1. Introduction
Among the greater discovery of humankind in the 20th century was the discovery of antibiotics. Antibacterial triumph changed contemporary biomedicine, and seeks to define, mold, and grow both its potential and its boundaries. Regrettably, the possibility for resistance to any therapeutic agent to evolve and limits its ability to be effective [
1,
2]. The next series of antibiotics must be developed since resistance compromises efficacy (therapeutic effect). A pathogen's enhanced resistance to the prescribed standard therapy to which it was previously vulnerable is referred to as tolerance to an antibacterial agent (in this case, an antibiotic) [
3,
4,
5].
The clinical effectiveness of the existing antibiotics and their recommended treatments are now seriously threatened by the epidemic of antimicrobial resistance [
6,
7]. The issue has several facets because it affects the social, financial, medicinal, and ecological sectors. To maintain the long-term accessibility of an effective therapy for infections, proper usage of currently available antibiotics is required in the lack of the advancement of new generations of antibiotic medicines [
8]. Antibiotics may lose their effectiveness, which could result in a rise in mortality, the use of healthcare resources, and early mortality from infectious diseases that are already well-established and still posing a threat [
9].
However, the increased utilization of antibiotics over the past 50 years has placed stress on bacteria that are more vulnerable to them and may have helped resistant bacteria, a few of which are resistance to several antibiotics, to survive. Since sensitive bacteria may be more "fit" than bacterial resistance [
10], it is anticipated that bacterial resistance may be replace by susceptible bacteria if overuse of antibiotics can be stopped [
11]. Many researches have been conducted on origin, evolutionary aspect of antibiotic resistance and mechanisms of antibiotics resistance, but its effect on future drug development is little understood. Therefore, the recent review will highlight the role of antibiotic resistance in drug designing and impacts of antibiotic resistance on drug development in future.
2. Origin of Antibiotics
The use of antimicrobial agents for healing and prevention of infections has been practiced since ancient civilization when varieties of natural extracts were utilized. Some plant extracts and molds have been utilized as healing agents due to their characteristic antibiotic properties [
12]. However, American microbiologist Selman Waksman and his research team firstly coined the term “antibiotics”, as they extracted chemical substances from microorganism which limits the proliferation and growth of other microbes [
13]. Extending the implication of antibiotics in clinical practice, the ancient Greece, China, Egypt and other regions practiced the use of microorganisms for management of microbial infections since middle of 19th century.
However, modern implication of antibiotics started from unexpected discovery of penicillin, extracted by Alexander Fleming, who grown the culture filtrate of
Penicillium notatum fungus [
14]. The first true antibiotic compounds were discovered by Alexander Fleming in 1924 named as penicillin [
14], which found to be linked with ancient knowledge of previous generations; for instance, the Egyptians used moldy bread for wound infections. The 1940s to 1960s, known as the "golden era" of antibiotic discovery, had seen discovery of the majority of antibiotic classes currently in use [
15]. As antibiotic of penicillin was initially developed in 1928, but its complete structure was not revealed since 1949 and confirmed after synthesis and crystallization studies of Dorothy Crowfoot Hodgkin by 1959.
When antibiotics were first revealed and especially how quickly they were found, it was widely believed that infections could soon be under control. Before penicillin’s discovery, the sulfonamides have been used as first effective antimicrobials and chemical in nature which were restricted in therapeutic use due to specific mechanisms of resistance [
16]. The same processes that caused sulfonamide resistance in the late 1930s still exist today, around 90 years later. Two individuals of the penicillin discovery team detected a bacterium penicillinase as penicillin [
17] was being developed as well as several years before penicillin was made available as a treatment [
18].
