1. Introduction
Computer vision has emerged as an essential technology in horticulture, revolutionising various aspects of crop production and management. By utilising visual signals such as 2D images, videos, multi-view videos, and 3D data, computer vision systems extract valuable knowledge about crop health, growth dynamics, and environmental conditions. These insights enable informed decision-making processes, leading to improved crop quality, better yield, and enhanced resource management. The success of computer vision in horticulture relies heavily on the quality of the visual signals used as input. High-quality data ensures accurate and reliable analysis, facilitating the development of robust and efficient computer vision models. However, the unique challenges of the horticultural domain pose significant obstacles in obtaining high-quality data for training and evaluation.
Horticulture environments exhibit diverse and complex characteristics that can impact the quality of visual signals. Factors such as varying lighting conditions, occlusions caused by foliage or trellis structures, background clutter, and phenotypic variations among plant species introduce significant variability and challenges in data acquisition. These challenges make it difficult to capture high-quality visual signals consistently. Furthermore, developing deep learning solutions for horticulture is hindered by the scarcity of high-quality training and evaluation datasets. Data-driven approaches such as deep learning models require large amounts of quality data to achieve optimal performance. However, creating such datasets for horticultural applications is a laborious and time-intensive process, requiring domain-specific expertise and manual annotations in some cases [
1].
In this paper, we explore the current state of vision systems and available data in horticulture, highlighting the critical role of data quality. We identify the specific data requirements to address the challenges in horticultural contexts and explore deep-learning approaches to enhance the quality of the visual signals used. Our aim is to provide insights and strategies to overcome the limitations imposed by data quality constraints in the development of efficient computer vision models for horticulture. By addressing these challenges, we can unlock the full potential of computer vision in horticultural practices, fostering advancements primarily in crop management, disease detection, yield estimation, and automated harvesting.
1.1. Current State of Computer Vision in Horticulture
Computer vision has emerged as an indispensable technology for horticulture applications with key advancements in plant growth monitoring, pest and disease detection, quality and yield estimation, and automated harvesting. In each of these use cases, computer vision techniques leverage various strategies for image processing: segmentation, object detection and tracking, feature extraction, and pattern recognition.
Figure 1 summarises the key applications of computer vision in horticulture, tasks undertaken by vision systems for each use case and the fundamental vision technologies used. To accomplish vision tasks such as classification and forecasting, deep learning models are frequently employed instead of conventional machine learning approaches and traditional computer vision techniques, owing to their automated ability to determine features of interest. The details of computer vision in each of the key horticulture use cases are discussed below.
Figure 1.
Overview of computer vision in horticulture, its applications and the tasks undertaken.
Figure 1.
Overview of computer vision in horticulture, its applications and the tasks undertaken.
1.1.1. Plant Growth Monitoring
Plant growth monitoring is an essential application of computer vision in horticulture. It is achieved through the utilisation of visual signals, such as 2D signals, either in the form of RGB, multispectral, hyperspectral, thermal or fluorescence, and 3D LiDAR data, to monitor and track the growth and development of plants. The scale of these signals can range from a single leaf to an entire tree to an aerial/satellite view of the farm canopy. Features such as leaf area, stem length, and branching patterns are extracted to develop knowledge about plant health, vigour, and response to environmental conditions. For instance, leaf area is extracted from 2D images by segmenting the plant leaves using image processing techniques such as thresholding or edge detection. The segmented leaf regions are then quantified to calculate the leaf area, which serves as an indicator of plant growth and photosynthetic activity. Similarly, stem length is determined by measuring the distance between key points on the stem, which can be identified through feature extraction algorithms like Harris corner detection or SIFT (Scale-Invariant Feature Transform) [
2,
3].
To effectively analyse the large datasets of visual signals by extracting valuable insights and information, deep learning models, such as convolutional neural networks (CNNs) or region-based convolutional neural networks (R-CNNs) [
4], have been applied to learn and extract meaningful features automatically with better outcomes [
5,
6]. These models capture complex patterns and variations in plant growth, enabling more accurate and robust growth tracking. Similarly, sequential models, using long short-term memory (LSTM) or recurrent neural networks (RNNs), capture temporal dependencies in plant growth using time sequence data. A recent survey [
7] has identified a total of 23 published works proposing deep learning models for plant monitoring applications, from which 15 were for horticulture crops, with most of the works published between 2017 to 2021 using either CNN/R-CNN with spatial data or convolutional LSTM/RNN with spatiotemporal data. All of these works were primarily focused on either of the three objective tasks: (i) plant growth classification, (ii) instance segmentation and object detection for growth stage identification, and (iii) regression for plant growth analysis. Images were the predominant form of input to these vision systems. However, another recent study [
8] conducted in the application of light detection and ranging (LiDAR) and ultrasonic sensors to high-throughput phenotyping and precision horticulture reported a steeply increasing trend in work done in this space. Interestingly, most of the works studied [
8] primarily focused on controlling the real-time variable rates of farm inputs such as agricultural sprays, with up to 11 instances of using ultrasonic signal-based vision systems saving up to 40% of agricultural spray inputs. Although these learning-based vision systems enable growers to make informed decisions regarding plant management practices, resource allocation, and optimisation of growth conditions, there is still a significant challenge in terms of the shortage of data for training and evaluation of these learning-based systems.
1.1.2. Pest and Disease Detection
Pest and disease detection are other important use cases in horticulture, where visual signals captured from plants are used to detect various symptoms and signs of pest or disease, which are sometimes challenging to detect without complex pattern recognition abilities [
9]. Specific patterns or anomalies associated with pests or diseases are identified through features like leaf discolouration, lesion patterns, the presence of pests, or characteristic patterns caused by pathogens, allowing for early intervention and effective disease management strategies. For instance, in the case of leaf disease detection, colour variations and texture patterns on the surface of leaves are studied with statistical measures, such as colour histograms or texture descriptors like local binary patterns, to quantify the disease symptoms and distinguish between healthy and infected regions. The traditional machine vision methods for plant diseases and pest detection depend on manual feature design for classifiers [
10]. Although carefully constructed imaging schemes can reduce design difficulty in these approaches, they increase costs and struggle to neutralise the impact of environmental changes on recognition results [
11]. In complex natural environments, detection of plant diseases and pests encounters challenges such as small differences between the foreground and background, low contrast, varied lesion scales and types, and noise, making traditional methods ineffective for optimal detection of pest or disease [
12].
With a proven ability to be more robust to certain real and complex natural environmental changes and automatic feature learning abilities, deep learning models are gaining interest in the detection of pests and diseases. These models can be primarily categorised into a classification network (e.g. to label lesions with a category), a detection network (e.g. to locate and categorise lesions) and a segmentation network (e.g. to identify and distinguish lesions from normal areas of the leaves) [
13]. The most common approach is to deploy these models with visualisation techniques such as feature maps, heat maps, saliency maps, activation maps, etc., for visual recognition of pests and diseases. Adopting transfer learning and other retraining techniques, popular deep learning models, such as VGG [
14], ResNet [
15] and AlexNet [
16], are being used to develop the classification and detection models in this space with a recent focus on using hyperspectral imaging [
17]. Despite the network architecture and visual signal used, the need for large datasets with good quality for training and evaluation remains a major shortcoming.
1.1.3. Yield Estimation
Yield estimation involves using computer vision techniques for pre-harvest estimation of crop yield, enabling effective decision-making and planning. Conventional machine learning approaches, mostly using artificial neural networks, support vector regression, regression trees and k-nearest neighbours, were developed for accurate yield estimations [
18]. The need for feature selection based on dimensionality reduction and feature selection based on agronomy and plant physiology posed significant challenges to the conventional machine learning models [
19]. However, with advances in automated feature extraction with deep learning, recent research works have explored the application of deep learning-based computer vision techniques, such as object detection and counting, to estimate yields for various horticultural crops. In a recent survey [
1], seven key works were identified in this domain, primarily focusing on yield estimation for banana, citrus, strawberry, mango and tomato crops. A Fast R-CNN [
20] model was developed to detect and estimate citrus fruit yields, while deep, multifaceted systems incorporating LSTM layers were developed for forecasting banana harvest yields with promising results [
21]. In addition, deep learning algorithms, including the Faster R-CNN model, have been employed for accurate banana detection and counting using high-resolution RGB images captured by UAVs. These techniques have demonstrated high detection rates and precise yield estimates. Similar advances have been made for vegetable crops, including automatic strawberry flower detection for yield prediction and simulated learning for tomato fruit counting.
Similarly, another study [
22] reviewing scientific reports on the use of deep learning models in the estimation of tree fruit number per image reported a total of 12 reports, with almost all the works developing or discussing either CNN, R-CNN, VGG, or You Only Look Once (YOLO) [
23,
24,
25,
26] based deep architectures. The development of these techniques holds great potential for practical implementation, providing valuable guidance for planting, harvesting, and marketing decisions in horticultural production. However, despite methodologically being a heavily data-driven domain, the impact of data availability and quality is underexplored in the literature.
1.1.4. Quality Assessment
Crop quality estimation is increasingly a critical aspect of horticulture, and computer vision techniques have proven valuable in this task. In this use-case, important quality attributes of crops, including size, shape, colour, and ripeness, are used to classify and grade the produce, ensuring consistent product quality and enabling effective sorting and distribution processes [
27]. Images of fruit are used to extract features, including colour histograms, shape descriptors (e.g., circularity or elongation ratio), and texture characteristics. These features are then correlated with predefined quality standards or ripeness indices to determine the overall fruit quality. There is a growing trend of developing non-destructive computer vision models based on hyperspectral images [
28]. Machine learning algorithms, such as support vector machines and partial least squares discriminant analysis, remain the major methods employed in quality estimation based on extracted features. Although advances in deep learning techniques, particularly CNNs, have shown remarkable success in automating the process of fruit quality assessment, conventional machine learning approaches account for the majority of work done in the past two decades [
29]. These methods are primarily focused on estimating quality parameters such as pigments, firmness and elasticity, moisture content and dry matter, soluble solid contents, acidity, physiological disorders, mechanical damage and bacterial/fungal infections.
