1. Introduction
Klebsiella pneumoniae is the second most common Gram-negative opportunistic pathogen and one of the most prevalent causes of community and hospital acquired infections [
1]. It is responsible for health care-associated pneumonia [
2] and bacterial neonatal sepsis in low- and middle-income countries [
3]. A serious public health threat is the emergence and dissemination of carbapenem-resistant
K. pneumoniae (CRKP) that is associated with high morbidity and mortality, increased medical costs and prolonged hospital stay [
4]. CRKP isolates have a variety of mechanisms, which may confer resistance to virtually all available β-lactam antibacterial drugs, including carbapenems. The main resistance molecular mechanism is the production of a range of carbapenemases, including KPC, NDM, VIM and OXA-48-like carbapenemases [
5,
6]. KPC-producing CRKP strains display the most extensive global distribution and represent a significant challenge due to their limited therapeutic options [
7].
A novel β-lactam/β-lactamase inhibitor (BL/BLIs) combination is effective against strains of non-metallo-β lactamase producing
Enterobacterales (Ambler class A, class C and some class D β-lactamases) [
8,
9]. Ceftazidime/avibactam (CAZ/AVI) [
10] has become an important first-line option for treating adult and pediatric (>3 months of age) patients with serious infections caused by carbapenem-resistant organisms, but not yet for neonates (IDSA) [
11]. It is indicated for the treatment of complicated intra-abdominal and urinary tract infections, and infections caused by carbapenem-resistant
Enterobacterales (CRE) or carbapenem-resistant
Pseudomonas aeruginosa, in patients with limited or no other treatment options [
12].
Although KPC producing
Enterobacterales strains are generally considered susceptible to CAZ/AVI, isolates resistant to this antimicrobial agent have been documented without the evidence of metallo-β-lactamases [
13]. Ιn 2018, rapid risk assessment conducted by ECDC identified CAZ-AVI resistance in CRE as public health threat that merits careful monitoring [
14]. CAZ/AVI resistance mechanisms include increased expression of the blaKPC gene product (acquisition of resistance was mostly associated with isolates harboring the substitution D179Y in KPC-3 or in KPC-2) [
15,
16], the presence of other genetic determinants of resistance against ESBL-producing
Enterobacterales (SHV-, CTX-M- or VEB-type β-lactamases) [
17,
18], changes in cell permeability (i.e. non-functional porins- OmpK35, OmpK36 and OmpK37) [
19] and expression of efflux pumps [
20].
VEB-type β-lactamases (Vietnamese extended-spectrum β-lactamase) are a group of Ambler class A enzymes inhibited by avibactam. VEB-25 differs from VEB-1 by a missense mutation (substitution of lysine by arginine at position 237 -K234R) [
21], which compromises the inhibitory efficiency of avibactam [
22].
Here in, we report a successful treatment of bloodstream infection associated with CAZ/AVI -resistant KPC-2 producing- K. pneumoniae carrying the VEB-25 gene in a preterm neonate hospitalized in the neonatal intensive care unit (NICU) of a tertiary hospital and the use of a customized active surveillance program in conjunction to infection control measures for early recognition and prevention of an outbreak.
2. Results
2.1. Index case
A favorable clinical and microbiological response was documented including defervescence and decrease of CRP within 48-72 hours, first negative blood culture within 4 days and discontinuation of invasive mechanical ventilation within 8 days since colistin and tigecycline initiation. Administration of both daptomycin and CAZ/AVI was discontinued, whereas ciprofloxacin was empirically added four days after the first positive blood culture for a total of 13 days. The neonate was successfully treated with colistin and tigecycline for a total of 18 days.
NGS report
A variety of genes conferring resistance to antimicrobial agents and heavy metals, as well as genes related with virulence, capsule, and efflux and regulator systems were detected (
Table 1). Only one serine-carbapenemase was detected, which was the KPC-2 gene. Other five β-lactamases (SHV-33, TEM-1B, VEB-25, DHA-1, OXA-10) were co-detected, including the gene VEB-25. Co-production of KPC-2 and VEB-25 in
K. pneumoniae has been associated with CAZ/AVI resistance in the absence of a metallo-β-lactamase [
17].
