1. Introduction
An enzyme called DNA polymerase I (also known as Pol I) is involved in bacterial DNA replication. For the first time, DNA polymerase was isolated from E. coli and studied by Arthur Kornberg in 1956. DNA polymerases are critical for DNA replication and repair because they produce complementary DNA strands from a DNA template. Most living organisms possess a variety of DNA polymerases, which are classified into the following 7 groups based on their basic structures: A, B, C, D, E, X, and Y (Kushida et al., 2019). A number of DNA-binding proteins work together during the tightly controlled process of DNA replication to create nascent DNA strands that match the template DNA sequences. Hub proteins in the DNA transaction machinery, such as the bacterial single-stranded binding protein (SSB), and eukaryotic and archaeal replication protein A (RPA), are essential for protecting transiently formed single-stranded DNA (ssDNA) during the process of unwinding double-stranded DNA (dsDNA), detecting DNA damage, and recruiting repair proteins (Nagata et al., 2019).
The thermostable DNA polymerase is known as Taq polymerase was isolated from the thermophilic bacterium Thermus aquaticus. It serves a major purpose in the polymerase chain reaction (PCR) method, automating the tedious process of amplifying particular DNA sequences. DNA molecules can be multiplied up to a billion times using the polymerase chain reaction. This results in the production of numerous particular genes that can be used in subsequent applications (Ishino and Ishino, 2014). Chien et al. (1976) published the report as their course project for their master's programme. Nobody anticipated how well-known this enzyme would eventually become at the time. The Klenow fragment of DNA polymerase I from Escherichia coli was used in PCR (polymerase chain reaction) technology, which was first published in 1985. (Saiki et al., 1985). It was simple to anticipate that this method of gene amplification might develop into a useful technology if a heat-stable DNA polymerase that is not inactivated at the denaturation stage from double-stranded to single-stranded DNA existed. Later, a straightforward and reliable PCR technique with Taq polymerase was published (Saiki et al., 1988).
The advantages of using Thermococcus kodakarensis (KOD) thermostable DNA polymerase for PCR amplification and subsequent detection through mass spectrometry were reported (Benson et al., 2003). Thermococcus kodakarensis DNApol was prepared using a technique developed by researchers, and purified DP1 and DP2 proteins formed a stable complex in solution. The N-terminal region of DP1 was found to contain an intrinsically disordered region, but the static light scattering analysis gave DP1 a reasonable molecular weight (Takashima et al., 2019). The molecular mechanism by which CMG (GAN–MCM–GINS)-like helicase cooperates with the DNApol in Thermococcus kodakarensis was mentioned in a report. The archaeal GINS contains two Gins51 subunits, the C-terminal domain of which (Gins51C) interacts with GAN. This is the first proof that replicase and helicase in Archaea have a functional relationship. These results imply that the direct interaction of DNA pol with CMG-helicase is crucial for synchronizing strand unwinding and nascent strand synthesis and might offer a piece of functional machinery for the effective development of the replication fork (Oki et al., 2022).
The long segment of DNA polymerase I from Geobacillus stearothermophilus GIM1.543 (Bst DNA polymerase) containing 5′-3′ DNA polymerase capability while lack of 5′-3′ exonuclease activity shows high heat stability and polymerase activity. The efficiency of polymerase activity was increased by replacing residues Gly310 and Asp540 with alanine or leucine (Ma et al., 2016). DNA polymerase developed from Geobacillus stearothermophilus has a strand-displacement activity and is employed in loop-mediated isothermal amplification (LAMP) for fast detection of COVID-19 (Agustriana et al., 2022).
Despite the applications of Taq pol, it has some serious limitations. The specificity of Taq DNA polymerase is lower than that of regular ones. Taq polymerase lacks the activity of the 3' to 5' exonuclease, which makes it unable to fix mismatched nucleotides. Huang et al. (1992) showed Taq polymerase's inability to stretch mismatches effectively seems to be an inherent trait of the enzyme rather than a result of its inability to bind to 3'-terminal mispairs. When compared to avian myeloblastosis reverse transcriptase and HIV-1 reverse transcriptase, which extend the majority of mismatched base pairs permissively, Taq polymerase demonstrates roughly 100 to 1000 times stronger discriminating against mismatch extension. Researchers assessed the family B DNA polymerase from Thermococcus kodakaraensis KOD1 (formerly Pyrococcus kodakaraensis KOD1)'s 3'-5'exonuclease (proofreading) activity and PCR performance (Kuroita et al., 2005). Christian et al. (2018) observed the assembly of proof-reading complexes and the migration of DNA pols of Geobacillus stearothermophilus along a DNA substrate by using smFRET.
In this work, a brief insilico analysis of DNApol sequences and structures from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus was done to investigate their differences and to find a more heat-stable DNApol rather than the ancient Taq polymerase. As Taq polymerase has some limitations, this study will enlighten the other polymerase enzyme which can overcome the limitations of Taq polymerase.
2. Materials and methods:
2.1. Dataset:
Uniprot, the largest database of proteins was used to all reviewed protein sequences of DNA-polymerase of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus. High-resolution crystal structures of DNA-polymerase of Thermus aquaticus (1TAU), Thermococcus kodakarensis (1WN7), and Geobacillus stearothermophilus (2HHU) were retrieved from the RCSB PDB database (Bittrich et al., 2023).
2.2. Physico-chemical analysis of sequences:
Amino acid compositions along with other sequence properties were calculated through the ProtParam server (Gasteiger et al., 2003; Mitra et al., 2021). To prepare the Block of sequence, MSA was done through Clustal Omega (Sievers and Higgins, 2014). A block of sequences was used to calculate the hydrophobicity, mutability, and polarity. It was done with the help of the ProtScale server (Gasteiger et al., 2003). Intrinsic disorder regions were identified by DisEMBL (Linding et al., 2003).
2.3. Analysis of protein structures:
All high-resolution crystal structures were minimized through Chimera (Pettersen et al., 2004) along with a force field in 1000 steps. As the chemical structures shown in the drawings are not energetically advantageous, it is crucial to minimize energy while establishing the ideal molecule arrangement in space. To enhance the model, iterative optimization can precisely target and improve low-quality regions that quality tests have identified (Roy et al., 2015). The secondary structure was predicted by the CFSSP server (Kumar, 2013). Intra-protein interactions were identified as earlier methods (Mitra and Mohapatra, 2022). Tunnels, cavities, and voids were determined by Mole 2.0 server (Sehnal et al., 2013) with modification of parameters. They are playing a crucial role in the attachment and transfer of different small molecules, ions, and drugs (Mitra et al., 2021).
2.4. Molecular dynamics simulations:
Understanding the interaction between a protein and a vaccination peptide requires the use of molecular dynamics simulations. Molecular dynamic simulations have various additional benefits over docking since they take into account the many physiological traits required to predict the real nature of interactions (Mitra et al., 2021). Molecular dynamic simulations were performed using GROMACS (Bjelkmar et al., 2010; Abraham et al., 2015) and the GROMOS96 43a1 forcefield. The steepest descent method was used to reduce energy on the solvated systems in 5000 stages and finished in a final production run of 100 ns molecular dynamic simulations at 300 K temperatures after equilibration. The radius of gyration (Rg), the solvent accessible surface area (SASA), the root mean square deviation (RMSD), and the root mean square fluctuation (RMSF), hydrogen bonds were all calculated using molecular dynamic simulations.
3. Results and discussions:
3.1. Analysis of protein sequences:
The biological activity of a protein is determined by the chemical characteristics of its amino acids. The rate of evolution in bacterial species is greatly influenced by amino acid composition, which is in turn influenced by GC content (Du et al., 2018).