A recent study conducted to review the application of hyper-spectral imaging and artificial intelligence (AI) in quality estimation identified a rapid increase in the amount of work done in the estimation of fruit quality using computer vision, with the majority of works using reflectance as an imaging mode for internal and external quality estimation [
30]. In regards to deep learning methodologies, commonly used deep model architectures, such as CNN and R-CNN, are used with RGB images for quality estimation. As with other applications within horticulture, popular deep models, such as VGG, ResNet and AlexNet, are likewise adopted for quality estimation. To combine the sensitivity-related benefits of hyper-spectral imaging and precision-related benefits of RGB imaging, multi-modal data are also being used with deep learning models. Similar to yield estimation, to our knowledge, there is little to no discussion in the literature about the availability of data and its quality for data-driven learning methods in quality estimation.
1.1.5. Automated Harvesting
Automated harvesting techniques analyse visual signals captured from plants, robotic systems, or other automated machines for identifying ripe fruit or vegetables and performing precise harvesting operations [
31]. Computer vision algorithms can detect specific visual cues such as colour, size, or texture to determine the ripeness of the produce. This enables automation in the harvesting process, reducing labour costs and increasing operational efficiency [
32] to significantly streamline the harvest workflow, improve productivity, and minimise post-harvest supply chain losses.
Object detection algorithms are one of the drivers for automated harvesting, such as Faster R-CNN or YOLO, which have been popularly employed to detect and localise target objects such as ripe fruit in real-time. For instance, in apple harvesting, specifically within narrow, accessible, and productive trellis-trained apple tree architectures, a vision-based shaking points detection technique using the segmented pixels of branches and trunks can be used along with deep models. A Deeplab v3+ResNet-18 [
33] model was proposed recently with an effective outcome to build such a shaking points detection vision system. Similar detection and segmentation-based robotic harvesting methods have also been proposed for tomato harvesting [
34]. These models can be trained on annotated datasets of various fruit or vegetable types, enabling them to generalise across different crops and adapt to different environmental conditions. However, the impact of illumination, environmental conditions and canopy density pose challenges in building these models. The task becomes particularly challenging if the pixel density and resolution of the captured visual signal inputs are inadequate for these models to learn segmentation and detection tasks efficiently.
1.2. Deep Learning Practices
Deep learning-based computer vision systems offer several advantages, including their ability to automatically learn and extract complex features from visual signals, their adaptability to different horticultural contexts, and their scalability to large datasets. However, these approaches often require substantial amounts of good-quality training data and demand computational resources during both training and inference. Despite these challenges, advances in deep learning have significantly improved the accuracy and efficiency of computer vision systems in horticulture, paving the way for more automated and intelligent agricultural practices.
Deep learning models for computer vision in horticulture have employed different types of learning, including supervised, unsupervised, and semi-supervised learning. Supervised learning is the most common approach, where models are trained on datasets, with each input paired with its corresponding ground truth in the process of classification (pattern recognition). The models learn to map the input visual signals to the desired output labels, allowing them to recognise and classify plants, detect diseases, or classify produce quality grades. Similarly, regression is another type of supervised learning where the goal is to predict continuous numerical values rather than discrete categories or labels. This involves training a model to learn the underlying patterns and relationships in the data and making predictions based on input features. A regression model is trained using ground-truth data, where each data instance is associated with a target value or output. During training, the model adjusts its parameters to minimise the difference between its predictions and the true target values, typically using optimisation techniques such as gradient descent.
Unsupervised learning is employed when ground truth data is scarce or unavailable. In this approach, models learn to extract meaningful representations from the unlabelled data without explicit annotations. These representations can then be utilised for various tasks, such as clustering similar plant images, identifying shared patterns, or discovering hidden structures within the data. Semi-supervised learning combines labelled and unlabelled data to train deep learning models. The limited availability of labelled data in horticulture can be supplemented with a larger pool of unlabeled data. These models leverage the unlabeled data to learn additional features and generalise better, leading to improved performance on the given task.
Supervised learning remains the most commonly used approach for training computer vision learning algorithms in horticulture, resulting in the need for training datasets with ground truths. The number of training instances and the quality of each training instance in the dataset determine the learning ability of these models. Given that the data used for computer vision systems in horticulture are predominantly visual signals, the spatial and spatiotemporal quality of these signals significantly dictate model training effectiveness. To understand more about the inherent nature of commonly used signal types and their attributes in relation to requirements for deep learning-based vision systems, we take a closer look at visual signal data and their datasets in the following section.
2. Materials and Methods
2.1. Visual Signal Data and Quality
The input data used for either training, evaluating or day-to-day use of the learning-based vision systems exhibit diverse geometry, characteristics and information content.
Table 1 provides an overview of various visual signal data sources and modalities frequently utilised in computer vision systems for horticulture and corresponding details in relation to their data representation. These data encompass a broad range of imaging methods, including 2D images, videos, hyperspectral imaging, 3D point clouds, etc., each offering unique characteristics and geometric formations that contribute to the learning capabilities of the models. To develop and evaluate vision systems, researchers and practitioners rely on datasets that are either publicly available or purpose-built for specific application requirements since the availability of relevant public quality datasets is a well-known challenge.
As shown in
Table 2, existing visual signal datasets in horticulture vary in terms of their data formats, coverage, and objectives. Some datasets focus on specific crops or plant species, while others cover a broader range of horticultural scenarios. These datasets may include annotations for classification, ground truth values for regression, or no ground truth for unsupervised (usually clustering) training. Irrespectively, the limited size and diversity of these datasets often hinder the generalisation and performance of computer vision models in real-world horticultural settings.
2D images remain the predominant form of visual input signal among the learning-based vision systems in horticulture, as shown in
Table 2. However, hyperspectral imaging and 3D point clouds are emerging formats, particularly for quality estimation and plant growth monitoring applications respectively. RGB images are also sometimes captured initially as videos and later extracted as individual image frames. Given the increasing demand and frequent usage of this format as the origin of visual signals, the remainder of the paper is primarily focused on 2D images/video frames.
Table 1.
Overview of Visual Signals Commonly Used in Horticulture.
Table 1.
Overview of Visual Signals Commonly Used in Horticulture.
| Imaging Method |
Characteristics |
Data and Representation |
| 2D Images |
Planar representations of objects captured from a single viewpoint or angle. These have height and width dimensions considered as resolution. |
Typically represented as matrices or tensors, with each element representing the pixel value at a specific location. Colour images have multiple channels (e.g., red, green, blue), while greyscale images have a single channel. |
| 2D Videos |
A sequence of images captured over time, creating a temporal dimension. Each frame is a 2D image. |
Composed of multiple frames, typically represented as a series of 2D image matrices or tensors. The temporal dimension adds an extra axis to represent time. |
| RGB-D Images |
Same 2D spatial dimensions as colour images, but include depth information. |
Consist of colour channels (red, green, blue) for visual appearance and a depth channel represented as distance or disparity values. Can be stored as multi-channel matrices or tensors. |
| 3D Point Clouds |
Represent the spatial coordinates of individual points in a 3D space. |
Consist of a collection of points, where each point is represented by its and z coordinates in the 3D space. |
| Thermal Imaging |
Same 2D spatial dimensions as regular images, but represent temperature values instead of colour or greyscale pixel values. |
Stored as matrices or tensors with scalar temperature values at each pixel location. |
| Fluorescence Imaging |
Captures and visualises the fluorescence emitted by objects, typically using specific excitation wavelengths. |
Represented as images or videos with intensity values corresponding to the emitted fluorescence signal. These can involve the use of specific fluorescence channels or spectra. |
| Transmittance Imaging |
Measures the light transmitted through objects, providing information about their transparency or opacity. |
Represented as images or videos with pixel values indicating the amount of light transmitted through each location. |
| Multi-spectral Imaging |
Captures images at multiple discrete wavelengths or narrow spectral bands. |
Represented as multi-dimensional arrays, where each pixel contains intensity values at different wavelengths or spectral bands. Provides detailed spectral information for analysis. |
| Hyperspectral Imaging |
Same 2D spatial dimensions as regular images, but each pixel contains a spectrum of reflectance values across the spectral bands. |
Represented as multi-dimensional arrays, where each pixel contains a spectrum of reflectance values across the spectral bands. Provides a more comprehensive spectral analysis compared to multi-spectral imaging. |
Despite being the most commonly used data format, the 2D visual signals available in the literature offer limited quality in terms of spatial resolution. It is evident from
Table 2 that most of the 2D datasets offer spatial resolutions below high-definition (HD). In a world where 4K and 8K spatial resolution capture is becoming the norm, high-quality data acquisition and processing present a major challenge in the horticulture domain due to environmental, human resource and hardware constraints.
Table 2.
Snapshot of Visual Signal Datasets for Computer Vision in Horticulture. Quality represents spatial resolution in pixels(px) and/or spectral resolution in nano-meters(nm). Quantity represents either the number of images, videos or objects (such as plants, leaves, etc.)
Table 2.
