Submitted:
26 June 2023
Posted:
28 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Anthropometric assessment of the pregnant women
2.3. Foetal body composition
2.4. Anthropometry and body composition of neonates
2.5. Sample collection and DNA extraction
2.6. Methylation analysis
2.7. Data analysis
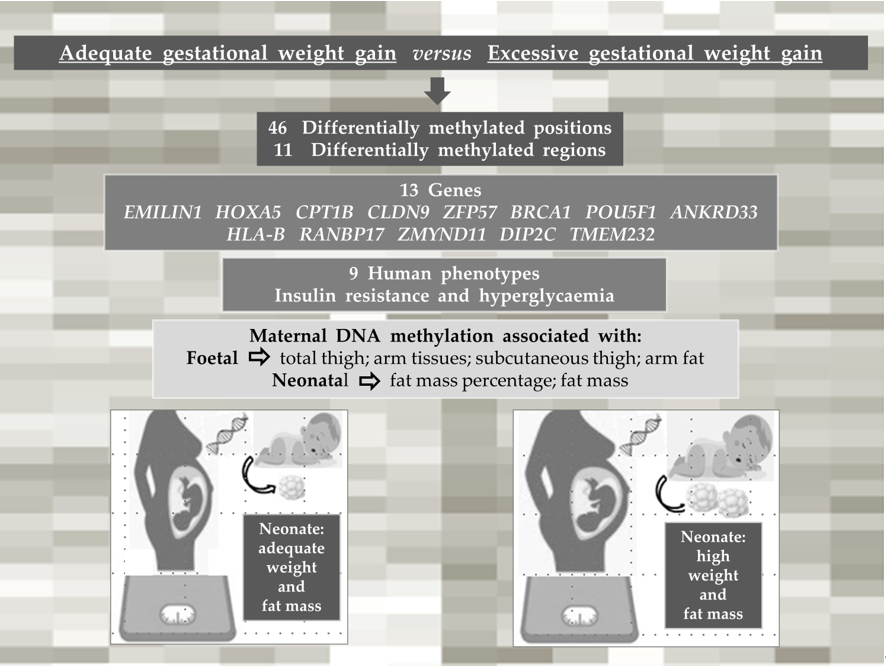
3. Results
3.1. Characteristics of the pregnant women and their neonates
3.2. Anthropometry and body composition of pregnant women and of their foetuses and neonates
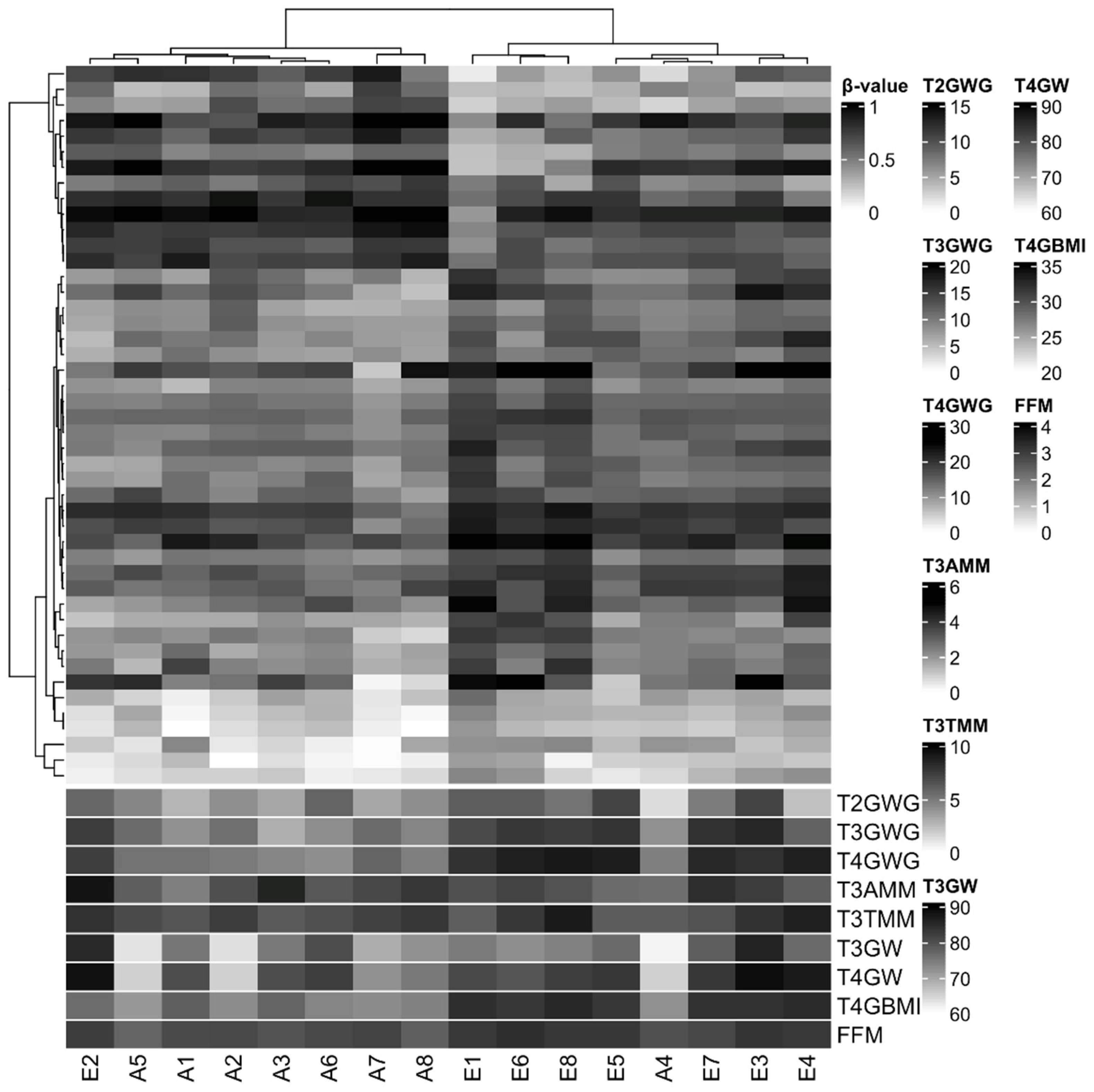
3.3. Characterization of DNA methylation
3.4. DNA methylation changes are associated with some foetal and neonatal outcomes
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of abbreviations
References
- Rasmussen, K.M.; Yaktine, A.L. Guidelines I of M (US) and NRC, Weight (US) Committee to Reexamine IOM Pregnancy. Weight Gain During Pregnancy. Weight Gain Dur Pregnancy Reexamining Guidel 2009.
- Guelinckx, I.; Devlieger, R.; Beckers, K.; Vansant, G. Maternal obesity: Pregnancy complications, gestational weight gain and nutrition. Obes. Rev, 2008, 9, 140-150. [CrossRef]
- Centers for disease control and prevention et al. QuickStats: Gestational Weight Gain* Among Women with Full-Term, Singleton Births, Compared with Recommendations - 48 States and the District of Columbia, 2015. MMWR Morb Mortal Wkly Rep. 2016, 3, 2018.
- Goldstein, R.F.; Abell, S.K.; Ranasinha, S.; Misso, M.L.; Boyle, J.A.; Harrison, C.L.; et al. Gestational weight gain across continents and ethnicity: systematic review and meta-analysis of maternal and infant outcomes in more than one million women. BMC Med [Internet]. BMC Medicine. 2018, 16, 1-14. [CrossRef]
- Carrilho, T.R.B.; Rasmussen, K.M.; Hutcheon, J.A.; Alves, R.F.S.; Farias, D.R.; Freitas-Costa, N.C.; et al. Prevalence and temporal trends in prepregnancy nutritional status and gestational weight gain of adult women followed in the Brazilian Food and Nutrition Surveillance System from 2008 to 2018. Matern & Child Nutr. 2022, 18, e13240. [CrossRef]
- Barker, D.J. In utero programming of chronic disease. Clin Sci (Lond). 1998, 95, 115-28.