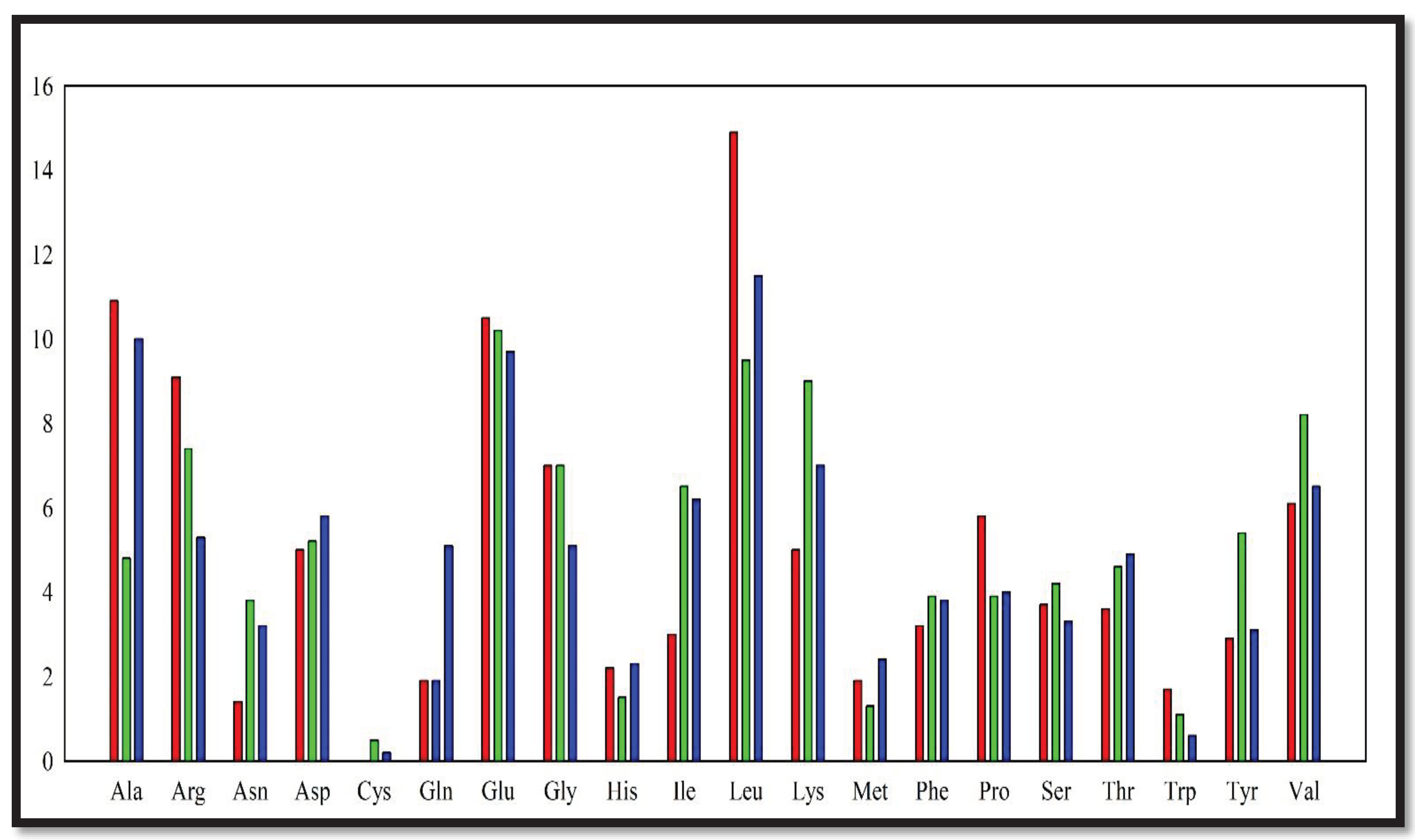
From the analysis of DNA polymerase (DNApol) of those 3 organisms, it was found that the charged polar residues have equal or slightly higher abundance in DNApol of Thermococcus kodakarensis, and Geobacillus stearothermophilus rather than Thermus aquaticus (
Figure 1). However, the number of uncharged polar residues have heavily higher abundance in
Thermococcus kodakarensis and
Geobacillus stearothermophilus. Hydrophobic amino acid residues have an almost equal propensity in DNApol of all these 3 organisms. The instability index of DNApol of
Thermococcus kodakarensis was higher than Thermus aquaticus DNApol.
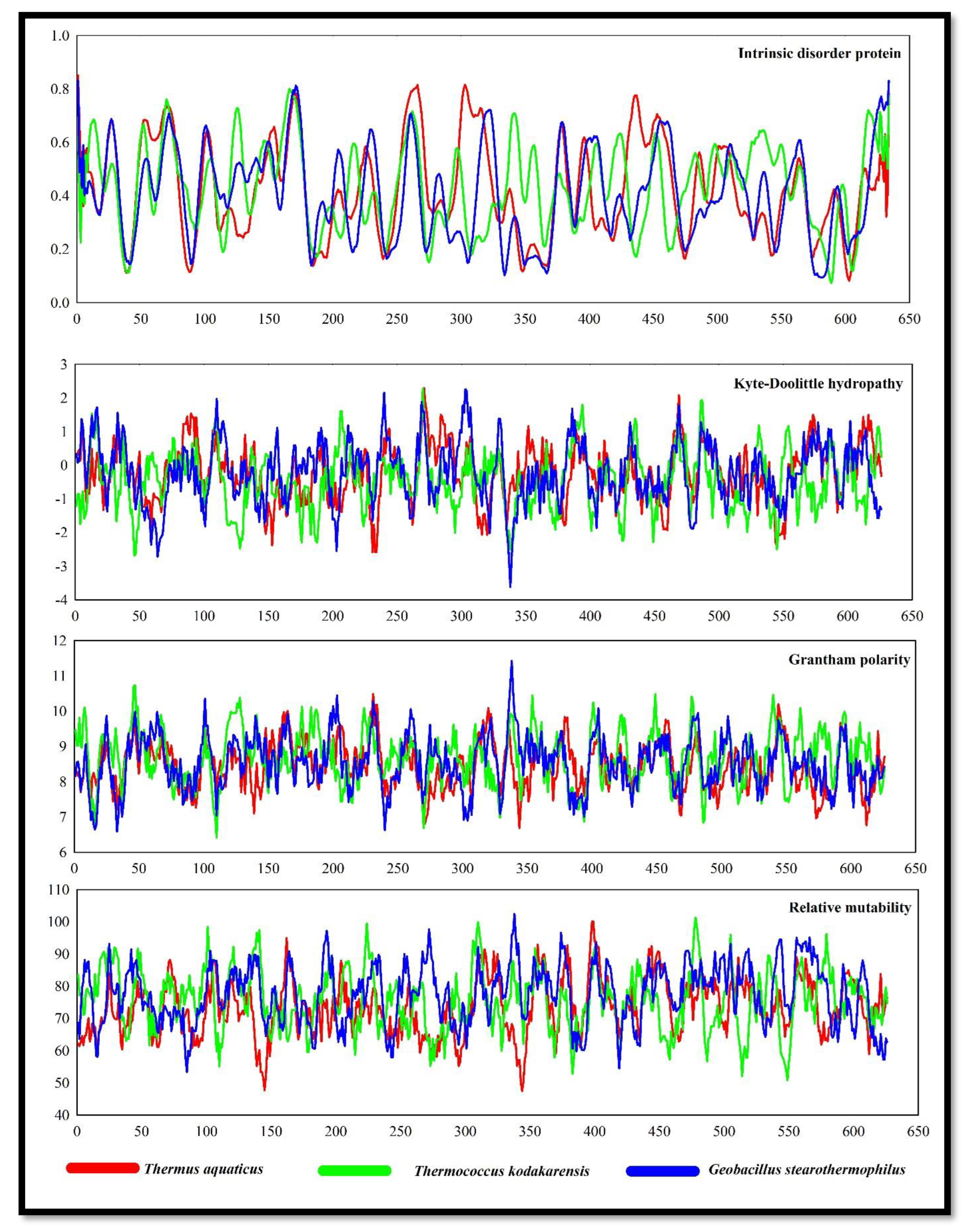
Intrinsically disordered proteins (IDPs) have biased amino acid compositions, poor sequence complexity, and large proportions of charged and hydrophilic amino acids compared to low quantities of bulky hydrophobic amino acids. IDPs have some dynamic and structural organization but lack a clear three-dimensional structure. (Wright et al., 2014). IDPs showed some noticeable pick points in Thermus aquaticus DNApol. However, the DNApol of
Geobacillus stearothermophilus also showed almost equal IDPs (
Figure 2).
Kyte-Doolittle hydrophobicity plot revealed DNApol of Thermococcus kodakarensis, and Geobacillus stearothermophilus were more hydrophilic rather than the DNApol of Thermus aquaticus. The polarity of DNApol from Thermococcus kodakarensis showed higher polarity followed by DNApol of Geobacillus stearothermophilus with some noticeable pick points. The rate of relative mutability was also high in DNApol of Thermococcus kodakarensis, and Geobacillus stearothermophilus. Calculating the relative mutability of the amino acids involves counting the number of times that each amino acid has changed over time and the number of times that it has appeared in sequences, resulting in mutation.