2.2. Molecular and phenotypic surveillance within the NICU and the hospital
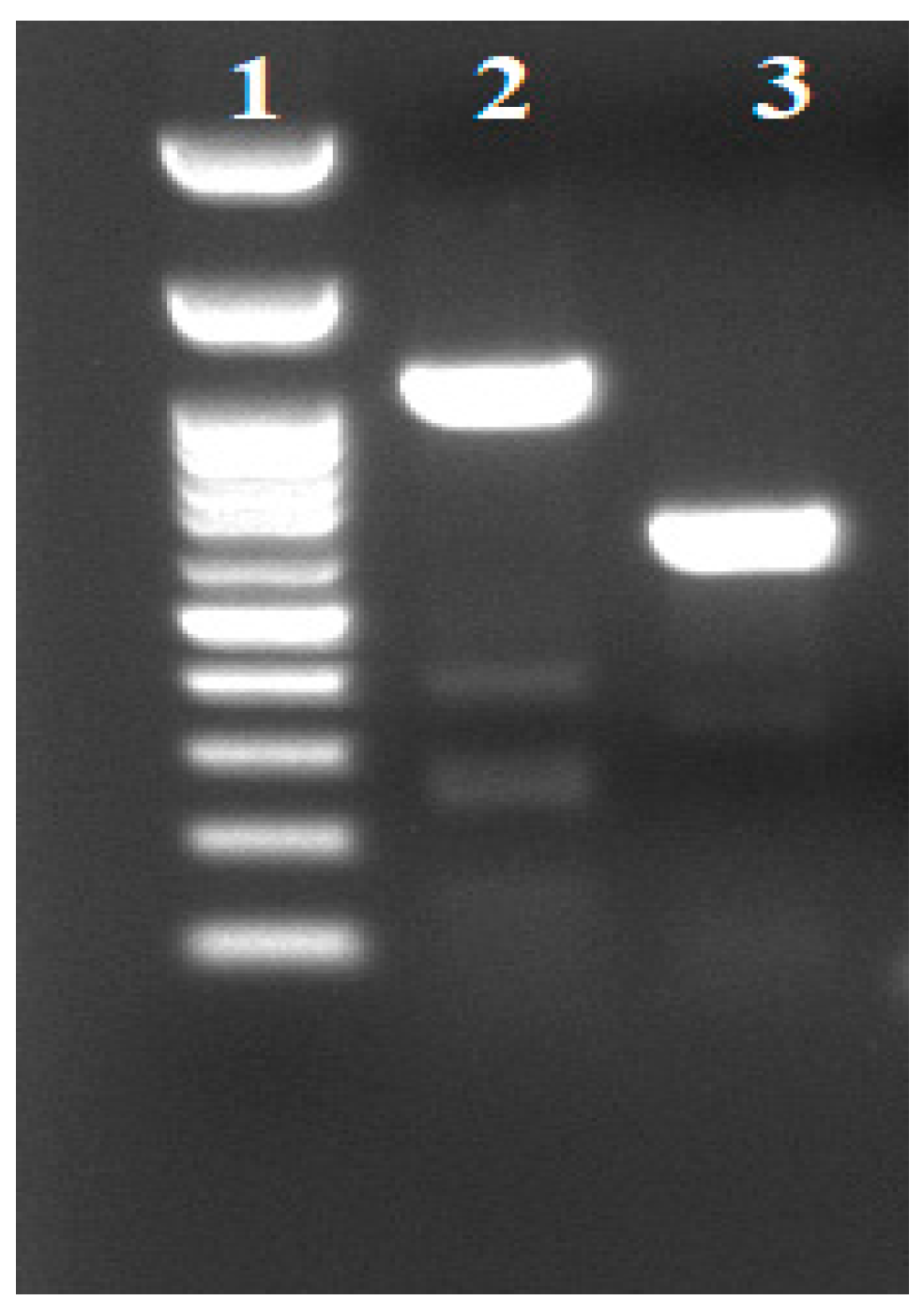
Thirteen
K. pneumoniae strains were isolated from stool samples from neonates hospitalized in NICU within a period of 3 months upon the recognition of the index case. Among these isolates only the index case was VEB-25 positive (
Figure 1), confirming the NGS result. Based on AST results, 24 additional carbapenem resistant
K. pneumoniae strains collected from various hospital sites were also analyzed by targeted PCR; although they contained blaVEB-1, they did not harbor blaVEB-25.
2.3. Overall assessment
This index case was the last neonate that was infected by Acinetobacter baumannii and colonized by Providencia stuartii within the NICU after the implementation of enhanced infection control measures targeting these two pathogens. Upon the recognition of the first K. pneumoniae producing KPC-2 and VEB-25, and combination of intensified and targeted infection control actions there were no other cases within the NICU for the next 6 months.