- Godfrey, K.M.; Sheppard, A.; Gluckman, P.D.; Lillycrop, K.A.; Burdge, G.C.; Mclean, C.; et al. Epigenetic gene promoter methylation at birth is associated with child’s later adiposity. Diabetes. 2011, 60, 1528-1534. [CrossRef]
- Portha, B.; Fournier, A.; Ah Kioon, M.D.; Mezger, V.; Movassat, J. Early environmental factors, alteration of epigenetic marks and metabolic disease susceptibility. Biochimie. 2014, 97, 1-15. [CrossRef]
- Sharp, G.C.; Lawlor, D.A,; Richmond, R.C.; Fraser, A.; Simpkin, A.; Suderman, M.; et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2015, 44, 1288-1304. [CrossRef]
- Olerup, O.; Zetterquist, H.; HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: An alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992, 39, 225-35. [CrossRef]
- Morris, T.J.; Butcher, L.M.; Feber, A.; Teschendorff, A.E.; Chakravarthy, A.R.; Wojdacz, T.K.; et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014, 30, 428-30. [CrossRef]
- Nordlund, J.; Bäcklin, C.L.; Wahlberg, P.; Busche, S.; Berglund, E.C.; Eloranta, M.L.; et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013, 14, 1-15. [CrossRef]
- Zhou, W.; Laird, P.W.; Shen, H. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 2017, 45, e22-e22. [CrossRef]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013, 29, 189-96. [CrossRef]
- Teschendorff, A.E.; Menon, U.; Gentry-Maharaj, A.; Ramus S,J.; Gayther, S.A.; Apostolidou, S.; et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009, 4, e8274. [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007, 8, 118-27. [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H,; et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012, 13, 1-16. [CrossRef]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004, 3, 1. [CrossRef]
- Jaffe, A.E.; Murakami, P.; Lee, H.; Leek, J.T.; Fallin, M.D.; Feinberg, A.P.; et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012, 41, 200-9. [CrossRef]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol [Internet]. 2010, 28, 495-501. [CrossRef]
- Venkataraman, H.; Ram, U.; Craik, S.; Arungunasekaran, A.; Seshadri, S.; Saravanan, P. Increased fetal adiposity prior to diagnosis of gestational diabetes in South Asians: more evidence for the ‘thin-fat’ baby. Diabetologia. 2017, 60, 399-405. [CrossRef]
- Yerlikaya-Schatten, G.; Feichtinger, M.; Stopp, T.; Huhn, E.A.; Chalubinski, K.; Husslein, P.; et al. Trajectories of Fetal Adipose Tissue Thickness in Pregnancies After Gastric Bypass Surgery. Obes Surg. 2020, 30, 96-101. [CrossRef]
- Favero, G.; Paini, A.; De Ciuceis, C.; Rodella, L.F.; Moretti, E.; Porteri, E.; et al. Changes in extracellular matrix in subcutaneous small resistance arteries of patients with essential hypertension. Blood Press [Internet]. 2018, 27, 231-239. [CrossRef]
- Holzman, M.A.; Ryckman, A.; Finkelstein, T.M.; Landry-Truchon, K.; Schindler, K.A.; Bergmann, J.M.; et al. HOXA5 Participates in Brown Adipose Tissue and Epaxial Skeletal Muscle Patterning and in Brown Adipocyte Differentiation. Front Cell Dev Biol. 2021, 9, 311. [CrossRef]