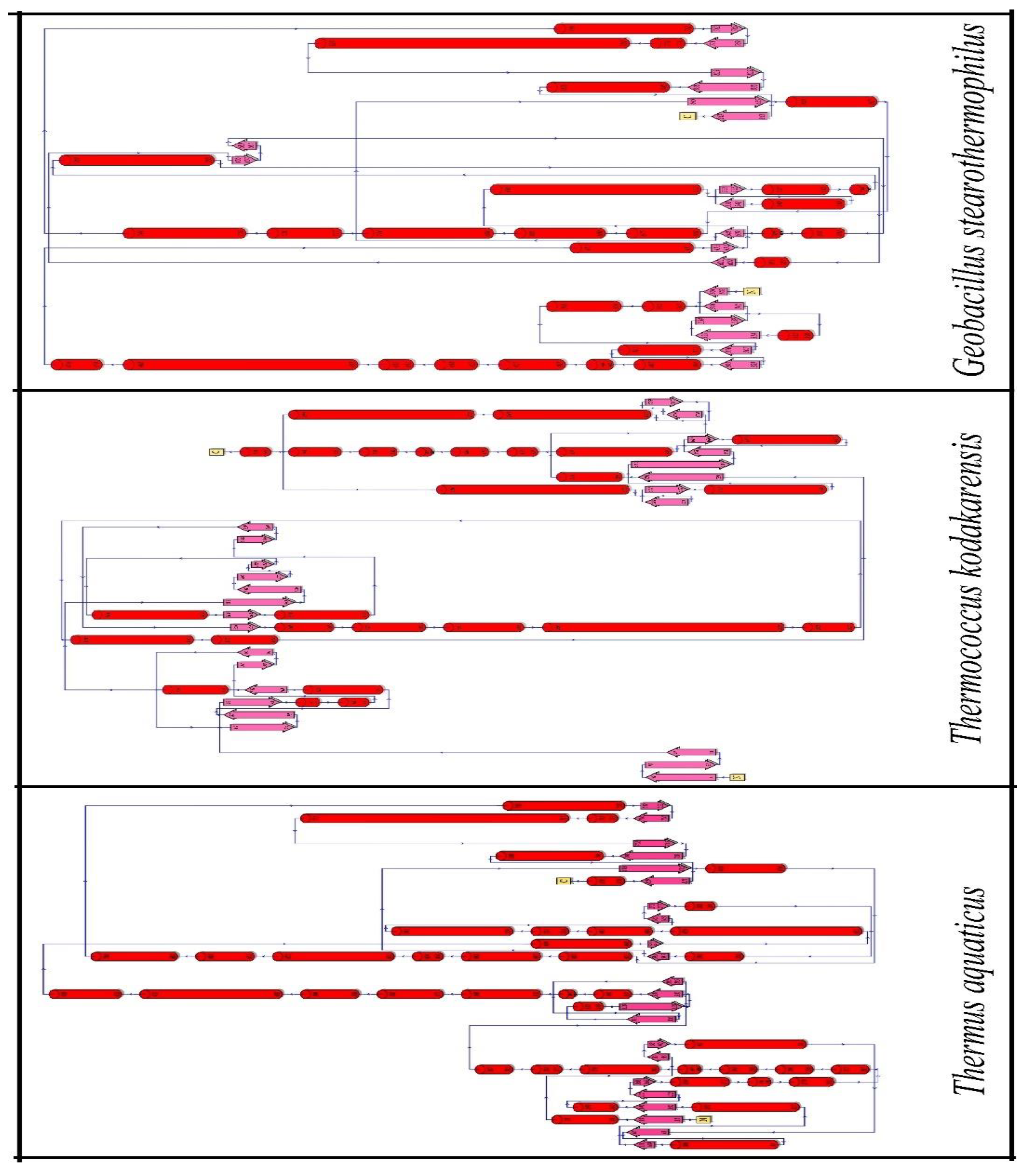
3.2. Analysis of protein secondary structure:
A polypeptide chain's adjacent amino acid residues are arranged in regular, recurrent patterns in space, which is known as a secondary structure. Both sequence-dependent side-chain contacts and sequence-independent backbone interactions help to stabilize early-stage secondary structure elements in proteins (Deller et al., 2016).
The highest amino acid abundance was found on the helix of all these three organisms (
Table 1 and
Figure 3). DNApol of
Thermococcus kodakarensis, and
Geobacillus stearothermophilus showed higher charged residue abundance in the helix of their proteins whereas DNApol of Thermus aquaticus jot down charged residues in coil and turn. Not only the charged residues but the higher propensity of uncharged polar residues was also noticed in the helix and sheet of DNApol from
Thermococcus kodakarensis, and
Geobacillus stearothermophilus.
3.3. Analysis of intra-protein interactions:
Minimized crystal structure of proteins was analysed to identify intra-protein interactions like salt bridges, aromatic-aromatic interactions, aromatic-sulphur interactions, cation-pi interactions, etc. An electrostatic attraction creates a noncovalent link known as a salt bridge between two oppositely charged residues. Generally, two types of salt bridges are found in protein salt bridges. However, nowadays a special type of salt bridge called cyclic salt bridge has been discovered in thermophilic protein (Mitra et al., 2021). DNApol of
Thermus aquaticus had 41 isolated and 12 network salt bridges whereas DNApol of
Thermococcus kodakarensis had 36 isolated and 19 network salt bridges (
Table 2). DNApol of
Geobacillus stearothermophilus had 25 isolated and 9 network salt bridges. Although, the length of
Geobacillus stearothermophilus DNApol is almost half of the other structures.
Pairs of interacting aromatic residues that meet some requirements like the two interacting residues must have their aromatic ring center at least 4.5 to 7 apart, and their dihedral angles must be between 30 and 90 are considered to be engaging in aromatic-aromatic interactions. Recently it has been discovered that bacteria possess aromatic-aromatic interactions in a long network formation (Mitra et al., 2021). DNApol in Thermus aquaticus had 11 isolated and 4 network aromatic-aromatic interactions whereas DNApol of
Thermococcus kodakarensis had 10 isolated and 7 network aromatic-aromatic interactions (
Table 3). Not only numbers, but
Thermococcus kodakarensis also had a very long network of aromatic-aromatic interaction where 8 intermediate bonds were formed. 10 isolated and 2 network aromatic-aromatic interactions were found in DNApol of
Geobacillus stearothermophilus.
The bulk of protein structures involve sulphur-aromatic interactions, yet little is understood about how these interactions function in ion channels. A protein can be stabilized by interactions between an aromatic and sulphur-containing amino acid (Gómez-Tamayo et al., 2016). DNApol of Thermus aquaticus had only 1 isolated and 2 network aromatic-sulphur interactions. 5 isolated and 1 network aromatic-sulphur interactions were formed in the DNApol of Thermococcus kodakarensis (
Table 4). Despite of small size, DNApol of Geobacillus stearothermophilus had 8 isolated and 1 network aromatic-sulphur interactions.
In addition to the hydrophobic effect, the hydrogen bond, and the ion pair in shaping the macromolecular structure and drug-receptor interactions, the chemistry community now considers the cation interaction as a major force for molecular recognition. DNApol of
Thermus aquaticus had 22 isolated and 2 network cation-pi interactions (
Table 5). DNApol of
Thermococcus kodakarensis increases its cation-pi interactions by forming 20 isolated and 6 network bonds. DNApol of
Geobacillus stearothermophilus had 9 isolated and 3 network cation-pi interactions.
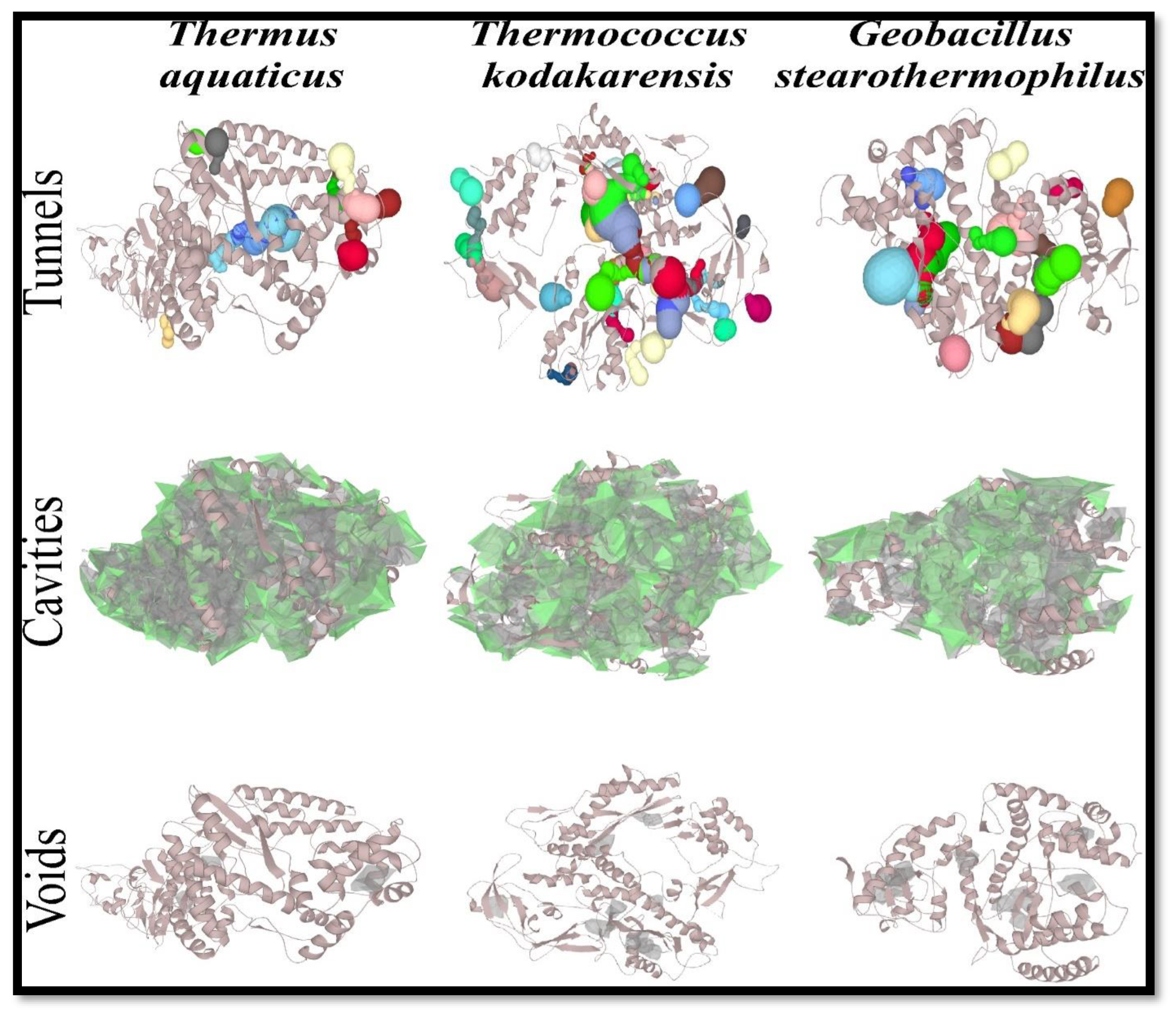
3.4. Study of tunnels, cavities, and voids:
The structural elements that control the exchange rates of ligands, ions, and aqueous solvents are represented by protein tunnels that link the functional buried cavities with bulk solvent and protein channels that enable transport over biological membranes (Brezovsky et al., 2018). The number of tunnels was high in DNApol of
Thermococcus kodakarensis i.e., 35 (
Table 6 and
Figure 4). Although, the length of DNApol from
Geobacillus stearothermophilus was shooter, it also showed a higher number of tunnels rather than DNApol of
Thermus aquaticus.