The antibiotics of modern era have been originated from microbial fermentation and semi-synthetic route by using the antibiotic raw material. The discovery of numerous antibiotics as new generation semi-synthetic drugs such as antibacterial and antifungal agents initially treated and managed the diseases or infections successfully [
19]. But, after emergence, spread and proliferation of infectious diseases by various bacterial species, the strains got resistance against all therapeutic antibiotics during last few decades which enlightened the limitation of those drugs [
20]. Although hypersensitivity and frequent antibiotic use is considered as a major contributing reason to the resistance, it is clear at this point that antimicrobial resistance does not involve antibiotic administration of the microorganisms. Also discovered is that millions of years ago antibiotics gained the importance due to therapeutic use, genes associated with the production of antibiotics and antibiotic resistance emerged [
21]. As a result, both antibiotics and the factors that determine their resistance play different roles in bacterial behavior. All across the evolution of antibiotics, research on mechanisms of action has revealed molecular details about ligands and targets, and using antibiotics as "phenotypic mutations" has been an effective strategy in cell physiology investigations [
22]. Before moving towards the impact of intensified antibiotic resistance on the drug development, there is a need to highlight the origin, mechanism, and effects of antibiotic resistance on human life.
3. Origin of Antibiotic Resistance
The survivability of pathogens against the exposure and administration of antibiotics that could kill them or restrict their proliferation is called antibiotic resistance. There are several factors behind expression of antibiotic resistance against antibiotics or antimicrobial agents include the degree of resistance expression of bacterial strain and its ability to survive through resistance mechanism [
23,
24]. Microbial strains may possess inherent resistance or react strongly by transgene expression from one bacterium to another via plasmids, transgenes, genetic elements, and phages, or through alterations in cell genes (chromatin instability) that result in cross-resistance [
25]. Resistant microorganisms may proliferate swiftly if resistance factors are present on plasmids. Biochemical mechanisms work behind the resistance to protect bacterial cell wall from various agents, which triggers the target alteration, enzymatic degradation, and reduced or increased uptake of efflux pump proteins. Thus, the first-generation antibiotics have been facing antibiotics resistance in various clinical practices through natural process [
26].
Antibiotic resistance has become global challenge for human health as it linked with high rates of mortality and morbidity. The gram-positive and gram-negative bacteria related infections become difficult to treat due to multidrug resistance and could not be treated with traditional antibiotics. Among both pre-antibiotic and antibiotic periods, the antibiotic resistance has badly affected the antibiotic efficacy in clinical practices [
27]. Since, the first antibiotic resistance has been reported as soon as sulphonamides were introduced in 1930; it predicted the occurrence of antibiotics resistance in natural environment in the pre-antibiotic era without supporting the existence of deadly resistant pathogens. The advancement of antibiotics and their application in other aspects of life led the beginning of antibiotic era. As an outcome of human activity, high concentrations of toxic doses of antibiotics were used, which caused a significant alteration in the inherent significant role and the rapid growth of bacteria that were antibiotic resistant.
The antibiotic resistance is ancient as the clinical use of antibiotics, as it become prevalent and resistive due to capability of inactivation of drug agents acting upon their cell walls. Research evidences suggested the chemical modification of antibiotics to restrict or limit the cleavage by penicillinases (β-lactamases) [
28]. However, the detection and inactivation of bacterial penicillinase can enhance the efficacy of antibiotics and these are genes or components of other useful microorganisms as the different research findings reported. Increased use of antibiotics, genetic modification in bacterial strains, and antibiotic resistance genes are involved in cellular mechanism of antibiotics resistance before human intervention. They have also been released from natural genomic sources, rapidly spreading to infectious and commensal bacteria with different taxonomic classifications.
The total absence of viable preventative measures, the difficulty in dealing with bacterial infections and their accompanying diseases, and the small number of new antibacterial drugs in progress in the clinic would then necessitate the advancement of novel treatment approaches and broad-spectrum antimicrobial treatments. In patients with bacteremia and other serious illnesses, the rapid recognition of pathogenic bacteria and their antimicrobial sensitivity trends is lacking. According to the researchers, a greater knowledge of how infectious disease-induced biochemical mechanisms and specific virulence strategies work offers new opportunities to attack and interact with critical pathogenicity variables or infectivity characteristics of bacteria without subjecting them to pressure from evolvement to acquire resistance.