3. Discussion
We report a neonatal case of bloodstream infection caused by a
K. pneumoniae strain co-producing KPC and VEB-25 β-lactamases and the use of precise medicine to customize infection control measures. Treatment options for infections caused by carbapenem-resistant bacteria are extremely limited in neonates. The use of either “last-line” antimicrobial agents (such as polymyxins and tigecycline) or the currently available newer β-lactam/ β-lactam inhibitor combinations, such as CAZ/AVI, meropenem-vaborbactam and imipenem-relebactam that are not licensed for neonates yet, for empirical treatment of neonatal sepsis in areas endemic for CRKP is still questionable due to limited pharmacokinetic data and local epidemiology of resistant genes [
31].
To date, there are three reports of CAZ/AVI resistant KPC-producing-
K. pneumoniae emergence in Greece, all in adults (six infected and five colonized patients) [
17,
32,
33]. Notably, the first CAZ/AVI resistant clinical isolate was detected in Greece before the introduction of CAZ/AVI in clinical practice. The resistance was due to the production of KPC-23 (variant differed from KPC-3 by one -V240A, and from KPC-2 by two amino acid substitutions -V240A and H274Y) [
32]. CAZ/AVI resistance due to VEB-25 production has been reported in additional two cases (one isolate from blood and one from the lower respiratory tract) from patients without prior CAZ/AVI exposure [
33]. Eight more CAZ/AVI resistant CRKP isolates were detected in patients not previously exposed to CAZ/AVI (two patients with catheter-related bloodstream infections, one with ventilator-associated pneumonia and five with colonization); the resistance was conferred by VEB-25 and VEB-14 production [
17]. After intense epidemiological and microbiological surveillance in our NICU, as well as in adult departments within our general hospital, we could not find the source of this resistant organism. However, our index case had been previously exposed to multiple courses of antimicrobial agents, including CAZ/AVI, as well as it had gut colonization with XDR Gram-negative bacteria, such as
A. baumannii and
P. stuartii.
This was the first premature neonate presenting with sepsis due to CAZ/AVI-resistant KPC-2 producing-