Many biological structures have cavities. Cavities have been found in virus capsids, multimeric protein aggregates, single-domain proteins, and even more complicated structures like ribosomes. Biological cavities may enclose an area. It is not known what conditions lead to solvent molecules being or not being able to fill the holes (Chwastyk et al., 2020). The number of cavities reflects the same result as tunnels.
3.5. Analysis through molecular dynamics simulations:
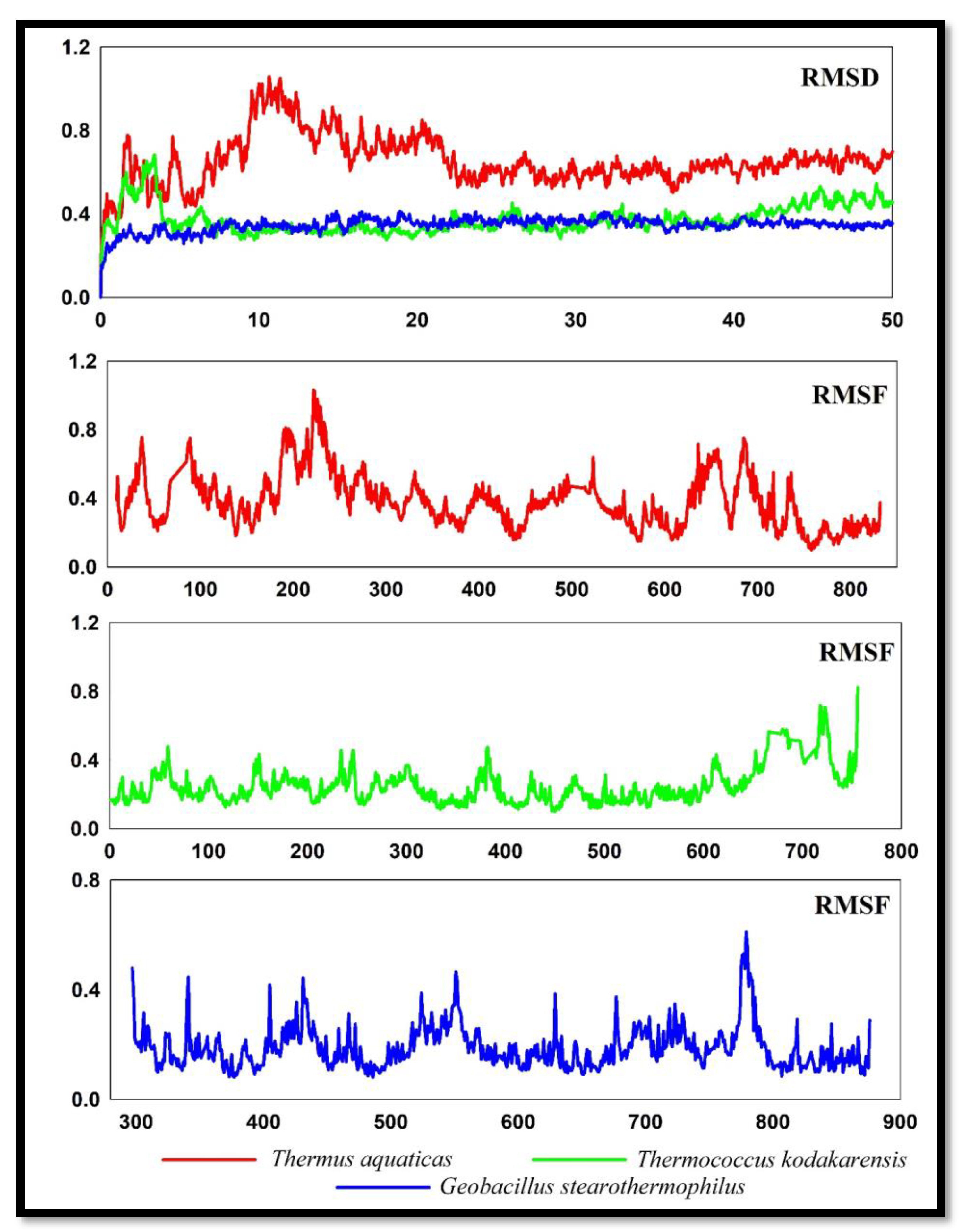
Now, a sophisticated method called molecular dynamics simulations can successfully comprehend the connections between macromolecular structure and function. A biologically significant stage is relatively close to the current simulation time. It is conceivable to change the standard paradigm of structural bioinformatics from researching single structures to analysing conformational ensembles using the information on the dynamic properties of macromolecules (Hospital et al., 2015; Abdalla et al., 2022). A simulation of these molecules' molecular dynamics lasting 50 ns was used to look at the stability of these DNApol enzymes from three organisms.
A root mean square deviation study revealed that the DNApol of
Thermus aquaticus displayed substantial deviations (
Figure 5) at 10ns to 22 ns, deviating up to 1 Å, after that reducing the deviation of RMSD. It started from 0.4 Å and finally stable at 0.6 Å. DNApol of
Thermococcus kodakarensis initially deviate at the very beginning but immediately stable after 5ns and remain stabilized throughout the path. At the of 50ns, it stables at 0.3 Å. DNApol of
Geobacillus stearothermophilus showed much lower RMSD than the other two organisms and it doesn’t have any larger deviation throughout the path. Starting from 0.2 Å and finally ending at the same o.2 Å.
The RMSF graph reveals the DNApol enzymes' stability. Low fluctuation or a low value in a plot denotes less distortion and well-structured sections in a complex, whereas high fluctuation or a high value in a plot reveals increased flexibility and unstable links. DNApol of Thermus aquaticus showed fluctuated plots throughout the path. The highest pick point was observed near residue number 220 at 1 Å. DNApol of Thermococcus kodakarensis showed very low RMSF and was almost stable throughout the path except at the end where it showed slight fluctuations. DNApol of Geobacillus stearothermophilus showed better RMSF than Thermus aquaticus DNApol. However, it also showed few fluctuations at some specific points like residue numbers 440, 550 and 780.