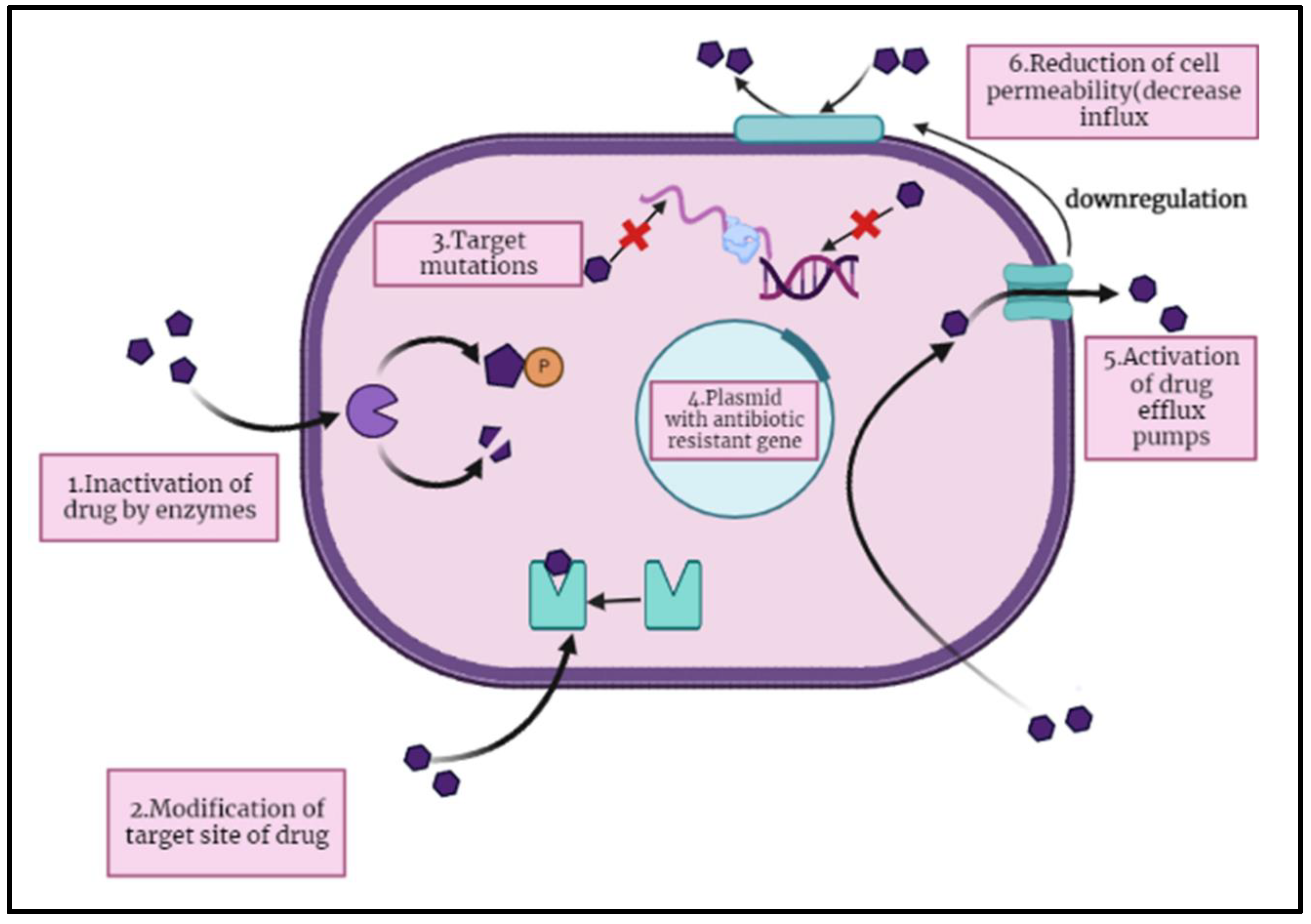
4. Mechanism of Antibiotic Resistance
The antibiotics mainly attacks on biochemistry and physiology of the microbial cells to reduce their growth or cause death. Some antibiotics destroy the cell walls or cell membranes of bacterial cell by dissolving the β-lactam and glycopeptides components and other antibiotics targets the protein synthetic machinery by combining with ribosomal units which stops the antibacterial activity of those microbes (
Figure 1). These cell wall targeting antibiotics includes the aminoglycosides, tetracycline, linezolid, chloramphenicol, and macrolides. The other cell machinery targeting antibiotics and nucleic acid synthesis interfering molecules include rifampin and fluoroquinolones (FQ). Remaining antibiotics are those which interfere with metabolic pathways and destroy membrane matrix include folic acid analog, daptomycin, polymyxins, and sulphonamides [
29].