K. pneumoniae carrying the VEB-25 gene that was successfully treated with non-conventional antimicrobial agents. Currently, available diagnostic platforms detect the presence of the most prevalent carbapenemases, such as KPC, VIM, NDM and OXA. Neonatologists and infectious disease specialists should be cautious when interpreting results from these molecular platforms for decision making in empiric and targeted treatment for neonatal sepsis. The mechanism of resistance, especially for the newer β-lactam/β-lactamase inhibitors, may differ in times and in different parts of the world and even within the same institution [
34]. In addition, various mechanisms of CAZ/AVI resistance emphasize the need for surveillance of CAZ/AVI-resistant pathogens, as well as for its judicious use.
4. Materials and Methods
4.1. Index case
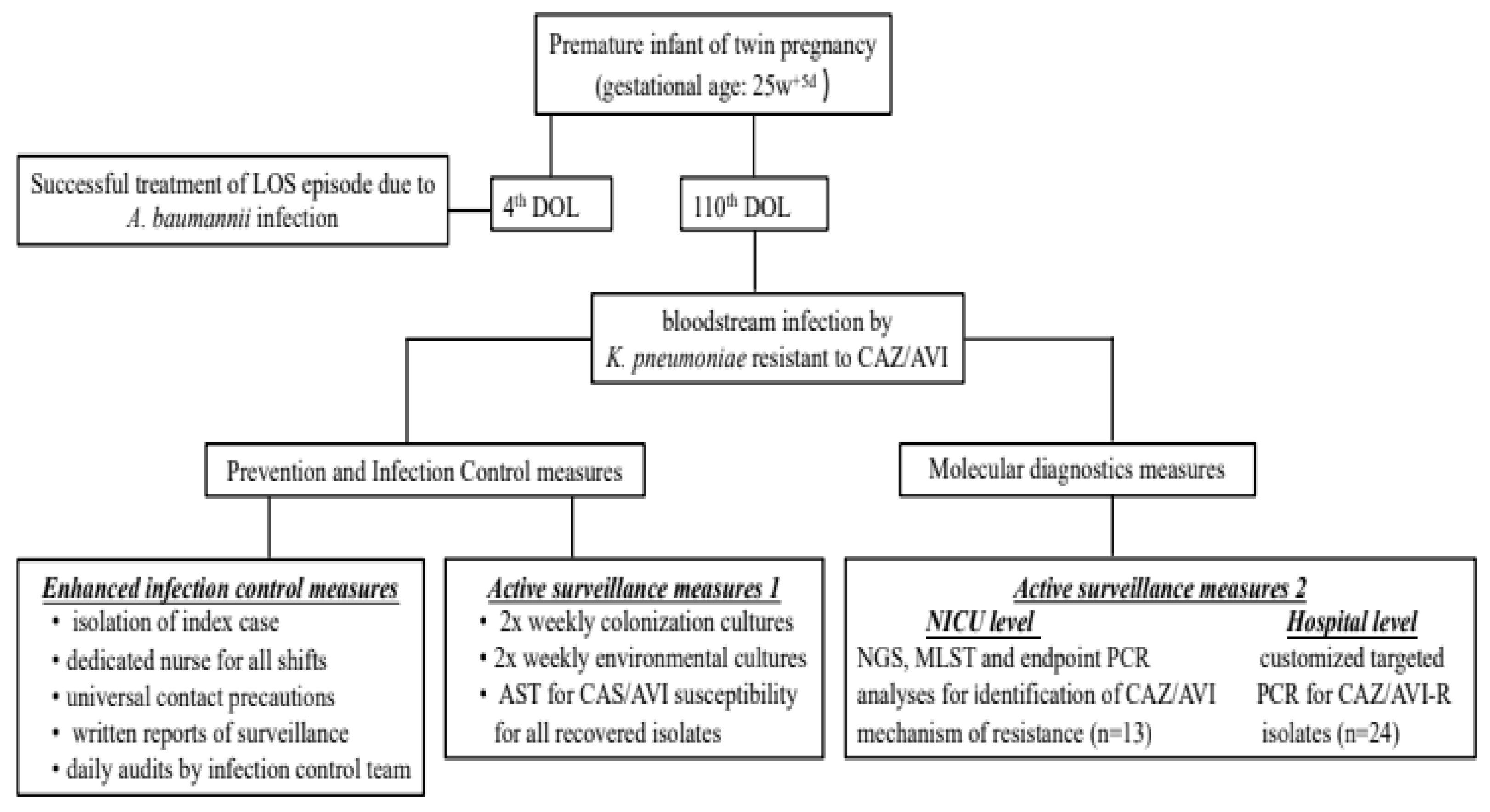
The index case was the first neonate of a twin pregnancy born to a 33-year-old healthy primigravida at gestational age of 25w+5d (birth weight=850 gr, appropriate for gestational age neonate) due to premature rapture of membranes and onset of labor. Postnatally, the patient presented respiratory distress syndrome, patent ductus arteriosus, severe bronchopulmonary dysplasia and need for prolonged mechanical ventilation, posthemorrhagic ventricular dilation, gastro-oesophageal reflux disease, retinopathy of prematurity and episodes of late onset sepsis (LOS). The first LOS occurred at 4th Day of life due to carbapenem resistant Acinetobacter baumannii, which was successfully treated. The patient was colonized with carbapenem resistant A. baumannii and P. stuartii between Day 4 and 25, respectively. During that time, the neonate had been exposed to multiple antibiotic regimens for prolonged time periods, including meropenem, aminoglycosides, colistin, tigecycline and ceftazidime-avibactam due to episodes of suspected LOS and colonization by CR Gram negative bacteria.
At Day 108, the neonate was on nasal continuous positive airway pressure due to chronic lung disease, and presented with fever, impaired peripheral perfusion and empiric antibiotic treatment with colistin (300,000 IU/kg/day every 8 hours), tigecycline (2 mg/kg/day every 12 hours) and daptomycin (10mg/kg/day once daily) was immediately initiated for suspected sepsis and the previous history of multiple antimicrobial regimens. Blood culture was positive for a Gram-negative rod within 24 hours since onset of symptoms. A multiplex PCR platform (Biofire® FilmArray®, Biomeriuex, Marcy-l’Étoile, France) was used within an hour from positive blood culture. A KPC producing K. pneumoniae was detected and CAZ/AVI at a reduced dose of 31 mg/kg/d every 8 hours was added to the antimicrobial regimen in attendance of the Antimicrobial Susceptibility Testing (AST).