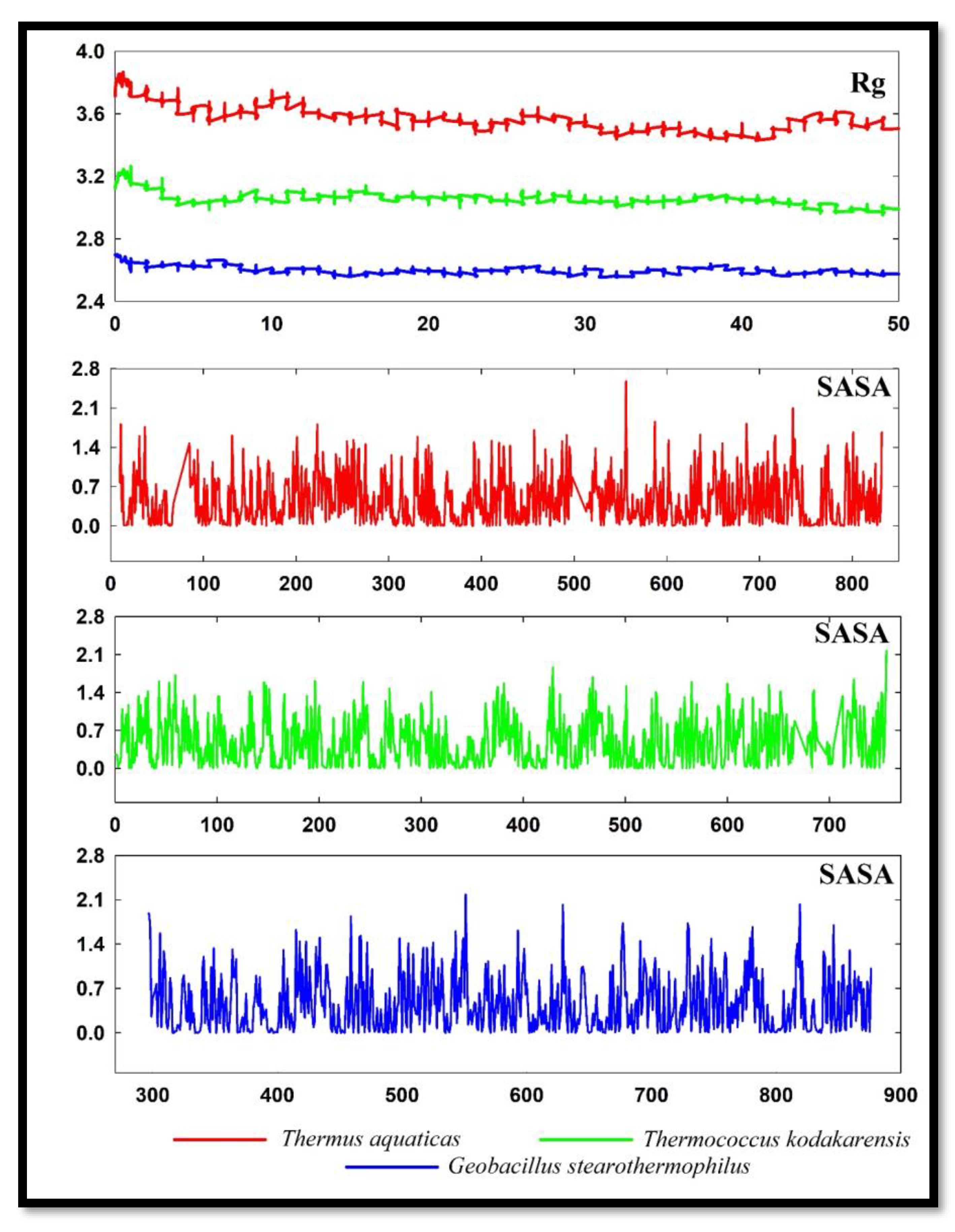
The distribution of a protein's atoms along its axis is known as the radius of gyration (Rg). Rg is the length that corresponds to the separation between the rotating point and the location where the energy transfer has the greatest impact (Sneha and Doss, 2016). The RG of DNApol from
Thermus aquaticus was very high and showed very lower value in other organisms (
Figure 6). DNApol of
Geobacillus stearothermophilus revealed the lowest Rg in molecular dynamics analysis.
Proteins' solvent-accessible surface area (SASA) has long been regarded as a key variable in research on protein folding and stability. A hypothetical solvent sphere's centre and the protein's van der Waals contact surface describe it as the surface around the protein (Ausaf et al., 2014). SASA of all three DNApol were almost the same with some minute differences. DNApol of Thermococcus kodakarensis and Geobacillus stearothermophilus has slightly lower SASA at some points.
4. Discussions:
The abundance of polar amino acids in protein sequences has a significant contribution to stability increasing (Mitra et al., 2022). Increase polar amino acids may provide higher stability DNApol of Thermococcus kodakarensis, and Geobacillus stearothermophilus and may appear as better than Thermus aquaticus DNApol. Lower instability index revealed that DNApol of Thermococcus kodakarensis, and Geobacillus stearothermophilus have higher stability. IDPs' structural adaptability suggests that entropy-driven motions are essential to how they work. By interacting with certain binding partners, many IDPs go through function-related disorder-to-order transitions. The high amount of IDPs may promote protein-protein interactions in Geobacillus stearothermophilus.
Kyte-Dolittle hydropathy showed a more hydrophilic nature of Thermococcus kodakarensis and Geobacillus stearothermophilus proteins which boost the protein interaction with aqueous solutions. The abundance of uncharged polar amino acids increases protein polarity which was revealed by Grantham polarity. Protein polarity increases its thermal stability (Mitra et al., 2022). A protein's polarity pattern plays a crucial role in both its structure and function. For instance, the burial of hydrophobic residues is one of the primary causes of protein folding (Manor et al., 2012).
The presence of charged residues tends to increase the stability of the helix which further affects the protein stability. One of the primary causes of the enhanced heat stability of proteins in thermophilic bacteria is their greater conformational stability. Moreover, the helix frequently participates in protein interactions with lipids found in cell membranes, nucleic acids, and other proteins. It is assumed that the sidechains are hydrophobic because the helical structure can internally satisfy all backbone hydrogen bonds, leaving no polar groups exposed to the membrane (Yakimov et al., 2016).
The presence of a higher number of salt bridges in the DNApol of Thermococcus kodakarensis and DNApol of Geobacillus stearothermophilus make them more stable than Taq polymerase. The presence of high-network salt bridges gives extra advantages to these two organisms (Mitra et al., 2021). A high abundance of network aromatic-aromatic interactions in Thermococcus kodakarensis and Geobacillus stearothermophilus have gained more thermal stability rather than the DNApol of Thermus aquaticus. The increasing number of aromatic-sulfur interactions of DNApol in Thermococcus kodakarensis, and Geobacillus stearothermophilus gives advantages over the DNApol of Thermus aquaticus. Several studies have demonstrated that in drug-receptor and protein-protein interactions, cation interactions can boost binding energies by 2 to 5 kcal/mol, making them competitive with hydrogen bonds and ion pairs (Dougherty, 2013). So, again DNApol of Thermus aquaticus lag by the other two organisms due to the lower formations of cation-pi interactions. Increasing tunnels help to increase ion transfer and increase catalytic activity.
From the molecular dynamic simulations, the lower value of RMSD in DNApol of Thermococcus kodakarensis, and Geobacillus stearothermophilus is more stable than DNApol of Thermus aquaticus. Lower RMSF showed lower fluctuations during 50ns molecular dynamics simulations of DNApol form Thermococcus kodakarensis, and Geobacillus stearothermophilus. Lower values in plots of Rg of Thermococcus kodakarensis, and Geobacillus stearothermophilus revealed that the DNApol of these two organisms was more tightly packed than the DNApol of Thermus aquaticus. Decreasing points of SASA from Thermococcus kodakarensis, and Geobacillus stearothermophilus indicate that folding mechanisms of these DNApol were more efficient than DNApol of Thermus aquaticus.
5. Conclusions:
Using an insilico structure-based methodology, this work investigated possible most heat-tolerable DNApol. DNApol of Thermus aquaticus is used in PCR techniques. But the DNApol of Thermococcus kodakarensis and Geobacillus stearothermophilus stand up as more stable and heat-tolerable DNApol by forming a high number of intra-protein interactions. The abundance of charged and uncharged polar residues affects helix stability. A higher number of tunnels increase the catalytic activity of DNApol of Thermococcus kodakarensis and Geobacillus stearothermophilus. Variations in amino acid compositions also a playing huge role in the increment of heat stability. Evidence suggests that DNApol of these two organisms can be used in PCR along with other heat-tolerable biological interactions.
Funding
This research received no external funding.