Snapshot of Visual Signal Datasets for Computer Vision in Horticulture. Quality represents spatial resolution in pixels(px) and/or spectral resolution in nano-meters(nm). Quantity represents either the number of images, videos or objects (such as plants, leaves, etc.)
| Dataset |
Imaging |
Quality |
Quantity |
Application |
| [35] |
RGB Image |
Varied Resolution |
587 images |
Detection of sweet pepper, rock melon, apple, mango, orange and strawberry. |
| [36] |
RGB Image |
px |
270 images |
Fruit maturity classification for pineapple. |
| [37] |
RGB Image |
|
1357 images |
Fruit maturity classification and defect detection for dates. |
| [38] |
RGB Image |
px |
764 leaves |
Growth classification for Okinawan spinach. |
| [39] |
Gray Image |
px |
13,200 |
Prediction of white cabbage seedling’s chances of success. |
| [40] |
Hyperspectral |
px px, 2.8nm |
45 plants |
Estimation of strawberry ripeness. |
| [41] |
2D Video |
px |
11 videos, >8000 frames |
Segmentation of apple and ripening stage classification. |
| [42] |
Timelapse Images |
px |
1218 images |
Monitoring growth of lettuce. |
| [43] |
RGB Images |
px |
1350 images |
Monitoring growth of lettuce. |
| [44] |
2D Video |
px |
20 videos |
Detection of apple and fruit counting. |
| [45] |
Timelapse Images |
px |
724 images |
Estimation of compactness, maturity and yield count for blueberry. |
| [45] |
RGB-D Images |
px |
123 images |
Detection of ripeness of tomato fruit. |
| [46] |
RGB-D Images |
px |
123 images |
Detection of ripeness of tomato fruit and counting. |
| [47] |
RGB Images |
px |
480 images |
Detection of growth stage of apple. |
| [48] |
RGB Images |
px, px |
108 images |
Classification of panicle stage of mango. |
| [49] |
RGB Images |
px, px |
3704 images |
Detection and segmentation of apple, mango and almond. |
| [50] |
RGB Images |
px |
8079 images |
Classification of fruit maturity and harvest decision. |
| [51] |
RGB-D and IRS |
px |
967 multimodal images |
Detection of Fuji apple. |
| [52] |
RGB Images |
px |
49 images |
Detection of mango fruit. |
| [53] |
Thermal Images |
px |
- |
Detection of bruise and its classification in pear fruit. |
| [54] |
RGB Images |
px |
1730 images |
Detection of mango fruit and orchard load estimation. |
| [55] |
RGB Images |
Varied Resolution |
2298 images |
Robotic harvesting and yield estimation of apple. |
| [56,57] |
RGB Images, Multiview Images and 3D Point Cloud |
px for images |
288 images |
Detection, localisation and segmentation of fuji apple. |
2.2. Data Requirement Analysis
The spatial resolution of 2D input data impacts deep learning models in terms of their ability to capture fine details and make precise predictions. Higher spatial resolution can provide more detailed information about crop characteristics, diseases, or quality attributes, leading to improved model performance. Therefore, the limited spatial resolution of 2D visual signal datasets in horticulture is a challenge that needs to be addressed for enhanced accuracy and applicability of computer vision models. To this end, we perform analysis to understand how the spatial resolution of 2D video frames impacts the quality of information contained within these frames and the features that a deep learning model can extract from the corresponding frames. We perform a statistical analysis in combination with deep feature extraction to gain insights into the information content and diversity using Shannon’s entropy [
58] analysis of the deep feature maps. We also perform a Fourier Transform analysis to highlight the variations in frequency details represented by varied spatial resolution.
2.2.1. Deep Feature Entropy Analysis
Based on
Section 1.1, it is evident that AlexNet and VGG16 are two of the most commonly used deep models to extract features in many different horticulture contexts. Hence, in this analysis, we study the impact of resolution on feature quality extracted from these two models. A video dataset developed for flower detection during flowering for apples is used for this analysis. The dataset was acquired for a growing season from the United States Department of Agriculture – Agriculture Research Service – Appalachian Fruit Research Station [
59]. The dataset consists of videos acquired on four different dates in April 2017 at the research station. The video for block 1A, row 2 of the orchard with the golden delicious apple variety and M.9 rootstock is considered in this analysis. Twenty frames were sampled from the middle of each video for the purpose of this analysis. In order to simulate low-resolution counterparts of the original camera-capture high-resolution video frames, Gaussian blur with a standard deviation of
and
down-sampling is used.
The low-resolution video frames and corresponding high-resolution frames are then fed to pre-trained VGG-16 and AlexNet to obtain feature maps from the final layer of these models. To analyse the feature quality extracted from low-resolution (LR) and camera-captured high-resolution (HR) frames, we make use of Shannon’s entropy analysis. In the context of deep learning feature maps, entropy is a measure of complexity in the distribution of feature values. Shannon’s entropy quantifies the level of information present represented as:
is the entropy of the feature map, n is the number of elements in the feature map, represents each element in the feature map and is the value of the element in the feature map. The entropy calculation is negated when dealing with probability distributions, where the values represent probabilities. The logarithm of a value between 0 and 1 will be a negative number. The entropy equation, when negated, ensures that the entropy value is positive. However, in the case of deep learning feature maps, the values are activations or responses, not necessarily in the range of 0-1, and thus the entropies in these cases might be a negative value. This still provides a meaningful measure of comparison of information present between the feature maps.
2.2.2. Fourier Transform Analysis
Fourier Transform analysis is utilised to examine the frequency details and variations present in different spatial resolutions. Converting video frames from the spatial domain to the frequency domain using Fourier Transform yields a frequency spectrum, providing insights into the distribution and intensity of different spatial frequencies of the original frame. By examining the magnitude of the spectrum of the Fourier Transform, we analyse the distribution of frequencies and identify the presence of high-frequency details. A higher concentration of high-frequency components indicates the presence of finer details and textures in the frame. Conversely, a lower concentration of high-frequency components suggests a loss of fine detail, which will typically be present in low-resolution frames.
The Fourier Transform decomposes the frame into a combination of sinusoidal components at different frequencies. This process yields a frequency spectrum that represents the amplitude and phase of each spatial frequency component in the frame. The magnitude spectrum is then computed from the Fourier Transform by taking the absolute value of the frequencies that represent the amplitude of each frequency component in the frame. A logarithm of magnitude spectrum is then taken to enhance the visibility of low-intensity frequency components. It provides information about the distribution and intensity of different spatial frequencies present in the frame and is used to compare different frames visually. This visualisation shows any differences in frequency content and the impact of spatial resolution on the distribution of frequencies.
2.3. Super-Resolution as a Generative AI Solution
Generative AI is the field of artificial intelligence that focuses on creating models and algorithms capable of generating new content, such as images, natural language texts or videos, based on existing data patterns. Superresolution, specifically in the context of image and video processing, involves generating high-resolution images or frames from low-resolution inputs [
60].
Superresolution, as a generative AI technique, is used to enhance and generate high-frequency details not present in the low-resolution input. These techniques leverage deep learning models to learn the mapping between low-resolution and high-resolution pairs. The process of generating a high-resolution (HR) frame with improved quality from a given low-resolution (LR) input in the context of videos is referred to as video super-resolution (VSR) [
61]. Assuming a high-resolution video frame has undergone the following operation:
Where is the low-resolution video frame when a high-resolution video frame is convoluted with a blur kernel k followed by downsampling operation d and the addition of noise , super-resolution of is then a task of estimating the blur kernel, downsampling operation, and the noise such that video frame can be obtained inversely from video frame.
Recent studies have adopted learning-based approaches that exploit spatiotemporal features present in an LR video in order to super-resolve it [
63,
64,
65,
66], gaining popularity due to their non-deterministic and data-driven nature, allowing them to generalise well over different videos and their ability to learn complex non-linear functions to map LR video to HR video. These models can provide a soft-computing solution to enhancing the spatial resolution of low-quality videos taken in the context of horticulture. The enhanced spatial resolution will impact the quality of the feature map that the deep models can extract and consequently their performance [
67]. To investigate how the information content and diversity within frames and corresponding feature maps change when a low-resolution video frame is enhanced with super-resolution, we make use of a state-of-the-art deep learning video super-resolution model.