The research evidences suggested that evolution of antibiotic resistance determinants induce the modification among other antibiotic-sensitive counterparts upon antibiotic exposure. Among bacterial strains, the multidrug resistance or related determinants induce the resistance for other chemical unrelated compounds rather than antibiotics and those include quaternary ammonium compounds [
30], the anionic detergent sodium dodecyl sulfate, ethidium bromide, the DNA-intercalating mutagen acridine, and uncouplers. Therefore, the multidrug resistance has greater impact on bacterial physiology. Additionally, they provide resistance to various metabolic products made by the organism, such as bile acids. It is hypothesized that some as-yet-unidentified biological functions of the factors in these bacteria lead to resistance to antibiotics [
31].
The antibiotic resistance also emerges through the genetic mutations encoded on bacterial chromosome which occurs due to evolution of genetic material against antibiotics and its transfer to next generation (
Figure 2). This mechanism, that can take place through a number of ways, including plasmid transfer, and that is likely the most crucial, can end up supplying the host organism and its offspring with additional genetic information expressing antibiotic resistance. Antibiotic have an impact on this by causing the transmission of resistant genes among microbes as well as by applying a selection pressure towards the development of resistant bacteria. Other antibiotic resistive strains transfer the genes encoding resistance for particular antibiotics which evolve through mutation [
32,
33] and those genes are associated with antibiotic biosynthesis pathway either in form of clusters or in form of single gene. As evidence from
Streptomyces coelicolor predicted that degree of antibiotic resistance is linked with antibiotic biosynthetic gene as it determines the role of antibiotic resistance determinants and postulates about the biosynthetic pathway of antibiotics [
34]. Even in isolates that don't generate, antibiotic-resistant genes are usually present. As a result, bacteria have developed a wide range of genes (known as the "resistome") that shield them from the effective dose of antibiotics. Clinically relevant resistant infections contain mobility genetic elements with genes orthologous to these on them. There is a chance that the genes that help compensate this external resistome will spread to pathogens, and there is some proof that at least many significant clinical gene mutations have their roots in environmental microorganisms [
35].
The antibiotic resistance genes and their epigenetic inheritance-based evolution has been found firstly in isolated strains of
Escherichia coli, that has been exposed to different concentration of antibiotics including tetracycline, nalidixic acid, and ampicillin [
36,
37]. Another study that used the data of de novo assembly of short-read sequences in conjugation with metagenomic approach suggested that lateral transfer of antibiotic resistance genes with non-coding region occurs due to the mobilization of modified sequences among clinical pathogens from environmental reservoir of soil bacteria. Randomized uncontrolled evolution really cannot explain the large probability of survival on lower antimicrobial agent. It was instead proposed that the genes for resistance to antibiotics were obtained through epigenetic mechanisms. Because of the maintaining of specific chromatin configurations or DNA methylation states, the high reversal of these resistive phenotypes further demonstrated that it was an instance of epigenetic mechanisms, which does not confer a durable phenotype [
38]. The lateral gene transfer or HGT contributes in transfer of resistivity, its evolution and maintenance among pathogenic bacteria and also have role in destruction of antibiotic resistance genes transferred from natural environment in clinical settings [
39].