During the first 48 hours of this sepsis episode the neonate deteriorated requiring mechanical ventilation and had high inflammatory indices (max CRP value of 394 mg/L) and thrombocytopenia. At day 110 the AST displayed high level of resistance to almost all antimicrobial agents: piperacillin/tazobactam, cefepime, cefoxitin, ceftazidime, ceftriaxone, imipenem, meropenem (MIC ≥16 mg/L), amikacin, gentamicin, ampicillin/sulbactam, aztreonam, ciprofloxacin, levofloxacin, fosfomycin, trimethoprim/ sulfamethoxazole. It was resistant to novel agents, like ceftolozane/tazobactam and CAZ-AVI, while it was susceptible to tigecycline and colistin. The isolate displayed a positive Phenyl Boronic Acid phenotypic test and the Lateral Flow ImmunoAssay and PCR method confirmed that the isolate carried KPC carbapenemase.
4.2. Risk assessment and bundle of actions taken after index case
That was the first case of a bloodstream infection caused by KPC producing
K. pneumoniae that was resistant to CAZ/AVI without the presence of a metalo-β-lactamase in the multiplex PCR platform in a neonate. The bundle of actions implemented is summarized in
Figure 2 and included: 1) enhanced infection control measures including strict isolation of the case index; 2) continuation of active surveillance for CRE and test for CAZ/AVI susceptibility report for all isolates recovered from surveillance; 3) application of next generation sequencing (NGS) and molecular testing for the index case to identify probable mechanism(s) of CAZ/AVI resistance; 4) targeted PCR analysis in all CRE isolates from all neonates in the ICU, independently to CAZ/AVI susceptibility, 5) targeted PCR analysis specifically for CAZ/AVI-R isolates from other departments of the hospital to identify potential sources and/ or burden of a potential outbreak.
4.2.1. Infection control measures
The NICU was already on strict infection control measures, including cohorting of all neonates colonized/infected with an XDR A. baumannii strain. Upon recognition of this index case extra measures were taken: isolation of index case, dedicated nurse for all shifts, universal application of contact precautions, written reports of active surveillance and daily audits by infection control team (with a dedicated IC nurse and a dedicated pediatric infectious disease specialist).
4.2.2. Active surveillance
Already in place with twice weekly colonization cultures. AST was applied to all isolates including CAZ/AVI susceptibility. Active surveillance included not only gut and pharyngeal colonization but also environmental cultures.
4.3. Microbiological methods - Antimicrobial susceptibility testing - Phenotypic analysis
CRKP was identified by VITEK 2 automated system (BioMérieux) using the GN ID according to the manufacturer’s instructions. AST of K. pneumoniae was performed using the AST 376 and XN10 cards; the interpretation of results was according to the EUCAST breakpoints of January 2022. Susceptibility testing to CAZ/AVI was performed using MIC test strips (Liofilchem srl, Roseto, Italy), while susceptibility testing to colistin was performed by broth microdilution method (Liofilchem srl, Roseto degli Abruzzi, Italy). Tigecycline was evaluated using the susceptibility breakpoints approved by the US Food and Drug Administration.
The isolate was phenotypically tested for KPC and metallo-β-lactamase (MBL) production using phenylboronic acid and ethylenediaminetetraacetic acid. Carbapenemase genes blaKPC, blaNDM, blaOXA-48-like, blaIMP, and blaVIM were screened by a multiplex lateral flow immunoassay (NG-Test CARBA 5, NG Biotech, France). The detection limits using purified recombinant enzymes for NDM, KPC, IMP, VIM and OXA-48-like were 150, 600, 200, 300 and 300 pg/mL, respectively.
4.4. Next generation sequencing (NGS)
DNA was extracted using the DNA extraction kit (Qiagen, Hilden, Germany). The Qubit double strand DNA HS assay kit (Q32851, Life Technologies Corporation, Grand Island, NY, USA) was used for measurement of dsDNA concentration. All procedures regarding shearing, purification, ligation, barcoding, size selection, library amplification and quantitation, emulsion PCR and enrichment were conducted according to manufacturer’s guidelines. After template enrichment, sequencing was performed on an Ion PGM™ semiconductor sequencer, using a Hi-Q View Sequencing Kit and a 316 Chip V2 BC (Thermo Fisher Scientific, Waltham MA, USA). The sequence reads were de novo assembled and annotatedusing Geneious Prime version 2021.2.1. The sequence of K. pneumoniae NTUH-K2044 strain (Accession number NC-012731) was used as reference.
