Tourbiez, D. and Pépin, J.F.,: Is horizontal transmission of the Ostreid herpesvirus OsHV-1 in Crassostrea gigas affected by unselected or selected survival status in adults to juveniles?. Aquaculture 2013, 408:51-57. [CrossRef]
- Hick PM, Evans, O., Rubio, A., Dhand, N.K. and Whittington, R.J.: Both age and size influence susceptibility of Pacific oysters (Crassostrea gigas) to disease caused by Ostreid herpesvirus-1 (OsHV-1) in replicated field and laboratory experiments. Aquaculture 2018, 489:110-120. [CrossRef]
- Bueno R, Perrott M, Dunowska M, Brosnahan C, Johnston C: In situ hybridization and histopathological observations during ostreid herpesvirus-1-associated mortalities in Pacific oysters Crassostrea gigas. Diseases of aquatic organisms 2016, 122(1):43-55. [CrossRef]
- de Lorgeril J, Lucasson A, Petton B, Toulza E, Montagnani C, Clerissi C, Vidal-Dupiol J, Chaparro C, Galinier R, Escoubas JM et al.: Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat Commun 2018, 9(1):4215. [CrossRef]
- Delisle L, Laroche O, Hilton Z, Burguin JF, Rolton A, Berry J, Pochon X, Boudry P, Vignier J: Understanding the Dynamic of POMS Infection and the Role of Microbiota Composition in the Survival of Pacific Oysters, Crassostrea gigas. Microbiol Spectr 2022, 10(6):e0195922. [CrossRef]
- Friedman CS, Estes RM, Stokes NA, Burge CA, Hargove JS, Barber BJ, Elston RA, Burreson EM, Reece KS: Herpes virus in juvenile Pacific oysters Crassostrea gigas from Tomales Bay, California, coincides with summer mortality episodes. Diseases of aquatic organisms 2005, 63(1):33-41. [CrossRef]
- Arzul I, Renault T, Thebault A, Gerard A: Detection of oyster herpesvirus DNA and proteins in asymptomatic Crassostrea gigas adults. Virus research 2002, 84(1-2):151-160. [CrossRef]
- Martenot C, Segarra A, Baillon L, Faury N, Houssin M, Renault T: In situ localization and tissue distribution of ostreid herpesvirus 1 proteins in infected Pacific oyster, Crassostrea gigas. Journal of invertebrate pathology 2016, 136:124-135. [CrossRef]
- Prado-Alvarez M, Darmody G, Hutton S, O'Reilly A, Lynch SA, Culloty SC: Occurrence of OsHV-1 in Crassostrea gigas Cultured in Ireland during an Exceptionally Warm Summer. Selection of Less Susceptible Oysters. Front Physiol 2016, 7:492. [CrossRef]
- Corbeil S, Faury N, Segarra A, Renault T: Development of an in situ hybridization assay for the detection of ostreid herpesvirus type 1 mRNAs in the Pacific oyster, Crassostrea gigas. Journal of virological methods 2015, 211:43-50. [CrossRef]
- Lipart C, Renault T: Herpes-like virus detection in infected Crassostrea gigas spat using DIG-labelled probes. Journal of virological methods 2002, 101(1-2):1-10. [CrossRef]
- Aurelian L: Herpesviruses, the immune system, and AIDS. Boston: Kluwer Academic Publishers; 1990.
- u J, Li D, Xia S, Song H, Dong Z, Chen F, Sun X, Tang Z: Absolute quantification of plasmid DNA by real-time PCR with genomic DNA as external standard and its application to a biodistribution study of an HIV DNA vaccine. Anal Sci 2009, 25(5):675-680. [CrossRef]
- Picot S, Faury N, Pelletier C, Arzul I, Chollet B, Degremont L, Renault T, Morga B: Monitoring Autophagy at Cellular and Molecular Level in Crassostrea gigas During an Experimental Ostreid Herpesvirus 1 (OsHV-1) Infection. Front Cell Infect Microbiol 2022, 12:858311. [CrossRef]
- Schikorski D, Faury N, Pepin JF, Saulnier D, Tourbiez D, Renault T: Experimental ostreid herpesvirus 1 infection of the Pacific oyster Crassostrea gigas: kinetics of virus DNA detection by q-PCR in seawater and in oyster samples. Virus research 2011, 155(1):28-34. [CrossRef]
- Evans O, Hick P, Whittington RJ: Detection of Ostreid herpesvirus-1 microvariants in healthy Crassostrea gigas following disease events and their possible role as reservoirs of infection. Journal of invertebrate pathology 2017, 148:20-33. [CrossRef]
- Reed AN, Putman T, Sullivan C, Jin L: Application of a nanoflare probe specific to a latency associated transcript for isolation of KHV latently infected cells. Virus research 2015, 208:129-135. [CrossRef]
- Slobedman B, Mocarski ES: Quantitative analysis of latent human cytomegalovirus. Journal of virology 1999, 73(6):4806-4812. [CrossRef]
- Kennedy PG, Grinfeld E, Gow JW: Latent varicella-zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc Natl Acad Sci U S A 1998, 95(8):4658-4662. [CrossRef]
- Shnayder M, Nachshon A, Krishna B, Poole E, Boshkov A, Binyamin A, Maza I, Sinclair J, Schwartz M, Stern-Ginossar N: Defining the Transcriptional Landscape during Cytomegalovirus Latency with Single-Cell RNA Sequencing. mBio 2018, 9(2). [CrossRef]
- Singh N, Tscharke DC: Herpes Simplex Virus Latency Is Noisier the Closer We Look. Journal of virology 2020, 94(4). [CrossRef]
- Martinez-Garcia MF, Grijalva-Chon JM, Castro-Longoria R, Re-Vega E, Varela-Romero A, Chavez-Villalba JE: Prevalence and genotypic diversity of ostreid herpesvirus type 1 in Crassostrea gigas cultured in the Gulf of California, Mexico. Diseases of aquatic organisms 2020, 138:185-194. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).