In some cases, the antibiotic resistance and tolerance of bacteria to certain antibiotic depends upon metabolic state. The phase of bacterium also influences the susceptibility against antibiotics, either sometimes high resistivity exhibited by stationary phase as compared to growing phase. The biofilm production in micro environment contributes to the resistance of antibiotics as the embedment of polysaccharide matrix which forms a protective layer. The reduction of nitrogen production also triggers the resistance to antibiotics which also called swarming [
31]. The swarming cells of bacteria posed resistance to variety of antibiotics. This data, along with the susceptibility seen in biofilm, strongly suggests that antimicrobial resistance is a result of bacterial social behavior [
40,
41]. Antimicrobial resistance has also been found to be modulated by alarm one (p) ppGpp formation. It is hypothesized that this alarming is responsible for endurance, which is resistance found in a tiny percentage of found naturally microorganisms. Phenotypic resistance, a kind of resistance, greatly worsens some therapeutic management challenges with some infectious diseases [
42].
5. Adverse Effects of Antibiotic Resistance
The antibiotics resistance has been considered as a global issue of public health nowadays due to increasing resistance development among microbes against traditional antibiotics drugs and poses a need for new drugs. It is believed that the emergence of resistance to new antibiotics is probable and could limit the duration of these medications’ therapeutic efficacy. The World Health Organization (WHO) have declared so-called “ESKAPE” pathogens on the basis of their clinical significance and resistance levels, which include (E: Enterococcus faecium, S: Staphylococcus aureus or Stenotrophomonas maltophilia, K: Klebsiella pneumoniae or C: Clostridioides difficile, A: Acinetobacter baumannii, P: Pseudomonas aeruginosa, E: Enterobacteriaceae). Resistance to antibiotics has an impact on every aspect of health, including the human, wildlife, and environmental sectors, affecting humanity as a whole. Antimicrobial are in fact frequently recommended in order to cure viral infections in both animals and humans as well as to improve meat consumption in the food sector.
Animal dung, industrial wastewater, treatment facilities, and organic wastes that are used in irrigation and fertilization of agricultural lands—all release a significant amount of antibiotics into the soil and water. Antimicrobial drugs that are released into the aquatic and soil ecosystem foster the growth of antifungal medication microorganisms and the emergence of antibiotic-resistant genes in the ecosystem [
43]. There are multiple factors related to human activities (such as non-therapeutic and therapeutic use of antibiotics and discard of antibiotic formulations in natural ecosystem) induce the resistance among natural microbial flora which in turn affects human health when antibiotics failed to treat infectious diseases [
44].
The antibiotic resistance has posed a major health concern due to the growing bacterial infection among human population as conventional antibiotics failed to control them. The antibiotic resistance has also been emerged due to the dose related issues, lack of accurate clinical disposal and less knowledge about amount of antibiotics to treat disease [
45]. As a result, distinct signs of pathogenic bacteria last until a sufficient amount is attained, at which point symptoms start to fade. Bacterial exposure to insufficient doses of these substances is thought to be the leading contributor to resistant pathogens, which may also be the root cause of bacterial development. In therapies, bacteria typically faced with an excessively high concentration of just one or a few medications, while soil-borne microorganisms live in a diverse milieu and deal with a variety of stressors at the same time. These reactions most probably cancel out one another, favoring the evolution of antibiotic sensitivity over bacterial resistance [
46]. Therefore, it is thought that humanistic variables are to blame for the evolution of antibiotic resistance in clinical settings, which differ greatly from found naturally susceptibility. By simulating these natural habitats, it may be possible to understand how antibiotic resistance spreads among the microbial community [
19].