4.5. MLST and detection of antimicrobial resistance genes and plasmids
MLST and antimicrobial resistance genes and plasmids were identified using the online databases at the Center for Genomic Epidemiology (MLST-2.0, Resfinder 4.1 and Plasmid finder) [
23,
24,
25,
26,
27,
28,
29,
30], and the Comprehensive Antibiotic Resistance Database (CARD Bait Capture Platform 1.0.0 (
https://card.mcmaster.ca/). Genes related to virulence, resistance to heavy metals, efflux and regulator systems and capsule were detected using Institut Pasteur website on
K. pneumoniae. (
https://bigsdb.pasteur.fr/klebsiella/).
4.6. Targeted PCR analysis
Molecular surveillance at NICU and Hospital level: After the recognition of VEB-25 as the mechanism of CAZ/AVI resistance in KPC
K. pneumoniae, targeted PCR protocol was started to investigate transmission within the NICU, but also to other carbapenem-resistant
K. pneumoniae isolated from other departments in the hospital. A total of 37
K. pneumoniae strains were tested for the presence of blaVEB-1. Thirteen of them were isolated from stool samples collected from neonates in the NICU where the VEB-25 index case was identified and 24 strains were isolated from different clinical sources (blood, urine, tracheal aspirate, trauma and central venous catheter) collected from several departments of the hospital to investigate potential sites of outbreak. Plasmid DNA was extracted using the alkaline lysis method, as described previously (H.C.Birnboim and J.Doly NAR 7: 1513-1523, 1979). For PCR amplification, VEB-F (5’-CGA CTT CCA TTT CCC GAT GC-3’) and VEB-B (5’-GGA CTC TGC AAC AAA TAC GC-3’) primers were used as diagnostic primers to amplify a 642 bp internal VEB-1 DNA segment, whereas the external primers VEBcas-F (5’-GTT AGC GGT AAT TTA ACC AGA TAG-3’) and VEBcas-B (5’-CGG TTT GGG CTA TGG GCA G-3’) were used to amplify the entire gene for DNA sequencing. For each PCR reaction, 50-70 ng of
K. pneumoniae plasmid DNA was used in a standard PCR reaction using Kapa Hi Fi DNA polymerase (KAPA Biosystems) with the following amplification program: 1 cycle of 95oC 3 min, 35 cycles of 20 sec at 94oC, 30 sec at 55oC, 30 sec at 72oC and a final extension step of 1 min at 72oC. The PCR products were Sanger sequenced. Nucleotide sequence analysis and pairwise alignments were performed using the National Center of Biotechnology Information website (
http://www.ncbi.nih.gov).
5. Conclusions
Applying the NGS technology is crucial for guiding the prediction of underlying resistance mechanisms facilitating the evolution and molecular epidemiology studies of the pathogens. The emergence of VEB-25 is a warning for horizontal transfer of plasmids at hospital facilities and it is of greatest concern for maintaining a sharp vigilance for surveillance of novel resistance mechanisms. Use of molecular diagnostics may guide appropriate antimicrobial therapy, early implementation of strict infection control measures and therefore play an important role in the fight against antimicrobial resistance.
Author Contributions
C.Z: Investigation, Formal analysis, Review and editing, Writing the initial draft. E.I: Investigation, Formal analysis, Review and editing, Writing the initial draft. M.S: Formal analysis, Review and editing. S.P: Investigation, Review and editing. A.K: Provision of study materials, Review and editing. E.R: Methodology, Data curation, Provision of study materials, Formal analysis, Review and editing, Supervision. A.P: Conceptualization, Methodology, Data curation, Formal analysis, Visualization, Reviewing and editing, Supervision, Funding acquisition. All authors have read and agreed to the published version of the manuscript. C.Z and E.I contributed equally to this work. In addition, E.R and A.P contributed equally to this work.
Funding
This work was supported by the European Union′s Horizon 2020 project VEO (grant number 874735).
Institutional Review Board Statement
This study was performed in line with the principles of the Declaration of Helsinki. The study was approved by the Ethics Committee of Aristotle’s University Medical Faculty (no. of approval 5.160/18-12-19). Since this is a retrospective analysis mainly microbiological of the bacteria isolated from the index case and during surveillance from other hospitalized patients according to the policy of the Infection Control and Prevention Committee of Hippokration General Hospital there was no need for informed consenting by the parents or the patients.
Informed Consent Statement
Informed consent for publication was signed by the father of the index patient.
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available due to these are the results of patient examinations carried out in a public hospital, but are available from the corresponding author on reasonable request.
Acknowledgments
The authors thank the personnel of the NICU that helped receiving surveillance samples from the patients, and the laboratory personnel who helped in the laboratory work. The help of Kostantinos Zarras in the experiments is also greatly acknowledged.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Accession Numbers
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under Biosample accession number SAMEA112484914.