Recent advancements in deep learning-based super-resolution have introduced several recurrent neural networks (RNN), such as BasicVSR [
65], Recurrent Residual Network (RRN) [
68], and Recurrent Structure-Detail Network (RSDN) [
63]. These models have demonstrated the capability to learn long-term inter-frame correlations within a given temporal radius, leading to improved super-resolution quality. Unlike alignment-based methods, RNN models leverage global information propagation, offering a more efficient and cost-effective alternative. While subsequent works like IconVSR [
65], BasicVSR++ [
69], and Global Omniscient Video Super-resolution [
70] have further refined recurrent modelling by adopting bi-directional approaches, they are not suitable for real-time applications. Horticulture applications of computer vision often involve scenarios where all frames are not simultaneously available such as robotic harvesting, making bi-directional models less applicable. A recent VSR model called Replenished Recurrency with Dual-Duct(R2D2) [
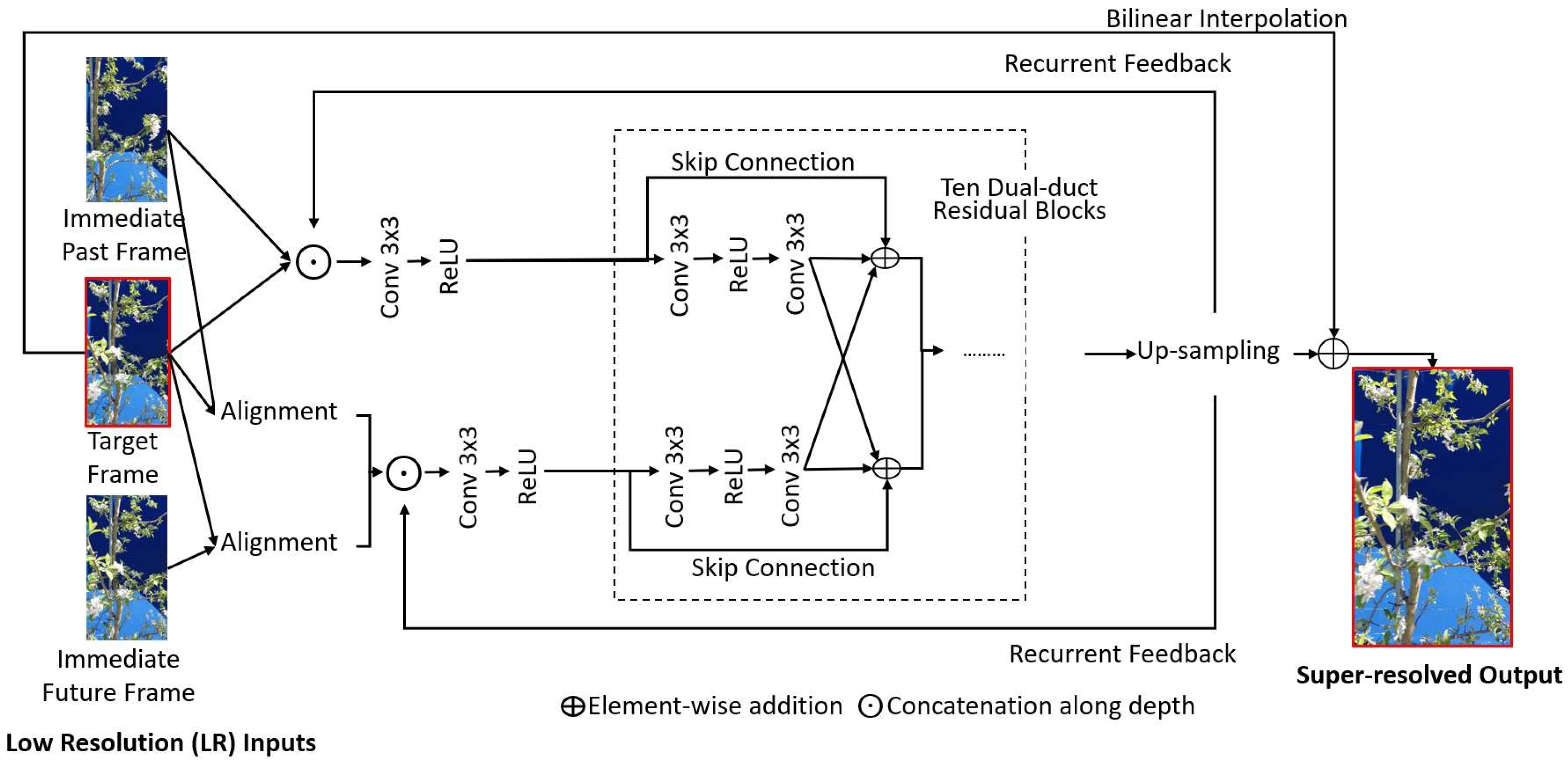
62] is aimed at enhancing the super-resolution performance of unidirectional recurrent models in real-time scenarios where limited frames are available for each timestamp. It incorporates a sliding-window-based local alignment and a dual-duct residual network resulting in a hybrid model with two information propagation pipelines for efficient super-resolution of each video frame, as shown in
Figure 2. R2D2 has demonstrated competitive performance and computational efficiency in the VSR task and is hence considered for this study.
Figure 2.
Overview of the architecture of the R2D2 [
62] super-resolution model.
Figure 2.
Overview of the architecture of the R2D2 [
62] super-resolution model.
3. Results and Discussion
3.1. Entropy of Deep Feature Maps
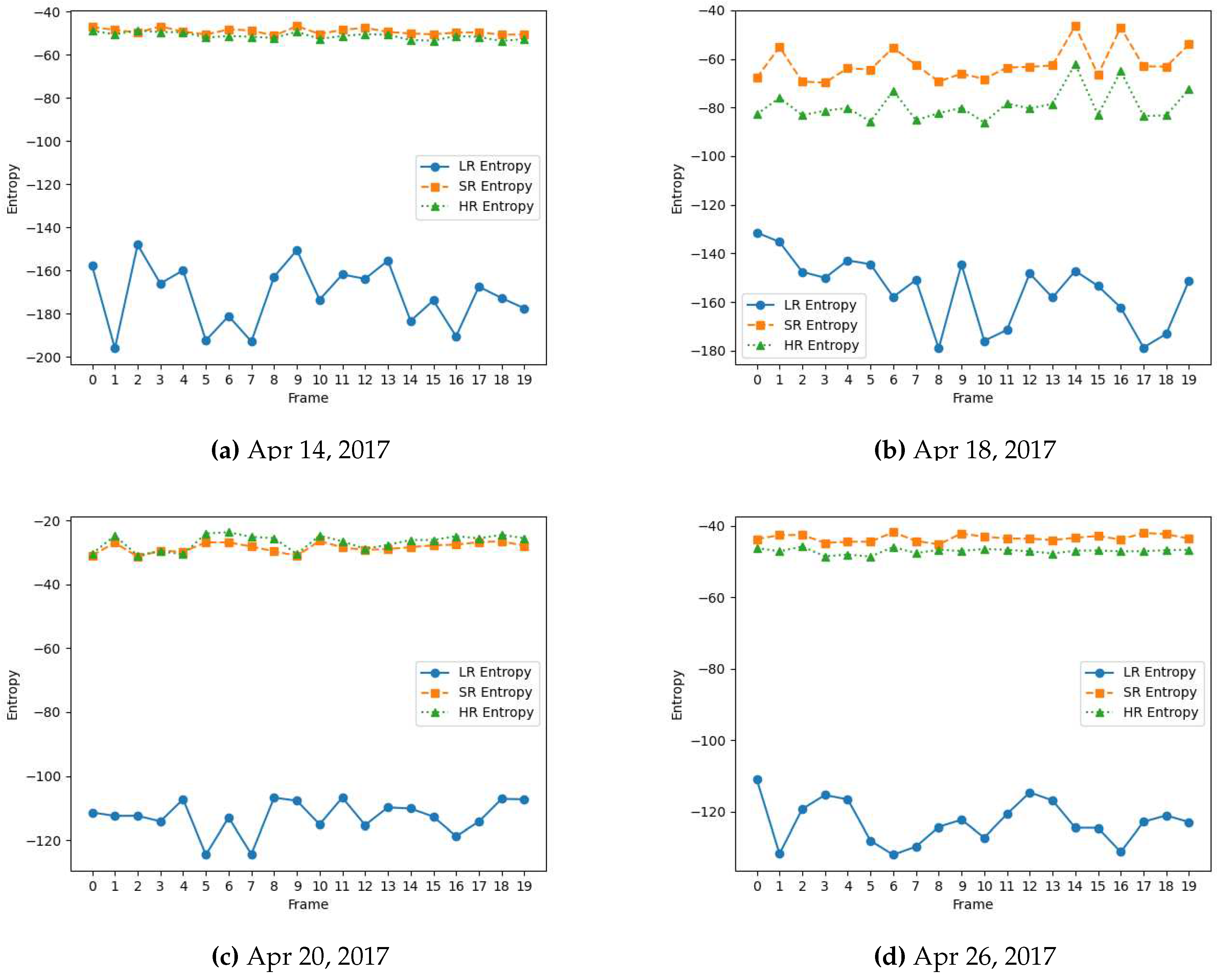
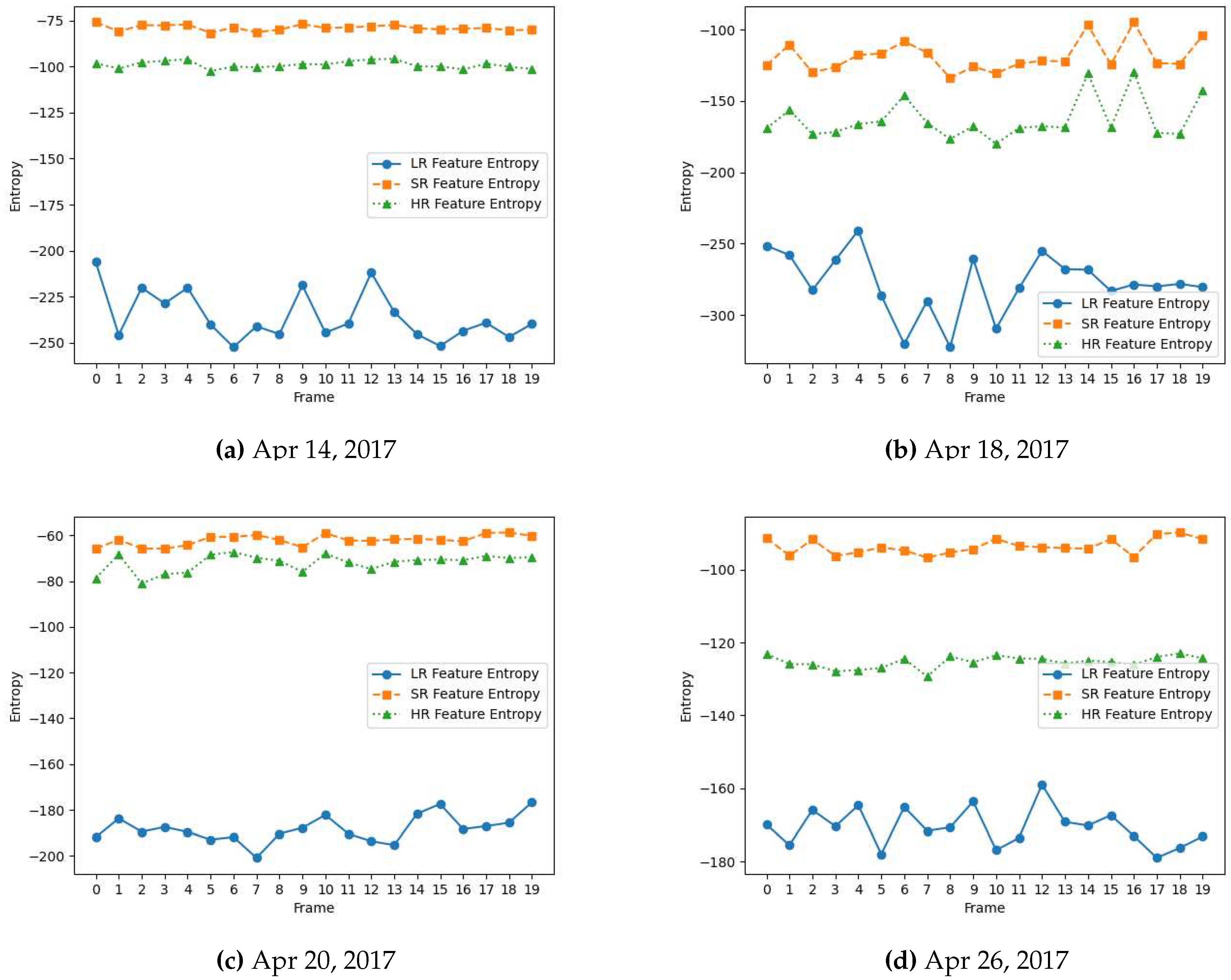
The entropy value reflects the diversity and variability of the feature values within the feature map. A higher entropy value indicates a more complex and diverse distribution of patterns or details in the input data that can benefit the vision task at hand. As shown in
Figure 4 and
Figure 5, high-resolution (HR) video frames originally captured from high-resolution cameras have higher entropy feature maps because they contain finer details and more complex structures. Conversely, low-resolution frames yield lower entropy feature maps as they often lack fine-grained information, making it more challenging for vision models to perform tasks effectively, highlighting the need for high-quality data for vision systems.