Since the discovery of antibiotics, the antibiotics resistance has challenging for the scientific effort made for humanity to save them from infectious diseases. In the beginning of antibiotics discovery, resistance was not too much challenging as in 21st century which identified the scientific research gaps and need of new drugs development [
47]. But recent developments in the profile of antibiotic resistance have forced the majority of drug companies to refocus their attempts on creating novel particles used in treating serious diseases [
48]. This strategy offers more financial advantages that ensure their preservation in difficult economic times. As a result, the pathway for antibiotic may stop flowing, leaving behind a few powerful compounds that may restrict the options for antibiotics used to treat particular illness situations. The rise and spread of antibiotic-resistant organisms is a new danger to public health, necessitating worldwide action and a multifaceted strategy to address the fundamental difficulties involved in reducing drug resistance and perfect the road forward [
49].
6. Effects of Antibiotic Resistance on Drug Development
The modern clinical and scientific advancements in drug development are related to therapeutic success and cope with different challenges of therapeutic fields. As many antibiotic advancements and developments are in queue to cope with public health emergency and issues of antibiotic resistance among population [
50]. Both biofilm and planktonic infections are main target of major therapeutic strategies imposed by antibiotics. The target of antibiotics is to stop the survival or growth of bacteria including the synthesis of DNA, and RNA or essential proteins, synthesis/maintenance of cell wall. Several medications originated from substances that microbes have used to fight against one another for thousands of years [
51]. Because bacteria have evolved the inherent ability to mutate and evade the damaging processes of many common antibiotics, the attacking instruments produced by microbes in this combat have triggered defense responses. Antibiotic used as a "last option" or in multiples or large dosage may be necessary to eradicate multidrug resistant (MDR) microorganisms [
52]. When microorganisms reside in biofilms, microbe resistance complicates the treatment challenge and frequently necessitates intense physical clearance of the biofilm via vigorous exfoliation, for instance, along with large doses of antibacterial therapy. The increased risks of adverse effects, antibiotic resistance and failure outcomes have posed cost challenges for drug development and treatments [
53].
According to reports of Pew trust (2017), the new drug development of Phases I to III will bring 39 new antibiotics whose development is in progress. However, more research shows that the present antimicrobial pipeline is insufficiently strong to meet the current and future clinical needs. First off, only 13 (33%) of the 39 antibiotics [
54] under research are likely to be developed into a medication that can be sold. This is based on the treatment outcomes of progressing an antibiotic through various clinical phases [
55]. Secondly, the majority of newly developed antibiotics lack the innovative modes of action or unique chemical compositions that attack well-validated targets required to considerably guarantee their efficacy against resistant pathogens [
56]. A large portion of the medicines in research are new formulations or mixtures of already existing substances. Finally, a number of these medications miss the most important infections that are resistant to antibiotics. According to the Pew Trust report, just 31% of medications under research would indeed be effective against an ESKAPE infection and 33% might be effective against that immediate danger pathogen identified by the US Centers for Disease Control [
26].
Significant capital firms have mostly abandoned the antibiotic market in favor of more lucrative therapeutic endeavors. Small and medium-sized businesses (SMEs) have made an effort to fill this gap, but they typically do not have the funds or capacity to embark in extensive and protracted study and development (R&D) [
57]. As a result, the cheap fruit of antimicrobial research, such as chemical combination and renovations, has been picked. The costly and difficult challenge of identifying and creating genuinely innovative actions mechanisms that are efficient in combating the most resilient diseases is now left [
58]. Antibodies, probiotics, lysins, and phages are a few examples of the antibiotic options that some businesses are choosing to concentrate their future development on [
59].