References
- Ssekatawa, K.; Byarugaba, D.K.; Nakavuma, J.L.; Kato, C.D.; Ejobi, F.; Tweyongyere, R.; Eddie, W.M. Prevalence of pathogenic Klebsiella pneumoniae based on PCR capsular typing harbouring carbapenemases encoding genes in Uganda tertiary hospitals. Antimicrob. Resist. Infect. Control 2021, 10, 57. [Google Scholar] [CrossRef]
- Juan, C.H.; Chuang, C.; Chen, C.H.; Li, L.; Lin, Y.T. Clinical characteristics, antimicrobial resistance and capsular types of community-acquired, healthcare-associated, and nosocomial Klebsiella pneumoniae bacteremia. Antimicrob. Resist. Infect. Control 2019, 8, 1. [Google Scholar] [CrossRef]
- Milton, R.; Gillespie, D.; Dyer, C.; Taiyari, K.; Carvalho, M.J.; Thomson, K.; Sands, K.; Portal, E.A.R.; Hood, K.; Ferreira, A.; et al. Neonatal sepsis and mortality in low-income and middle-income countries from a facility-based birth cohort: An international multisite prospective observational study. Lancet. Glob. Health 2022, 10, e661–e672. [Google Scholar] [CrossRef]
- Agyeman, A.A.; Bergen, P.J.; Rao, G.G.; Nation, R.L.; Landersdorfer, C.B. Mortality, clinical and microbiological response following antibiotic therapy among patients with carbapenem-resistant Klebsiella pneumoniae infections (a meta-analysis dataset). Data Brief 2020, 28, 104907. [Google Scholar] [CrossRef] [PubMed]
- Ainoda, Y.; Aoki, K.; Ishii, Y.; Okuda, K.; Furukawa, H.; Manabe, R.; Sahara, T.; Nakamura-Uchiyama, F.; Kurosu, H.; Ando, Y.; et al. Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae ST258 isolated from a Japanese patient without a history of foreign travel - a new public health concern in Japan: A case report. BMC Infect. Dis. 2019, 19, 20. [Google Scholar] [CrossRef]
- Mathers, A.J.; Vegesana, K.; German-Mesner, I.; Ainsworth, J.; Pannone, A.; Crook, D.W.; Sifri, C.D.; Sheppard, A.; Stoesser, N.; Peto, T.; et al. Risk factors for Klebsiella pneumoniae carbapenemase (KPC) gene acquisition and clinical outcomes across multiple bacterial species. J. Hosp. Infect. 2020, 104, 456–468. [Google Scholar] [CrossRef]
- Jean, S.S.; Harnod, D.; Hsueh, P.R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- Sherry, N.; Howden, B. Emerging Gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam - epidemiology, laboratory detection and treatment implications. Expert Rev. Anti-Infect. Ther. 2018, 16, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Coskun, Y.; Atici, S. Successful Treatment of Pandrug-resistant Klebsiella pneumoniae Infection With Ceftazidime-avibactam in a Preterm Infant: A Case Report. Pediatr. Infect. Dis. J. 2020, 39, 854–856. [Google Scholar] [CrossRef] [PubMed]
- Administration., U.S.F.a.D., FDA approves new antibacterial drug Avycaz. FDA news release; U.S. FDA, Silver Spring. 26 February 2015.
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum beta-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2022. [Google Scholar] [CrossRef]
- van Duin, D.; Bonomo, R.A. Ceftazidime/Avibactam and Ceftolozane/Tazobactam: Second-generation beta-Lactam/beta-Lactamase Inhibitor Combinations. Clin. Infect. Dis. 2016, 63, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, S.; Giacobbe, D.R.; Maraolo, A.E.; Viaggi, V.; Luzzati, R.; Bassetti, M.; Luzzaro, F.; Principe, L. Resistance to ceftazidime/avibactam in infections and colonisations by KPC-producing Enterobacterales: A systematic review of observational clinical studies. J. Glob. Antimicrob. Resist. 2021, 25, 268–281. [Google Scholar] [CrossRef] [PubMed]
- (ECDC), E.C.f.D.P.a.C., Emergence of resistance to ceftazidime-avibactam in carbapenem-resistant Enterobacteriaceae - 12 June 2018. 2018. Stockholm; ECDC; 2018.