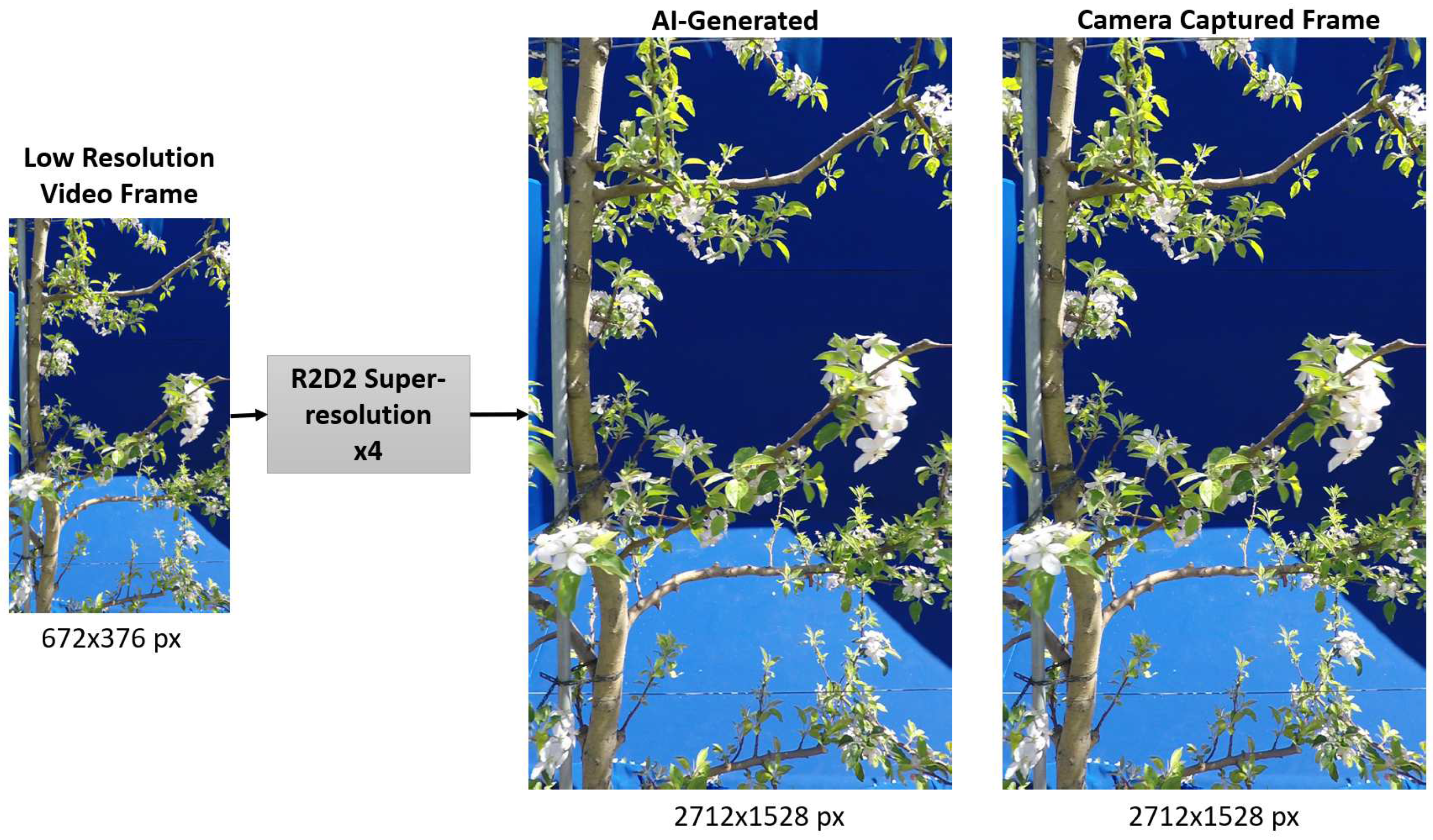
With advances in generative AI, the availability of such high-quality data does not always have to be hardware constrained. We show the potential of generative AI in creating high-quality counterparts of low-quality input data, providing a contemporary data acquisition pathway ahead and a potential to enhance the performance of learning-based vision systems. As shown in
Figure 3, a low-resolution video frame is super-resolved to
spatial dimension using the pre-trained R2D2 model. The enhanced frame is highly similar visually to its HR counterpart captured originally from a high-resolution camera. This signifies the data restorative and enhancing capabilities of the R2D2 learning-based super-resolution model. We further compare the spatial quality improvement in terms of the entropy of the feature maps extracted from these frames by VGG-16 and AlexNet. As shown in
Figure 4 and
Figure 5, the entropy of the super-resolved frame generated by R2D2 is highly similar to that of the original camera-captured frame indicating a similar level of information and details, while the low-resolution frame has significantly lower entropy.
Figure 3.
Super-resolution of LR frame using pre-trained R2D2 super-resolution model and corresponding output comparison with camera captured reference frame.
Figure 3.
Super-resolution of LR frame using pre-trained R2D2 super-resolution model and corresponding output comparison with camera captured reference frame.
Figure 4.
Entropy for feature maps of low-resolution (LR), super-resolved (SR) and camera-captured high-resolution (HR) frames extracted from VGG-16 model for flower-detection video data captured across the growing season in April 2017.
Figure 4.
Entropy for feature maps of low-resolution (LR), super-resolved (SR) and camera-captured high-resolution (HR) frames extracted from VGG-16 model for flower-detection video data captured across the growing season in April 2017.
Figure 5.
Entropy for feature maps of low-resolution (LR), super-resolved (SR) and camera-captured high-resolution (HR) frames from AlexNet model for flower-detection video data captured across the growing season in April 2017.
Figure 5.
Entropy for feature maps of low-resolution (LR), super-resolved (SR) and camera-captured high-resolution (HR) frames from AlexNet model for flower-detection video data captured across the growing season in April 2017.