References
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015 Sep 1;1:19-25. [CrossRef]
- Agustriana E, Nuryana I, Laksmi FA, Dewi KS, Wijaya H, Rahmani N, Yudiargo DR, Ismadara A, Helbert, Hadi MI, Purnawan A. Optimized expression of large fragment DNA polymerase I from Geobacillus stearothermophilus in Escherichia coli expression system. Preparative Biochemistry & Biotechnology. 2022 Jul 4:1-0. [CrossRef]
- Ausaf Ali S, Hassan I, Islam A, Ahmad F. A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Current Protein and Peptide Science. 2014 Aug 1;15(5):456-76. [CrossRef]
- Benson LM, Null AP, Muddiman DC. Advantages of Thermococcus kodakaraenis (KOD) DNA polymerase for PCR-mass spectrometry based analyses. Journal of the American Society for Mass Spectrometry. 2003 Jun 1;14(6):601-4. [CrossRef]
- Bittrich S, Bhikadiya C, Bi C, Chao H, Duarte JM, Dutta S, Fayazi M, Henry J, Khokhriakov I, Lowe R, Piehl DW. RCSB Protein Data Bank: Efficient Searching and Simultaneous Access to One Million Computed Structure Models Alongside the PDB Structures Enabled by Architectural Advances. Journal of Molecular Biology. 2023 Feb 2:167994. [CrossRef]
- Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. Journal of chemical theory and computation. 2010 Feb 9;6(2):459-66. [CrossRef]
- Brezovsky J, Kozlikova B, Damborsky J. Computational analysis of protein tunnels and channels. Protein Engineering: Methods and Protocols. 2018:25-42. [CrossRef]
- Chien A, Edgar DB, Trela JM. Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. JouDNAl of bacteriology. 1976 Sep;127(3):1550-7. [CrossRef]
- Christian TV, Konigsberg WH. Single-molecule FRET reveals proofreading complexes in the large fragment of Bacillus stearothermophilus DNA polymerase I. AIMS biophysics. 2018;5(2):144. [CrossRef]
- Chwastyk M, Panek EA, Malinowski J, Jaskólski M, Cieplak M. Properties of Cavities in Biological Structures—A Survey of the Protein Data Bank. Frontiers in Molecular Biosciences. 2020 Nov 6;7:591381. [CrossRef]
- Dougherty DA. The cation− π interaction. Accounts of chemical research. 2013 Apr 16;46(4):885-93.
- Du MZ, Liu S, Zeng Z, Alemayehu LA, Wei W, Guo FB. Amino acid compositions contribute to the proteins’ evolution under the influence of their abundances and genomic GC content. Scientific Reports. 2018 May 9;8(1):7382. [CrossRef]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic acids research. 2003 Jul 1;31(13):3784-8.
- Gómez-Tamayo JC, Cordomí A, Olivella M, Mayol E, Fourmy D, Pardo L. Analysis of the interactions of sulfur-containing amino acids in membrane proteins. Protein Science. 2016 Aug;25(8):1517-24. [CrossRef]
- Huang MM, Arnheim N, Goodman MF. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic acids research. 1992 Sep 11;20(17):4567-73. [CrossRef]
- Ishino S, Ishino Y. DNA polymerases as useful reagents for biotechnology–the history of developmental research in the field. Frontiers in microbiology. 2014 Aug 29;5:465.
- Kumar TA. CFSSP: Chou and Fasman secondary structure prediction server. Wide Spectrum. 2013;1(9):15-9. [CrossRef]
- Kuroita T, Matsumura H, Yokota N, Kitabayashi M, Hashimoto H, Inoue T, Imanaka T, Kai Y. Structural mechanism for coordination of proofreading and polymerase activities in archaeal DNA polymerases. Journal of molecular biology. 2005 Aug 12;351(2):291-8. [CrossRef]
- Kushida T, Narumi I, Ishino S, Ishino Y, Fujiwara S, Imanaka T, Higashibata H. Pol B, a family B DNA polymerase, in Thermococcus kodakarensis is important for DNA repair, but not DNA replication. Microbes and environments. 2019;34(3):316-26. [CrossRef]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003 Nov 1;11(11):1453-9. [CrossRef]
- Ma Y, Zhang B, Wang M, Ou Y, Wang J, Li S. Enhancement of polymerase activity of the large fragment in DNA polymerase I from Geobacillus stearothermophilus by site-directed mutagenesis at the active site. BioMed research international. 2016 Nov 17;2016.
- Manor J, Feldblum ES, Zanni MT, Arkin IT. Environment polarity in proteins mapped noninvasively by FTIR spectroscopy. The journal of physical chemistry letters. 2012 Apr 5;3(7):939-44. [CrossRef]
- MC, Kong L, Rupp B. Protein stability: a crystallographer's perspective. Acta Crystallographica Section F: Structural Biology Communications. 2016 Feb 1;72(2):72-95.
- Mitra D, Dey A, Biswas I, Das Mohapatra PK. Bioactive compounds as a potential inhibitor of colorectal cancer; an insilico study of Gallic acid and Pyrogallol. Iranian Journal of Colorectal Research. 2021 Mar 1;9(1):32-9. [CrossRef]
- Mitra D, Das Mohapatra PK. Discovery of novel cyclic salt bridge in thermophilic bacterial protease and study of its sequence and structure. Applied Biochemistry and Biotechnology. 2021 Jun;193(6):1688-700. [CrossRef]
- Mitra D, Das Mohapatra PK. Cold adaptation strategy of psychrophilic bacteria: an in-silico analysis of isocitrate dehydrogenase. Systems Microbiology and Biomanufacturing. 2021 Oct;1(4):483-93. [CrossRef]
- Mitra D, Pal AK, Das Mohapatra PK. Intra-protein interactions of SARS-CoV-2 and SARS: a bioinformatic analysis for plausible explanation regarding stability, divergency, and severity. Systems Microbiology and Biomanufacturing. 2022 Oct;2(4):653-64.
- Mitra D, Paul M, Thatoi H, Mohapatra PK. Study of potentiality of dexamethasone and its derivatives against Covid-19. Journal of Biomolecular Structure and Dynamics. 2022 Nov 29;40(20):10239-49. [CrossRef]
- Mitra D, Das Mohapatra PK. In silico comparative structural and compositional analysis of glycoproteins of RSV to study the nature of stability and transmissibility of RSV A. Systems Microbiology and Biomanufacturing. 2023 Apr;3(2):312-27. [CrossRef]
- Nagata M, Ishino S, Yamagami T, Ishino Y. Replication protein A complex in Thermococcus kodakarensis interacts with DNA polymerases and helps their effective strand synthesis. Bioscience, Biotechnology, and Biochemistry. 2019 Apr 3;83(4):695-704. [CrossRef]
- Oki K, Nagata M, Yamagami T, Numata T, Ishino S, Oyama T, Ishino Y. Family D DNA polymerase interacts with GINS to promote CMG-helicase in the archaeal replisome. Nucleic acids research. 2022 Apr 22;50(7):3601-15. [CrossRef]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004 Oct;25(13):1605-12. [CrossRef]
- Roy K, Kar S, Das RN. Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment. Academic press; 2015 Mar 3. [CrossRef]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350-4. [CrossRef]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. 1985. Biotechnology (Reading, Mass.). 1992 Jan 1;24:476-80.
- Sehnal D, Svobodová Vařeková R, Berka K, Pravda L, Navrátilová V, Banáš P, Ionescu CM, Otyepka M, Koča J. MOLE 2.0: advanced approach for analysis of biomacromolecular channels. Journal of cheminformatics. 2013 Dec;5:1-3. [CrossRef]
- Sievers F, Higgins DG. Clustal omega. Current protocols in bioinformatics. 2014 Dec;48(1):3-13.
- Sneha P, Doss CG. Molecular dynamics: new frontier in personalized medicine. Advances in protein chemistry and structural biology. 2016 Jan 1;102:181-224.
- Takashima N, Ishino S, Oki K, Takafuji M, Yamagami T, Matsuo R, Mayanagi K, Ishino Y. Elucidating functions of DP1 and DP2 subunits from the Thermococcus kodakarensis family D DNA polymerase. Extremophiles. 2019 Jan 22;23:161-72. [CrossRef]
- UniProt Consortium. UniProt: a hub for protein information. Nucleic acids research. 2015 Jan 28;43(D1):D204-12. [CrossRef]
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nature reviews Molecular cell biology. 2015 Jan;16(1):18-29. [CrossRef]
- Yakimov AP, Afanaseva AS, Khodorkovskiy MA, Petukhov MG. Design of stable α-helical peptides and thermostable proteins in biotechnology and biomedicine. Acta Naturae (англoязычная версия). 2016;8(4 (31)):70-81. [CrossRef]
Figure 1.
Amino acid abundance in DNApol from Thermus aquaticus (red), Thermococcus kodakarensis (green), and Geobacillus stearothermophilus (blue).
Figure 1.
Amino acid abundance in DNApol from Thermus aquaticus (red), Thermococcus kodakarensis (green), and Geobacillus stearothermophilus (blue).
Figure 2.
Intrinsic disorder protein, Kyte-Dollitle hydropathy, Grantham polarity and relative mutability of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 2.
Intrinsic disorder protein, Kyte-Dollitle hydropathy, Grantham polarity and relative mutability of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 3.
Secondary structure of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 3.
Secondary structure of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 4.
Tunnels, cavities and voids of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 4.
Tunnels, cavities and voids of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 5.
RMSD and RMSF of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 5.
RMSD and RMSF of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 6.
Rg and SASA of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Figure 6.
Rg and SASA of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 1.
Amino acids propensity in secondary structures of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 1.