- Lefort, N.; Glancy, B.; Bowen, B.; Willis, W.T.; Bailowitz, Z.; De Filippis, E.A.; et al. Increased reactive oxygen species production and lower abundance of complex I subunits and carnitine palmitoyltransferase 1B protein despite normal mitochondrial respiration in insulin-resistant human skeletal muscle. Diabetes. 2010, 59, 2444-52. [CrossRef]
- Walley, A.J.; Jacobson, P.; Falchi, M.; Bottolo, L.; Andersson, J.C.; Petretto, E.; et al. Differential coexpression analysis of obesity-associated networks in human subcutaneous adipose tissue. Int J Obes. 2012, 36, 137-47. [CrossRef]
- Tulyeu, J.; Kumagai, H.; Jimbo, E.; Watanabe, S.; Yokoyama, K.; Cui, L.; et al. Probiotics prevents sensitization to oral antigen and subsequent increases in intestinal tight junction permeability in juvenile–young adult rats. Microorganisms. 2019, 7, 463. [CrossRef]
- Mackay, D.J.G.; Callaway, J.L.A.; Marks, S.M.; White, H.E.; Acerini, C.L.; Boonen, S.E.; et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet. 2008, 40, 949-51. [CrossRef]
- Parkkola, A.; Turtinen, M.; Härkönen, T.; Ilonen, J.; Knip, M.; Knip, M.; et al. Family history of type 2 diabetes and characteristics of children with newly diagnosed type 1 diabetes. Diabetologia. 2021, 64, 581-90. [CrossRef]
- Iyengar, N.M.; Zhou, X.K.; Mendieta, H.; El-Hely, O.; Giri, D.D.; Winston, L.; et al. Effects of obesity on breast aromatase expression and systemic metabo-inflammation in women with BRCA1 or BRCA2 mutations. NPJ Breast Cancer [Internet]. 2021, 7, 18. [CrossRef]
- 31. Sun, T.; Han, X. Death versus dedifferentiation: The molecular bases of beta cell mass reduction in type 2 diabetes. Semin Cell Dev Biol [Internet]. 2020, 103, 76-82. [CrossRef]
- Jin, X.; Li, Y.; Guo, Y.; Jia, Y.; Qu, H.; Lu, Y.; et al. ERα is required for suppressing OCT4-induced proliferation of breast cancer cells via DNMT1/ISL1/ERK axis. Cell Prolif. 2019, 52, e12612. [CrossRef]
- Wang, J.; Gao, F.; Zhao, X.; Cai, Y.; Jin, H. Integrated analysis of the transcriptome- wide m6A methylome in preeclampsia and healthy control placentas. PeerJ. 2020, 8, e9880. [CrossRef]
- Rastogi, D.; Johnston, A.D.; Nico, J.; Loh, L.N.; Jorge, Y.; Suzuki, M.; et al. Functional genomics of the pediatric obese asthma phenotype reveal enrichment of rho-gtpase pathways. Am J Respir Crit Care Med. 2020, 202, 259-74. [CrossRef]
- Shen, J.; Guo, T.; Wang, T.; Zhen, Y.; Ma, X.; Wang, Y.; et al. HLA-B∗07, HLA-DRB1∗07, HLA-DRB1∗12, and HLA-c∗03:02 strongly associate with BMI: Data from 1.3 million healthy Chinese adults. Diabetes. 2018, 67, 861-71. [CrossRef]
- Karlsson, T.; Rask-Andersen, M.; Pan, G.; Höglund, J.; Wadelius, C.; Ek, W.E.; et al. Contribution of genetics to visceral adiposity and its relation to cardiovascular and metabolic disease. Nat Med [Internet]. 2019, 25, 1390-5. [CrossRef]
- Boyson, S.P.; Gao, C.; Quinn, K.; Boyd, J.; Paculova, H.; Frietze, S.; et al. Functional roles of bromodomain proteins in cancer. Cancers (Basel). 2021, 13, 3606. [CrossRef]
- Deane, C.S.; Willis, C.R.G.; Phillips, B.E.; Atherton, P.J.; Harries, L.W.; Ames, R.M.; et al. Transcriptomic meta-analysis of disuse muscle atrophy vs. resistance exercise-induced hypertrophy in young and older humans. J Cachexia Sarcopenia Muscle. 2021, 12, 629-45. [CrossRef]