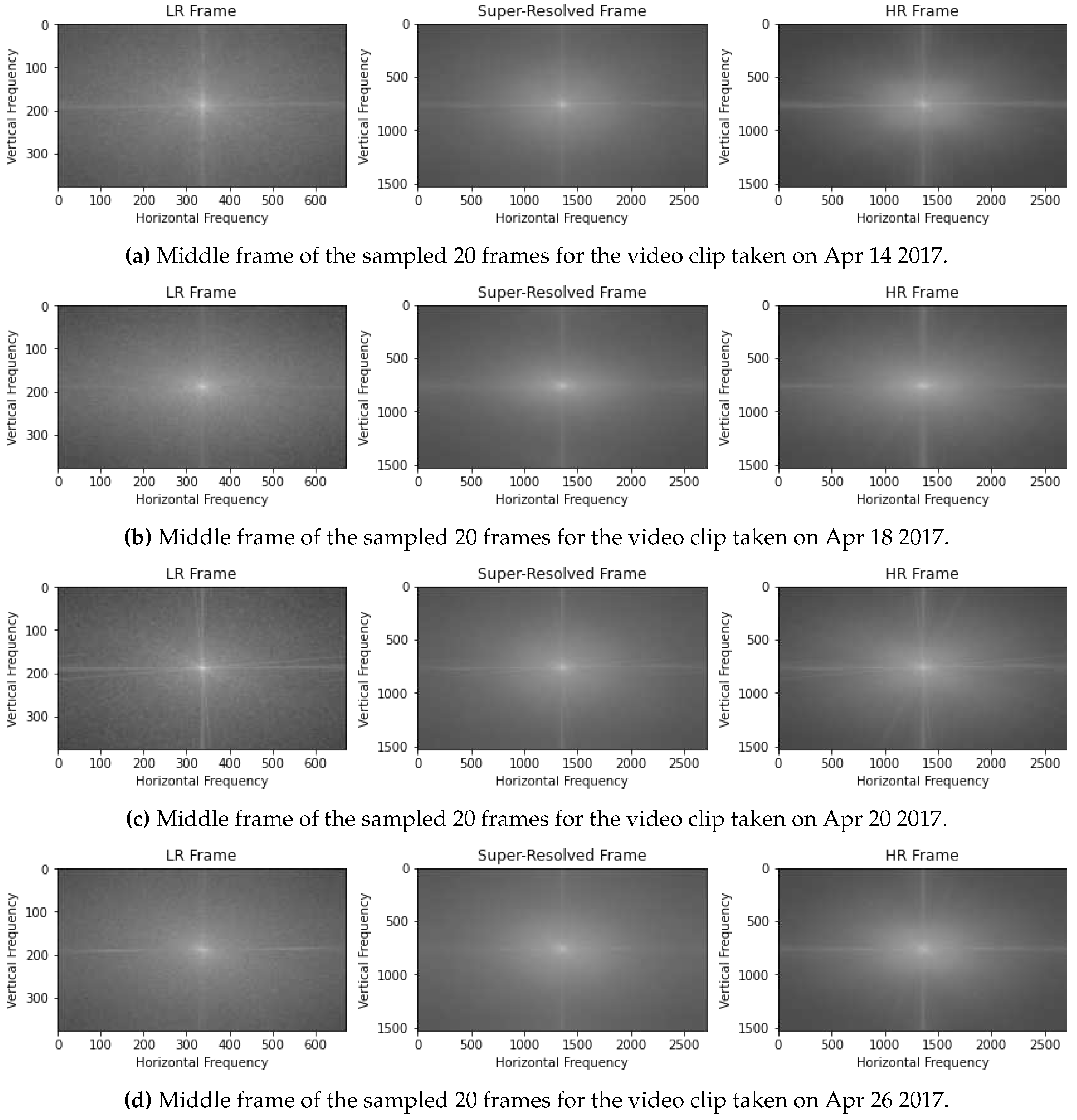
3.2. Magnitude Spectra
The horizontal and vertical frequency in magnitude spectra, as shown in
Figure 6 of the original camera-captured frame, is significantly higher than the low-resolution frame. This represents specific high-frequency components or patterns present in the frame that may correspond to edges, textures, or other prominent features in the case of the original camera-captured frame. In the super-resolved frame, we can observe the same range of high-frequencies present, indicating that the super-resolution has been successful in capturing and generating the high-frequency details similar to the camera-captured frame, suggesting that the super-resolution process has been effective in recovering fine details from the low-resolution frames. Super-resolution not only increases spatial resolution but also removes noise and artefacts from low-resolution input frames. The presence of blur noise represented by granular magnitude spectra in low-resolution frames is completely eliminated in super-resolved frames, and the concentration of frequency details is very similar to that of camera-captured HR frames.
This comparison of the magnitude spectra between the low-resolution, super-resolved, and original camera-captured frames provides insights into the quality and fidelity of the spatial information contained within each frame. It demonstrates the ability of the super-resolution to improve the representation of high-frequency details and recover the fine features present in high-resolution frames from low-resolution frames, leading to more accurate and high-quality data generation.
Figure 6.
Comaprison of the magnitude spectra of the frequency domain using Fourier transform of the video frames.
Figure 6.
Comaprison of the magnitude spectra of the frequency domain using Fourier transform of the video frames.
3.3. AI Generated vs Camera Captured – Quality and Cost
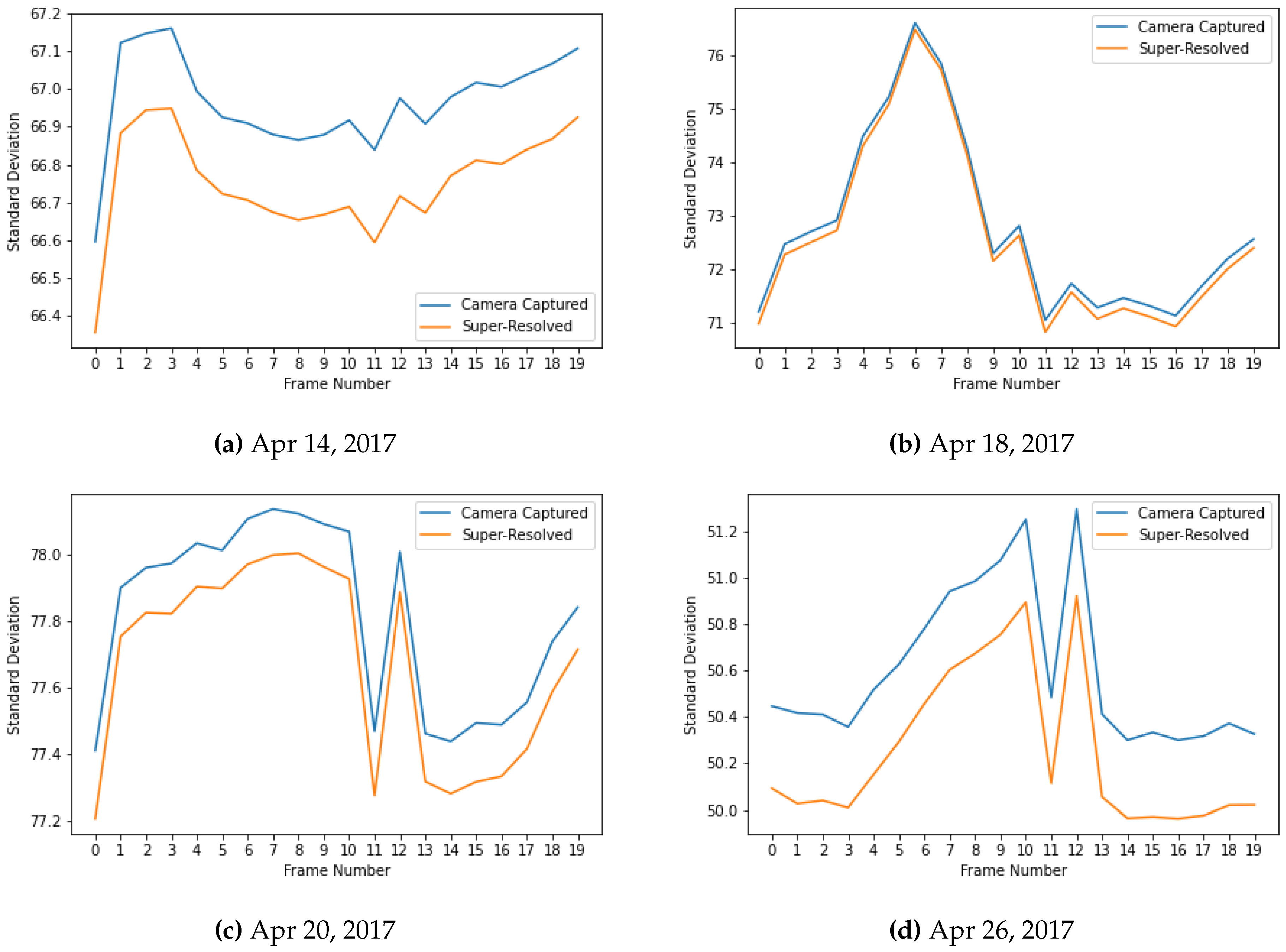
The analysis of pixel statistics, specifically the standard deviation, as shown in
Figure 7 across the generative AI-based super-resolved video frames and camera-captured frames reveals that the quality of the generated frames is comparable to that of the original high-resolution frames. The standard deviation is a measure of the spread or variability of pixel values within a frame. If the standard deviation is similar between the two sets of frames, it suggests that both sets exhibit comparable levels of variability and fine detail. The generative AI-based frames closely match that of the camera-captured high-resolution frames implying that the generative model successfully reproduces the fine detail and subtle variation that contribute to the overall quality of the image. It also indicates that the generative AI model is capable of producing frames with similar levels of richness and complexity as the original high-resolution camera-captured frames.
With no added cost, apart from the computational cost, and using an open-access generative AI-based VSR model (R2D2), the approach of using super-resolution to enhance the quality of 2D visual signals to meet the growing data demand for learning-based vision systems could be a game changer. This can also lead to significantly improved results from the subsequent computer vision models, especially where legacy low-resolution vision capture hardware is still in use. To provide a brief overview of hardware costs in obtaining high-quality data instead of using generative AI, we compare popular camera models in
Table 3, which shows that to obtain a
better resolution, a higher cost by
$1159 in case of Canon and
$2102 in case of Nikon is expected [
71]. Given the large areas that orchards and farms cover, with higher resolution requirements, the cost of high-resolution signal capture hardware can easily accumulate.
Figure 7.
Comparison of the standard deviation of pixel values in the super-resolved video frames with that of camera-captured frames, showcasing close similarity.
Figure 7.
Comparison of the standard deviation of pixel values in the super-resolved video frames with that of camera-captured frames, showcasing close similarity.
Table 3.
Comparison of Approximate Costs to Capture Higher Resolution Video for Popular Camera Brands [71]. Indicative Cost Differences (2023 prices) represent the difference between the high-resolution and low-resolution models for the same camera manufacturer.
Table 3.