Amino acids propensity in secondary structures of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
| Types |
Thermus aquaticus |
Thermococcus kodakarensis |
Geobacillus stearothermophilus |
| Charged polar |
Uncharged polar |
Hydrophobic |
Charged polar |
Uncharged polar |
Hydrophobic |
Charged polar |
Uncharged polar |
Hydrophobic |
| Helix |
22.45 |
4.44 |
31.21 |
24.51 |
6.10 |
23.74 |
24.53 |
8.98 |
33.51 |
| Sheet |
3.96 |
6.00 |
15.97 |
6.36 |
10.38 |
19.58 |
3.80 |
6.56 |
13.99 |
| Coil |
2.40 |
2.40 |
5.28 |
0.91 |
1.17 |
0.52 |
1.38 |
2.25 |
1.21 |
| Turn |
3.12 |
0.72 |
2.04 |
3.11 |
0.78 |
2.08 |
1.21 |
1.73 |
0.86 |
Table 2.
Isolated salt bridges and network (color pair) salt bridges in DNApol of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 2.
Isolated salt bridges and network (color pair) salt bridges in DNApol of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
| Thermus aquaticus |
Thermococcus kodakarensis |
Geobacillus stearothermophilus |
| Isolated |
Network |
Isolated |
Network |
Isolated |
Network |
| K53-E57 |
E434-R726 |
E10-R32 |
D6-R17 |
K315-E458 |
K298-E445 |
| D60-K260 |
E434-K762 |
K21-D204 |
D6-K253 |
E340-H341 |
E445-R449 |
| D67-R85 |
E465-R469 |
E22-K27 |
K20-E22 |
E364-K367 |
D305-R347 |
| D91-R94 |
E466-R469 |
D31-K124 |
K20-E29 |
D372-R375 |
R347-E440 |
| 0K100-E112 |
R559-E601 |
R58-D92 |
E49-K52 |
K374-E489 |
E325-K431 |
| 1E101-R275 |
R559-E790 |
K70-E81 |
E49-K53 |
K415-E420 |
E325-R435 |
| 4D104-R110 |
R573-E615 |
R78-E426 |
R17-D235 |
R459-D463 |
E321-K450 |
| 0D120-K247 |
R573-D785 |
R101-D108 |
E251-K253 |
E464-R467 |
D425-H446 |
| 7K127-E130 |
K709-E712 |
K118-D343 |
E35-K66 |
R466-D471 |
D425-K450 |
| 8K128-E132 |
K709-E713 |
E130-R335 |
E35-R67 |
R472-E476 |
E478-R769 |
| 4D144-R183 |
R726-D759 |
E154-K225 |
R67-E69 |
K548-D559 |
E478-K805 |
| 5R175-D177 |
D759-K762 |
E165-K324 |
D113-K371 |
R615-E658 |
R517-E520 |
| 3R183-D188 |
K354-D355 |
E187-R222 |
D113-R503 |
E667-R859 |
R517-E569 |
| 5H235-D238 |
K354-E445 |
R196-E200 |
D202-R234 |
E673-R677 |
R660-D678 |
| 7D237-K240 |
R362-E363 |
D212-R346 |
D202-R255 |
D680-R702 |
D678-K863 |
| 9R249-D251 |
E363-R556 |
K220-E224 |
D204-R234 |
K684-D688 |
K863-D865 |
| 5D265-R268 |
E601-R778 |
D246-R247 |
E376-R379 |
K730-E734 |
D646-K838 |
| 6R266-E289 |
R778-E790 |
K287-D315 |
E376-R380 |
E756-K760 |
K838-E839 |
| 0R270-E274 |
E296-R334 |
E294-R307 |
K526-E527 |
R769-D802 |
K417-E464 |
| 3H283-E284 |
R334-E401 |
R310-E314 |
K526-E530 |
K806-D810 |
E464-R467 |
| 3R313-E315 |
R349-D371 |
K317-E321 |
E527-K531 |
R814-E818 |
|
| 4K314-E388 |
D371-R435 |
K360-E363 |
E527-K570 |
H823-E835 |
| 0K340-D344 |
E423-R727 |
R364-D455 |
E530-K531 |
E840-R843 |
| 3R343-E363 |
E721-R727 |
E391-K591 |
R641-E645 |
E631-K635 |
| 1D381-R393 |
|
E393-K535 |
K644-E645 |
R637-E831 |
| 0E400-R405 |
R406-E578 |
E648-K649 |
|
| 0R450-E602 |
E458-K462 |
E648-K652 |
| 2D452-R596 |
E459-R482 |
R188-E189 |
| 3E473-K531 |
D480-R484 |
E189-K192 |
| 0H480-D496 |
E554-K558 |
R169-D182 |
| 7R487-E491 |
K557-E599 |
D182-R193 |
| 7E537-K540 |
K559-E562 |
E398-R585 |
| 2K542-D547 |
E621-K659 |
E584-R585 |
| 3R563-D578 |
E628-K632 |
E111-R119 |
| 4E634-R636 |
D635-K638 |
R119-D123 |
| 7D637-R659 |
E742-R746 |
E430-K443 |
| 1E641-R651 |
|
D432-K443 |
| 7R677-E681 |
D164-K201 |
| 4E694-R704 |
E166-K201 |
| 5R715-E745 |
H439-E511 |
| 4E734-R741 |
K507-E511 |
Table 3.
Isolated and network (color pair) aromatic-aromatic interactions in Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 3.
Isolated and network (color pair) aromatic-aromatic interactions in Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
| Type of interaction |
Thermus aquaticus |
Thermococcus kodakarensis |
Geobacillus stearothermophilus |
| Position |
Residue |
Position |
Residue |
Position |
Residue |
Position |
Residue |
Position |
Residue |
Position |
Residue |
| Isolated |
45 |
TYR |
278 |
PHE |
7 |
TYR |
116 |
PHE |
301 |
PHE |
345 |
PHE |
| 47 |
PHE |
66 |
PHE |
19 |
PHE |
26 |
PHE |
357 |
PHE |
360 |
TRP |
| 134 |
TYR |
258 |
PHE |
75 |
PHE |
110 |
TYR |
382 |
TRP |
490 |
PHE |
| 161 |
TYR |
167 |
TRP |
152 |
PHE |
218 |
TYR |
392 |
PHE |
461 |
PHE |
| 428 |
TRP |
724 |
PHE |
173 |
TRP |
299 |
TRP |
519 |
TYR |
526 |
PHE |
| 475 |
PHE |
482 |
PHE |
214 |
PHE |
218 |
TYR |
539 |
PHE |
554 |
TYR |
| 598 |
PHE |
827 |
TRP |
279 |
TYR |
283 |
PHE |
640 |
PHE |
872 |
TRP |
| 647 |
PHE |
692 |
PHE |
441 |
PHE |
516 |
TRP |
690 |
PHE |
735 |
PHE |
| 667 |
PHE |
671 |
TYR |
448 |
PHE |
504 |
TRP |
710 |
PHE |
714 |
TYR |
| 706 |
TRP |
749 |
PHE |
653 |
TYR |
727 |
TYR |
762 |
TYR |
772 |
TYR |
| 719 |
TYR |
729 |
TYR |
|
| Network |
24 |
TYR |
92 |
PHE |
34 |
PHE |
116 |
PHE |
650 |
PHE |
866 |
TYR |
| 27 |
PHE |
92 |
PHE |
34 |
PHE |
120 |
TYR |
650 |
PHE |
868 |
TYR |
| 146 |
TYR |
172 |
TYR |
34 |
PHE |
37 |
TYR |
739 |
TYR |
740 |
PHE |
| 146 |
TYR |
179 |
TRP |
37 |
TYR |
39 |
TYR |
739 |
TYR |
743 |
PHE |
| 172 |
TYR |
179 |
TRP |
37 |
TYR |
86 |
TYR |
|
| 172 |
TYR |
182 |
TYR |
38 |
PHE |
112 |
TYR |
| 179 |
TRP |
182 |
TYR |
38 |
PHE |
87 |
PHE |
| 306 |
PHE |
413 |
PHE |
140 |
PHE |
194 |
PHE |
| 413 |
PHE |
417 |
TRP |
140 |
PHE |
216 |
PHE |
| 417 |
TRP |
430 |
TYR |
194 |
PHE |
230 |
PHE |
| 645 |
TRP |
700 |
PHE |
216 |
PHE |
230 |
PHE |
| 696 |
TYR |
697 |
PHE |
209 |
TYR |
261 |
TYR |
| 696 |
TYR |
700 |
PHE |
261 |
TYR |
273 |
TYR |
| |
356 |
PHE |
493 |
TYR |
| 493 |
TYR |
496 |
TYR |
| 496 |
TYR |
497 |
TYR |
| 496 |
TYR |
499 |
TYR |
| 402 |
TYR |
545 |
PHE |
| 538 |
TYR |
545 |
PHE |
| 538 |
TYR |
588 |
PHE |
| 538 |
TYR |
594 |
TYR |
| 545 |
PHE |
583 |
TYR |
| 545 |
PHE |
588 |
PHE |
| 545 |
PHE |
594 |
TYR |
| 588 |
PHE |
594 |
TYR |
| 532 |
TYR |
563 |
PHE |
| 532 |
TYR |
566 |
TYR |
| 563 |
PHE |
566 |
TYR |
Table 4.