Antibiotics resistance has posed challenge to modern medicine, but has also evolved the world and maintained the survival rates of children, youngsters, and elders. According to World Health Organization (2011) reports, that antibiotic resistance has increased too much that if action has not taken against it, could be incurable issue [
60]. The disappearance of antibiotic drug development raises the threat of incurable infections. Antimicrobial Response is a new effort that has been started in response to the urgent need for action to avert this calamity. This action plan aims to develop and improve academic alliances, brought about modification of expensive and time-consuming processes of regulations of antimicrobial drugs, and identify the economic growth of new antibiotics by trying to bring around each other populations that would need these substances with academic research, health practitioners, and drug industry (cost of use vs profit) [
61]. A framework for these activities would be provided by a global partnership for the research and development of antimicrobial drugs [
62].
The drug companies no longer view antibiotic research as a prudent financial venture. Antimicrobial are not nearly as valuable as medications that manage serious diseases like diabetes, mental disorders, asthmatic, or gastric reflux since they are frequently therapeutic and only taken for brief periods of time [
23]. The net present value (NPV) of a new medicine is really only around
$50 million, according to a cost-benefit analysis conducted by the Department of Health Care in London, as opposed to nearly
$1 billion for a medicine used to cure a neurodegenerative illness. Drug corporations prefer to make investments in chronic illness medications since they are more valuable. The comparably low cost of antibiotic is another aspect that makes developing antibiotics unattractive from a business standpoint. In comparison to chemotherapeutic drugs [
63], which can cost hundreds of thousands of dollars each course, modern antibiotics are typically priced at a minimum of
$1,000 to
$3,000 per course. Additionally, funders and the general populace consider antibiotic to be of low value because to their accessibility, simplicity of use, and usually inexpensive cost [
64].
In contrast, experts in bacterial infections and microbiology have cautioned against overusing antibiotics. Due to their concern about fostering drug resistance, doctors frequently retain new antibiotics for use in the most severe instances rather than instantly giving them. Instead, they continue to use older antibiotics that have demonstrated equivalent efficacy. New medicines therefore are frequently used as "last-line" treatments to cure severe illnesses [
65]. This technique results in a decreased return on investments and a decrease in the quantity of antimicrobial drugs. Resistant can ultimately develop when local agents are introduced, and this is almost certain. However, because microbial evolution is random, it is difficult to forecast when resistance may emerge [
50]. Thus, when resistance arises to a new antibiotic, a company who has spent a lot of money on antibacterial drugs research may find that their earnings are unexpectedly reduced. End consumers of antibiotic have also been restrained by the Great Recession's effects on the economy. The population now has access to inexpensive and usually effective medications, which is advantageous; on the other hand, many payers anticipate that all antibiotic will be charged similarly, including novel treatments that target multi drug resistance (MDR) infections [
66].
Due to extreme antibiotic resistance and multi drug resistance have challenged the large pharmaceutical companies and badly affected the drug development which needs millions of U.S. dollars investment to design new antibiotics for future. According to reports of Infectious Diseases Society of America (IDSA) (2013), there are few antibiotics in phase 2 or 3 of development. The IDSA specifically cited the development of insufficiently few medicines that were active against developing, extremely resistant gram-negative pathogens such
Enterobacteriaceae,
Pseudomonas aeruginosa, and
Acinetobacter baumannii. Additionally, drug manufacturers are now more actively focused on creating medicines for gram-negative infections than methicillin-resistant
Staphylococcus aureus (MRSA) [
67]. The most probable reason of this disparity is that while MRSA is a significant issue globally, the market for treating gram-negative bacteria is less and slightly more unexpected due to the quick spread of resistance [
68].
The process of discovering novel antibiotics and turning them into medicines is time- as well as money-consuming. A new drug must be developed costing between 800 million and 1 billion dollars, and it often takes more than ten years for it to reach the laboratory [
69]. Examining alternative to antibiotic therapy is a new strategy given the urgency with which we must now combat antibiotic resistance. Another approach to combating antibiotic resistance is to prevent illnesses from taking place in the first place.