Comparison of Approximate Costs to Capture Higher Resolution Video for Popular Camera Brands [71]. Indicative Cost Differences (2023 prices) represent the difference between the high-resolution and low-resolution models for the same camera manufacturer.
| Model |
Video Resolution (px) |
≈Price (USD) |
≈Cost (USD) |
| Canon EOS 2000D |
1920×1080 |
440 |
1159 |
| Canon EOS 90D |
3840×2160 |
1599 |
|
| Nikon D5200 |
1920×1080 |
695 |
2102 |
| Nikon D780 |
3840×2160 |
2797 |
|
| Fujifilm XF1 |
1920×1080 |
499 |
1750 |
| Fujifilm XT5 |
6240×3510 |
2249 |
|
Figure 8.
Magnified visual inspection of profiled frame contents to compare the visual quality of the LR vs super-resolved vs camera captured frames.
Figure 8.
Magnified visual inspection of profiled frame contents to compare the visual quality of the LR vs super-resolved vs camera captured frames.
Therefore, super-resolution can be used as a data pre-processing technique within computer vision systems in horticulture to enhance the spatial resolution and quality of input data allowing deep models to extract high-frequency details and features beneficial for vision tasks such as classification, detection and regression. This can also help reduce the cost associated with installing newer high-quality data capture hardware and produce better outcomes using lower-resolution legacy hardware. With faster and optimised VSR models emerging as the research in the super-resolution domain rapidly progresses, the computational constraints associated with using super-resolution as a pre-processing technique can be significantly circumscribed [
62,
68]. Super-resolution can also be extended using transfer-learning to enhance other forms of input visual signals, such as 3D point clouds, to enhance the number of points in 3D space and their quality from a lower to higher space [
72]. The cost difference in capturing dense vs spare 3D point clouds from LiDAR is significantly different, and the potential for a generative AI-based model to automatically produce a high-resolution 3D point cloud from a low-resolution 3D point cloud is significant.
4. Conclusions
The critical role of input data quality in computer vision systems in horticulture is explored in this work. Horticultural environments present diverse and complex characteristics, posing challenges in consistently capturing high-quality visual signals and retarding the development of robust computer vision models. Our analysis of the current state of visual signal datasets in horticulture domains highlights the limited spatial resolution of 2D images as an example constraint to applying more contemporary deep machine learning methods. To investigate the impact of resolution on feature quality, deep learning models, namely AlexNet and VGG16, are used on a video dataset for flower detection during Spring blooming in an apple orchard. The results demonstrated that higher spatial resolution led to higher entropy feature maps, indicating the capture of finer quality detail and more complex feature structures. Furthermore, Fourier transform analysis, pixel analysis, and visual inspections confirm the attainment of improved detail and more intricate feature structures.
To address data quality limitations, super-resolution is introduced as a generative AI solution in this paper. The potential of super-resolution using the R2D2 model is demonstrated to improve the spatial resolution of low-quality video inputs, with the enhanced video frames exhibiting higher entropy feature maps comparable to those captured by high-resolution cameras. This highlights the restorative and enhancing capabilities of learning-based super-resolution models on input visual signals. Incorporating super-resolution as a data pre-processing technique can enable deep learning models to extract high-frequency details, leading to improved performance in learning-based vision tasks. Furthermore, this approach provides a cost-effective alternative to hardware upgrades for obtaining high-quality visual signals from low-resolution inputs. Addressing the data quality requirements in horticulture through generative AI techniques such as super-resolution enables the full potential of computer vision systems and holds significant promise for advancement in overall horticultural practices, particularly in high-value horticultural crops.
Author Contributions
Conceptualization, A.A.B; methodology, A.A.B; software, A.A.B; validation, A.A.B, T-K.L., P.W.E, S.A.; formal analysis, A.A.B; investigation, A.A.B; data curation, A.A.B; writing—original draft preparation, A.A.B; writing—review and editing, A.A.B, T-K.L, P.W.E. and S.A.; visualization, A.A.B, T-K.L, P.W.E; supervision, T-K.L, P.W.E. and S.A. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yang, B.; Xu, Y. Applications of deep-learning approaches in horticultural research: A review. Horticulture Research 2021, 8. [Google Scholar] [CrossRef]
- Spalding, E.P.; Miller, N.D. Image analysis is driving a renaissance in growth measurement. Curr. Opin. Plant Biol. 2013, 16, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; He, L.; Schupp, J.; Lyons, D.; Heinemann, P. Green fruit segmentation and orientation estimation for robotic green fruit thinning of apples. Comput. Electron. Agric. 2023, 207, 107734. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition, 2014, pp. 580–587.
- Yang, X.; Sun, M. A survey on deep learning in crop planting. IOP conference series: Materials science and engineering. IOP Publishing, 2019, Vol. 490, p. 062053.
- Pound, M.P.; Atkinson, J.A.; Townsend, A.J.; Wilson, M.H.; Griffiths, M.; Jackson, A.S.; Bulat, A.; Tzimiropoulos, G.; Wells, D.M.; Murchie, E.H.; others. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 2017, 6, gix083. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.S.; Tou-Hong, L.; Yen, K.S. Deep Learning for Image-Based Plant Growth Monitoring: A Review. Int. J. Eng. Technol. Innov. 2022, 12, 225. [Google Scholar] [CrossRef]
- Colaço, A.F.; Molin, J.P.; Rosell-Polo, J.R.; Escolà, A. Application of light detection and ranging and ultrasonic sensors to high-throughput phenotyping and precision horticulture: Current status and challenges. Horticulture Research 2018, 5, 35. [Google Scholar] [CrossRef]
- Pujari, J.D.; Yakkundimath, R.; Byadgi, A.S. Image processing based detection of fungal diseases in plants. Procedia Comput. Sci. 2015, 46, 1802–1808. [Google Scholar] [CrossRef]
- Tsaftaris, S.A.; Minervini, M.; Scharr, H. Machine learning for plant phenotyping needs image processing. Trends Plant Sci. 2016, 21, 989–991. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Park, D.S. Deep learning-based techniques for plant diseases recognition in real-field scenarios. Advanced Concepts for Intelligent Vision Systems: 20th International Conference, ACIVS 2020, Auckland, New Zealand, February 10–14, 2020, Proceedings 20. Springer, 2020, pp. 3–14.
- Indrakumari, R.; Poongodi, T.; Khaitan, S.; Sagar, S.; Balamurugan, B. A review on plant diseases recognition through deep learning. Handb. Deep Learn. Biomed. Eng.
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 1–18. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. Arxiv Prepr. 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant disease detection and classification by deep learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef] [PubMed]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- He, L.; Fang, W.; Zhao, G.; Wu, Z.; Fu, L.; Li, R.; Majeed, Y.; Dhupia, J. Fruit yield prediction and estimation in orchards: A state-of-the-art comprehensive review for both direct and indirect methods. Comput. Electron. Agric. 2022, 195, 106812. [Google Scholar] [CrossRef]
- Girshick, R. Fast r-cnn. Proceedings of the IEEE international conference on computer vision, 2015, pp. 1440–1448.
- Rebortera, M.; Fajardo, A. Forecasting Banana Harvest Yields using Deep Learning. 2019 IEEE 9th International Conference on System Engineering and Technology (ICSET), 2019, pp. 380–384. [CrossRef]
- Koirala, A.; Walsh, K.B.; Wang, Z.; McCarthy, C. Deep learning–Method overview and review of use for fruit detection and yield estimation. Comput. Electron. Agric. 2019, 162, 219–234. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 779–788.
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 7263–7271.
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. Arxiv Prepr. 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. Yolov4: Optimal speed and accuracy of object detection. Arxiv Prepr. 2020, arXiv:2004.10934. [Google Scholar]
- Nturambirwe, J.F.I.; Opara, U.L. Machine learning applications to non-destructive defect detection in horticultural products. Biosystems engineering 2020, 189, 60–83. [Google Scholar] [CrossRef]
- Arendse, E.; Fawole, O.A.; Magwaza, L.S.; Opara, U.L. Non-destructive prediction of internal and external quality attributes of fruit with thick rind: A review. J. Food Eng. 2018, 217, 11–23. [Google Scholar] [CrossRef]
- Wieme, J.; Mollazade, K.; Malounas, I.; Zude-Sasse, M.; Zhao, M.; Gowen, A.; Argyropoulos, D.; Fountas, S.; Van Beek, J. Application of hyperspectral imaging systems and artificial intelligence for quality assessment of fruit, vegetables and mushrooms: A review. Biosystems Engineering 2022, 222, 156–176. [Google Scholar] [CrossRef]
- Lu, Y.; Saeys, W.; Kim, M.; Peng, Y.; Lu, R. Hyperspectral imaging technology for quality and safety evaluation of horticultural products: A review and celebration of the past 20-year progress. Postharvest Biol. Technol. 2020, 170, 111318. [Google Scholar] [CrossRef]
- Zujevs, A.; Osadcuks, V.; Ahrendt, P. Trends in Robotic Sensor Technologies for Fruit Harvesting: 2010-2015. Procedia Comput. Sci. 2015, 77, 227–233, ICTE in regional Development 2015 Valmiera. [Google Scholar] [CrossRef]
- Barbashov, N.; Shanygin, S.; Barkova, A. Agricultural robots for fruit harvesting in horticulture application. IOP Conference Series: Earth and Environmental Science. IOP Publishing, 2022, Vol. 981, p. 032009.