- Hemarajata, P.; Humphries, R.M. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J. Antimicrob. Chemother. 2019, 74, 1241–1243. [Google Scholar] [CrossRef]
- Haidar, G.; Clancy, C.J.; Shields, R.K.; Hao, B.; Cheng, S.; Nguyen, M.H. Mutations in blaKPC-3 That Confer Ceftazidime-Avibactam Resistance Encode Novel KPC-3 Variants That Function as Extended-Spectrum beta-Lactamases. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Galani, I.; Karaiskos, I.; Souli, M.; Papoutsaki, V.; Galani, L.; Gkoufa, A.; Antoniadou, A.; Giamarellou, H. Outbreak of KPC-2-producing Klebsiella pneumoniae endowed with ceftazidime-avibactam resistance mediated through a VEB-1-mutant (VEB-25), Greece, September to October 2019. Euro Surveill. : Bull. Eur. Sur Les Mal. Transm. = Eur. Commun. Dis. Bull. 2020, 25. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum beta-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Humphries, R.M.; Hemarajata, P. Resistance to Ceftazidime-Avibactam in Klebsiella pneumoniae Due to Porin Mutations and the Increased Expression of KPC-3. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Nelson, K.; Hemarajata, P.; Sun, D.; Rubio-Aparicio, D.; Tsivkovski, R.; Yang, S.; Sebra, R.; Kasarskis, A.; Nguyen, H.; Hanson, B.M.; et al. Resistance to Ceftazidime-Avibactam Is Due to Transposition of KPC in a Porin-Deficient Strain of Klebsiella pneumoniae with Increased Efflux Activity. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Protonotariou, E.; Meletis, G.; Pilalas, D.; Mantzana, P.; Tychala, A.; Kotzamanidis, C.; Papadopoulou, D.; Papadopoulos, T.; Polemis, M.; Metallidis, S.; et al. Polyclonal Endemicity of Carbapenemase-Producing Klebsiella pneumoniae in ICUs of a Greek Tertiary Care Hospital. Antibiotics 2022, 11. [Google Scholar] [CrossRef]
- Lahiri, S.D.; Alm, R.A. Identification of Novel VEB beta-Lactamase Enzymes and Their Impact on Avibactam Inhibition. Antimicrob. Agents Chemother. 2016, 60, 3183–3186. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef]
- Clausen, P.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Ponten, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.; Wisplinghoff, H.; Rodriguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Griffiths, D.; Fawley, W.; Kachrimanidou, M.; Bowden, R.; Crook, D.W.; Fung, R.; Golubchik, T.; Harding, R.M.; Jeffery, K.J.; Jolley, K.A.; et al. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 2010, 48, 770–778. [Google Scholar] [CrossRef]
- Jaureguy, F.; Landraud, L.; Passet, V.; Diancourt, L.; Frapy, E.; Guigon, G.; Carbonnelle, E.; Lortholary, O.; Clermont, O.; Denamur, E.; et al. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom. 2008, 9, 560. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Pestel-Caron, M.; Lemeland, J.F.; Pons, J.L. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 2004, 42, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Zusman, O.; Altunin, S.; Koppel, F.; Dishon Benattar, Y.; Gedik, H.; Paul, M. Polymyxin monotherapy or in combination against carbapenem-resistant bacteria: Systematic review and meta-analysis. J. Antimicrob. Chemother. 2017, 72, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Galani, I.; Antoniadou, A.; Karaiskos, I.; Kontopoulou, K.; Giamarellou, H.; Souli, M. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin. Microbiol. Infect. 2019, 25, e763–e765. [Google Scholar] [CrossRef]
- Voulgari, E.; Kotsakis, S.D.; Giannopoulou, P.; Perivolioti, E.; Tzouvelekis, L.S.; Miriagou, V. Detection in two hospitals of transferable ceftazidime-avibactam resistance in Klebsiella pneumoniae due to a novel VEB beta-lactamase variant with a Lys234Arg substitution, Greece, 2019. Euro Surveill. : Bull. Eur. Sur Les Mal. Transm. = Eur. Commun. Dis. Bull. 2020, 25. [Google Scholar] [CrossRef]
- Evans, S.R.; Tran, T.T.T.; Hujer, A.M.; Hill, C.B.; Hujer, K.M.; Mediavilla, J.R.; Manca, C.; Domitrovic, T.N.; Perez, F.; Farmer, M.; et al. Rapid Molecular Diagnostics to Inform Empiric Use of Ceftazidime/Avibactam and Ceftolozane/Tazobactam Against Pseudomonas aeruginosa: PRIMERS IV. Clin. Infect. Dis. 2019, 68, 1823–1830. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).