Isolated and network (color pair) aromatic-sulphur interactions in DNApol of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 4.
Isolated and network (color pair) aromatic-sulphur interactions in DNApol of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
| Thermus aquaticus |
Thermococcus kodakarensis |
Geobacillus stearothermophilus |
| Position |
Residue |
Position |
Residue |
D(centroid-centroid) |
Dihedral Angle |
Position |
Residue |
Position |
Residue |
D(centroid-centroid) |
Dihedral Angle |
Position |
Residue |
Position |
Residue |
D(centroid-centroid) |
Dihedral Angle |
| 564 |
PHE |
444 |
MET |
4.7 |
32.27 |
180 |
TYR |
313 |
MET |
4.78 |
174.6 |
344 |
PHE |
311 |
MET |
4.67 |
147.5 |
| 611 |
TYR |
761 |
MET |
5 |
102.1 |
230 |
PHE |
223 |
CYS |
4.19 |
27.63 |
461 |
PHE |
416 |
MET |
5.12 |
76.57 |
| 611 |
TYR |
807 |
MET |
5 |
157.4 |
431 |
TYR |
428 |
CYS |
5.18 |
32.63 |
650 |
PHE |
845 |
CYS |
5.28 |
90.85 |
| 647 |
PHE |
646 |
MET |
5.1 |
133.8 |
579 |
TYR |
561 |
MET |
4.64 |
149.2 |
654 |
TYR |
852 |
MET |
5.29 |
158 |
| 647 |
PHE |
658 |
MET |
4.8 |
44.51 |
445 |
PHE |
442 |
CYS |
5.11 |
54.37 |
690 |
PHE |
701 |
MET |
5.11 |
27.71 |
| |
497 |
TYR |
506 |
CYS |
5.08 |
11.36 |
749 |
TYR |
750 |
MET |
4.75 |
69.29 |
| 497 |
TYR |
509 |
CYS |
4.11 |
16.76 |
786 |
PHE |
790 |
MET |
4.91 |
5.61 |
| 441 |
PHE |
509 |
CYS |
5.06 |
104.4 |
866 |
TYR |
845 |
CYS |
4.99 |
110.8 |
| |
868 |
TYR |
841 |
MET |
4.09 |
147.4 |
| 868 |
TYR |
845 |
CYS |
4.81 |
114.3 |
Table 5.
Isolated and network (color pair) cation-pi interactions in DNApol of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 5.
Isolated and network (color pair) cation-pi interactions in DNApol of Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
| Thermus aquaticus |
Thermococcus kodakarensis |
Geobacillus stearothermophilus |
| Position |
Residue |
Position |
Residue |
D(centroid-centroid) |
Dihedral Angle |
Position |
Residue |
Position |
Residue |
D(centroid-centroid) |
Dihedral Angle |
Position |
Residue |
Position |
Residue |
D(centroid-centroid) |
Dihedral Angle |
| 66 |
PHE |
100 |
LYS |
5.33 |
34.34 |
19 |
PHE |
17 |
ARG |
4.92 |
99.34 |
360 |
TRP |
368 |
LYS |
5.04 |
169.9 |
| 169 |
TRP |
175 |
ARG |
5.29 |
25.28 |
26 |
PHE |
234 |
ARG |
4.32 |
42.98 |
382 |
TRP |
596 |
ARG |
5.11 |
30.57 |
| 172 |
TYR |
143 |
LYS |
5.69 |
91.18 |
34 |
PHE |
124 |
LYS |
5.87 |
107.2 |
429 |
TYR |
435 |
ARG |
5.84 |
55.14 |
| 179 |
TRP |
183 |
ARG |
5.27 |
121.1 |
39 |
TYR |
73 |
LYS |
5.06 |
137.2 |
461 |
PHE |
417 |
LYS |
5.37 |
12.55 |
| 258 |
PHE |
128 |
LYS |
4.83 |
72.72 |
120 |
TYR |
124 |
LYS |
5.66 |
50.1 |
675 |
PHE |
660 |
ARG |
4.75 |
57.74 |
| 272 |
PHE |
275 |
ARG |
5.37 |
122.5 |
152 |
PHE |
221 |
LYS |
4.89 |
117.3 |
710 |
PHE |
706 |
LYS |
5.75 |
130.2 |
| 306 |
PHE |
349 |
ARG |
3.95 |
49.24 |
279 |
TYR |
317 |
LYS |
5.71 |
115.5 |
781 |
PHE |
784 |
ARG |
5.99 |
61.14 |
| 318 |
TRP |
313 |
ARG |
5.43 |
48 |
299 |
TRP |
174 |
LYS |
5.24 |
11.24 |
786 |
PHE |
789 |
ARG |
5.91 |
114.6 |
| 339 |
TYR |
362 |
ARG |
5.92 |
80.71 |
320 |
TYR |
324 |
LYS |
5.62 |
118.6 |
872 |
TRP |
637 |
ARG |
4.07 |
153.2 |
| 378 |
TYR |
726 |
ARG |
5.57 |
55.84 |
342 |
TRP |
346 |
ARG |
4.35 |
142.7 |
327 |
TYR |
374 |
LYS |
5.52 |
94.34 |
| 394 |
TYR |
393 |
ARG |
5.61 |
134.3 |
356 |
PHE |
360 |
LYS |
4.99 |
30.09 |
327 |
TYR |
375 |
ARG |
5.15 |
90.64 |
| 413 |
PHE |
349 |
ARG |
5.02 |
6.79 |
362 |
TYR |
119 |
ARG |
5.93 |
123.2 |
762 |
TYR |
770 |
ARG |
4.06 |
24.62 |
| 417 |
TRP |
431 |
ARG |
5.00 |
100.8 |
431 |
TYR |
440 |
ARG |
4.18 |
173.5 |
772 |
TYR |
770 |
ARG |
5.96 |
127.9 |
| 430 |
TYR |
435 |
ARG |
5.7 |
40.47 |
445 |
PHE |
425 |
ARG |
4.79 |
115.8 |
873 |
TYR |
635 |
LYS |
5.99 |
57.01 |
| 455 |
TYR |
596 |
ARG |
4.85 |
153.1 |
499 |
TYR |
501 |
ARG |
5.98 |
71.83 |
873 |
TYR |
876 |
LYS |
5.37 |
141.4 |
| 604 |
TRP |
778 |
ARG |
4.21 |
36.86 |
505 |
TYR |
380 |
ARG |
4.1 |
144.8 |
|
| 632 |
PHE |
617 |
ARG |
4.14 |
27.91 |
532 |
TYR |
559 |
LYS |
5.26 |
57.96 |
| 645 |
TRP |
630 |
ARG |
5.29 |
125 |
566 |
TYR |
570 |
LYS |
4.9 |
58.84 |
| 647 |
PHE |
695 |
ARG |
5.79 |
132 |
582 |
PHE |
557 |
LYS |
4.11 |
148.9 |
| 671 |
TYR |
677 |
ARG |
4.7 |
110.1 |
731 |
TYR |
713 |
ARG |
5.29 |
144.2 |
| 697 |
PHE |
704 |
ARG |
4.96 |
71.63 |
37 |
TYR |
119 |
ARG |
5.51 |
59.3 |
| 827 |
TRP |
595 |
ARG |
5.46 |
143.8 |
37 |
TYR |
84 |
LYS |
4.09 |
165.5 |
| 398 |
TRP |
314 |
LYS |
5.81 |
117 |
86 |
TYR |
67 |
ARG |
4.06 |
157.4 |
| 398 |
TRP |
405 |
ARG |
4.14 |
32.56 |
86 |
TYR |
84 |
LYS |
4.57 |
58.46 |
| 719 |
TYR |
717 |
ARG |
4.16 |
164 |
112 |
TYR |
101 |
ARG |
5.67 |
132.8 |
| 719 |
TYR |
727 |
ARG |
4.58 |
34.49 |
112 |
TYR |
97 |
ARG |
5.34 |
100.7 |
| |
261 |
TYR |
265 |
ARG |
5.45 |
56.59 |
|
| 273 |
TYR |
265 |
ARG |
4.87 |
149.3 |
| 291 |
TYR |
289 |
LYS |
5.99 |
30.03 |
| 311 |
TYR |
287 |
LYS |
4.55 |
65.15 |
| 311 |
TYR |
289 |
LYS |
4.36 |
120.1 |
| 481 |
TYR |
477 |
LYS |
4.61 |
53.28 |
| 481 |
TYR |
484 |
ARG |
5.64 |
54.44 |
Table 6.
Number of tunnels, cavities and voids of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
Table 6.
Number of tunnels, cavities and voids of DNApol from Thermus aquaticus, Thermococcus kodakarensis, and Geobacillus stearothermophilus.
| Table 13. |
Thermus aquaticus |
Thermococcus kodakarensis |
Geobacillus stearothermophilus |
| Tunnels |
13 |
35 |
19 |
| cavities |
15 |
17 |
18 |
| voids |
3 |
9 |
9 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).