- Zhang, X.; Karkee, M.; Zhang, Q.; Whiting, M.D. Computer vision-based tree trunk and branch identification and shaking points detection in Dense-Foliage canopy for automated harvesting of apples. J. Field Robot. 2021, 38, 476–493. [Google Scholar] [CrossRef]
- Benavides, M.; Cantón-Garbín, M.; Sánchez-Molina, J.; Rodríguez, F. Automatic tomato and peduncle location system based on computer vision for use in robotized harvesting. Applied Sciences 2020, 10, 5887. [Google Scholar] [CrossRef]
- Sa, I.; Ge, Z.; Dayoub, F.; Upcroft, B.; Perez, T.; McCool, C. Deepfruits: A fruit detection system using deep neural networks. sensors 2016, 16, 1222. [Google Scholar] [CrossRef] [PubMed]
- Azman, A.A.; Ismail, F.S. Convolutional neural network for optimal pineapple harvesting. Elektr. -J. Electr. Eng. 2017, 16, 1–4. [Google Scholar]
- Nasiri, A.; Taheri-Garavand, A.; Zhang, Y.D. Image-based deep learning automated sorting of date fruit. Postharvest Biol. Technol. 2019, 153, 133–141. [Google Scholar] [CrossRef]
- Hao, X.; Jia, J.; Khattak, A.M.; Zhang, L.; Guo, X.; Gao, W.; Wang, M. Growing period classification of Gynura bicolor DC using GL-CNN. Comput. Electron. Agric. 2020, 174, 105497. [Google Scholar] [CrossRef]
- Perugachi-Diaz, Y.; Tomczak, J.M.; Bhulai, S. Deep learning for white cabbage seedling prediction. Comput. Electron. Agric. 2021, 184, 106059. [Google Scholar] [CrossRef]
- Gao, Z.; Shao, Y.; Xuan, G.; Wang, Y.; Liu, Y.; Han, X. Real-time hyperspectral imaging for the in-field estimation of strawberry ripeness with deep learning. Artif. Intell. Agric. 2020, 4, 31–38. [Google Scholar] [CrossRef]
- Sabzi, S.; Abbaspour-Gilandeh, Y.; García-Mateos, G.; Ruiz-Canales, A.; Molina-Martínez, J.M.; Arribas, J.I. An automatic non-destructive method for the classification of the ripeness stage of red delicious apples in orchards using aerial video. Agronomy 2019, 9, 84. [Google Scholar] [CrossRef]
- Lu, J.Y.; Chang, C.L.; Kuo, Y.F. Monitoring growth rate of lettuce using deep convolutional neural networks. 2019 ASABE Annual International Meeting. American Society of Agricultural and Biological Engineers, 2019, p. 1.
- Reyes-Yanes, A.; Martinez, P.; Ahmad, R. Real-time growth rate and fresh weight estimation for little gem romaine lettuce in aquaponic grow beds. Comput. Electron. Agric. 2020, 179, 105827. [Google Scholar] [CrossRef]
- Gao, F.; Fang, W.; Sun, X.; Wu, Z.; Zhao, G.; Li, G.; Li, R.; Fu, L.; Zhang, Q. A novel apple fruit detection and counting methodology based on deep learning and trunk tracking in modern orchard. Comput. Electron. Agric. 2022, 197, 107000. [Google Scholar] [CrossRef]
- Ni, X.; Li, C.; Jiang, H.; Takeda, F. Deep learning image segmentation and extraction of blueberry fruit traits associated with harvestability and yield. Horticulture research 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Afonso, M.; Fonteijn, H.; Fiorentin, F.S.; Lensink, D.; Mooij, M.; Faber, N.; Polder, G.; Wehrens, R. Tomato fruit detection and counting in greenhouses using deep learning. Front. Plant Sci. 2020, 11, 571299. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, G.; Wang, Z.; Wang, H.; Li, E.; Liang, Z. Apple detection during different growth stages in orchards using the improved YOLO-V3 model. Comput. Electron. Agric. 2019, 157, 417–426. [Google Scholar] [CrossRef]
- Koirala, A.; Walsh, K.B.; Wang, Z.; Anderson, N. Deep learning for mango (Mangifera indica) panicle stage classification. Agronomy 2020, 10, 143. [Google Scholar] [CrossRef]
- Bargoti, S.; Underwood, J. Deep fruit detection in orchards. 2017 IEEE international conference on robotics and automation (ICRA). IEEE, 2017, pp. 3626–3633.
- Altaheri, H.; Alsulaiman, M.; Muhammad, G. Date fruit classification for robotic harvesting in a natural environment using deep learning. IEEE Access 2019, 7, 117115–117133. [Google Scholar] [CrossRef]
- Gené-Mola, J.; Vilaplana, V.; Rosell-Polo, J.R.; Morros, J.R.; Ruiz-Hidalgo, J.; Gregorio, E. Multi-modal deep learning for Fuji apple detection using RGB-D cameras and their radiometric capabilities. Comput. Electron. Agric. 2019, 162, 689–698. [Google Scholar] [CrossRef]
- Kestur, R.; Meduri, A.; Narasipura, O. MangoNet: A deep semantic segmentation architecture for a method to detect and count mangoes in an open orchard. Eng. Appl. Artif. Intell. 2019, 77, 59–69. [Google Scholar] [CrossRef]
- Zeng, X.; Miao, Y.; Ubaid, S.; Gao, X.; Zhuang, S. Detection and classification of bruises of pears based on thermal images. Postharvest Biol. Technol. 2020, 161, 111090. [Google Scholar] [CrossRef]
- Koirala, A.; Walsh, K.; Wang, Z.; McCarthy, C. Deep learning for real-time fruit detection and orchard fruit load estimation: Benchmarking of ‘MangoYOLO’. Precision Agriculture 2019, 20, 1107–1135. [Google Scholar] [CrossRef]
- Bhusal, S.; Karkee, M.; Zhang, Q. Apple Dataset Benchmark from Orchard Environment in Modern Fruiting Wall 2019.
- Gené-Mola, J.; Sanz-Cortiella, R.; Rosell-Polo, J.R.; Morros, J.R.; Ruiz-Hidalgo, J.; Vilaplana, V.; Gregorio, E. Fruit detection and 3D location using instance segmentation neural networks and structure-from-motion photogrammetry. Comput. Electron. Agric. 2020, 169, 105165. [Google Scholar] [CrossRef]
- Gené-Mola, J.; Gregorio, E.; Cheein, F.A.; Guevara, J.; Llorens, J.; Sanz-Cortiella, R.; Escolà, A.; Rosell-Polo, J.R. LFuji-air dataset: Annotated 3D LiDAR point clouds of Fuji apple trees for fruit detection scanned under different forced air flow conditions. Data Brief 2020, 29, 105248. [Google Scholar] [CrossRef]
- Leung, L.W.; King, B.; Vohora, V. Comparison of image data fusion techniques using entropy and INI. 22nd Asian Conference on Remote Sensing, 2001, Vol. 5, pp. 152–157.
- Tabb, A.; Medeiros, H. Video Data of Flowers, Fruitlets, and Fruit in Apple Trees during the 2017 Growing Season at USDA-ARS-AFRS. Ag Data Commons, 2018. [CrossRef]
- Wang, X.; Chan, K.C.; Yu, K.; Dong, C.; Change Loy, C. Edvr: Video restoration with enhanced deformable convolutional networks. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, 2019, pp. 0–0.
- Haris, M.; Shakhnarovich, G.; Ukita, N. Recurrent back-projection network for video super-resolution. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, pp. 3897–3906.
- Baniya, A.A.; Lee, T.K.; Eklund, P.W.; Aryal, S.; Robles-Kelly, A. Online Video Super-Resolution using Information Replenishing Unidirectional Recurrent Model. Neurocomputing 2023, 126355. [Google Scholar] [CrossRef]
- Isobe, T.; Jia, X.; Gu, S.; Li, S.; Wang, S.; Tian, Q. Video Super-Resolution with Recurrent Structure-Detail Network. Computer Vision – ECCV 2020; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.M., Eds.; Springer International Publishing: Cham, 2020; pp. 645–660. [Google Scholar]
- Isobe, T.; Li, S.; Jia, X.; Yuan, S.; Slabaugh, G.; Xu, C.; Li, Y.L.; Wang, S.; Tian, Q. Video Super-Resolution With Temporal Group Attention. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2020.
- Chan, K.C.; Wang, X.; Yu, K.; Dong, C.; Loy, C.C. BasicVSR: The Search for Essential Components in Video Super-Resolution and Beyond. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2021, pp. 4947–4956.
- Baniya, A.A.; Lee, T.K.; Eklund, P.W.; Aryal, S. Omnidirectional Video Super-Resolution using Deep Learning. IEEE Trans. Multimed. 2023. [Google Scholar] [CrossRef]
- Yamamoto, K.; Togami, T.; Yamaguchi, N. Super-resolution of plant disease images for the acceleration of image-based phenotyping and vigor diagnosis in agriculture. Sensors 2017, 17, 2557. [Google Scholar] [CrossRef]
- Isobe, T.; Zhu, F.; Jia, X.; Wang, S. Revisiting temporal modeling for video super-resolution. Arxiv Prepr. 2020, arXiv:2008.05765. [Google Scholar]
- Chan, K.C.; Zhou, S.; Xu, X.; Loy, C.C. BasicVSR++: Improving video super-resolution with enhanced propagation and alignment. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2022, pp. 5972–5981.
- Yi, P.; Wang, Z.; Jiang, K.; Jiang, J.; Lu, T.; Tian, X.; Ma, J. Omniscient video super-resolution. Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021, pp. 4429–4438.
- Product Chart. Product Chart. Online, 2023. https://www.productchart.com/cameras/.
- Shan, T.; Wang, J.; Chen, F.; Szenher, P.; Englot, B. Simulation-based lidar super-resolution for ground vehicles. Robot. Auton. Syst. 2020, 134, 103647. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).