- Descipio, C.; Conlin, L.; Rosenfeld, J.; Tepperberg, J.; Pasion, R.; Patel, A.; et al. Subtelomeric deletion of chromosome 10p15.3: Clinical findings and molecular cytogenetic characterization. Am J Med Genet Part A. 2012, 158, 2152-61. [CrossRef]
- Devarbhavi, P.; Telang, L.; Vastrad, B.; Tengli, A.; Vastrad, C.; Kotturshetti, I. Identification of key pathways and genes in polycystic ovary syndrome via integrated bioinformatics analysis and prediction of small therapeutic molecules. Reprod Biol Endocrinol. 2021, 19, 31. [CrossRef]
- Yang, I.V.; Lozupone, C.A.; Schwartz, D.A. The environment, epigenome, and asthma. J Allergy Clin Immunol [Internet]. 2017, 140, 14-23. [CrossRef]
- Voerman, E.; Santos, S.; Golab, B.P.; Amiano, P.; Ballester, F.; Barros, H.; et al. Maternal body mass index, gestational weight gain, and the risk of overweight and obesity across childhood: An individual participant data meta-analysis. PLoS Med. 2019, 16, e1002744. [CrossRef]
- Castillo, H.; Santos, I.S.; Matijasevich, A. Relationship between maternal pre-pregnancy body mass index, gestational weight gain and childhood fatness at 6-7 years by air displacement plethysmography. Matern Child Nutr. 2015, 11, 606-17. [CrossRef]
- Rzehak, P.; Covic, M.; Saffery, R.; Reischl, E.; Wahl, S.; Grote, V.; et al. DNA-Methylation and Body Composition in Preschool Children: Epigenome-Wide-Analysis in the European Childhood Obesity Project (CHOP)-Study. Sci Rep [Internet]. 2017, 7, 14349. [CrossRef]
- Bigaard, J.; Frederiksen, K.; Tjønneland, A.; Thomsen, B.L.; Overvad, K.; Heitmann, B.L.; et al. Body fat and fat-free mass and all-cause mortality. Obes Res. 2004, 12, 1042-9. [CrossRef]

| Variables | Excessive gestational weight gain (n=30) | Adequate gestational weight gain (n=45) | P |
| Pregnant women | |||
| Age (years) | 25.9±6.0 | 29.0±6.4 | 0.064 |
| Ethnicity | |||
| White | 10 (33.3%) | 15 (33.3%) | 0.351 |
| Black | 03 (10.0%) | 10 (22.2%) | |
| Brown | 17 (56.7%) | 20 (44.4%) | |
| Marital status | |||
| Single/without partner | 00 (0.0%) | 04 (8.8%) | 0.245 |
| Married/with partner | 30 (100.0%) | 41 (91.1%) | |
| Education | |||
| Elementary school | 01 (3.3%) | 09 (20.0%) | 0.115 |
| High school degree | 24 (80.0%) | 30 (66.7%) | |
| University degree | 05 (16.7%) | 06 (13.3%) | |
| Parity | |||
| 0 | 05 (16.7%) | 07 (15.6%) | 0.991 |
| 1 | 19 (63.3%) | 29 (64.4%) | |
| 2 a 4 | 06 (20.0%) | 09 (20.0%) | |
| Neonates | |||
| Age (weeks) | 39.9±1.1 | 39.2±1.4 | 0.162 |
| Sex | |||
| Female | 17 (56.6%) | 16 (35.6%) | 0.071 |
| Male | 13 (43,3%) | 29 (64.4%) | |
| Variables | Excessive gestational weight gain (n=30) | Adequate gestational weight gain (n=45) | p | ||
| Pregnant women | |||||
| Anthropometry | |||||
| T1 Pre-pregnancy weight (kg) | 60.54 | 6.37 | 57.52 | 7.17 | 0.682 |
| T1 Height (cm) | 163.71 | 6.56 | 161.39 | 6.93 | 0.785 |
| T1 Pre-pregnancy BMI (kg/m2) | 22.68 | 1.74 | 22.06 | 1.79 | 0.405 |
| T4 BMI (kg/m2) | 29.90 | 2.01 | 27.15 | 1.84 | 0.010 |
| T4 Total gestational weight gain (kg) | 19.60 | 2.43 | 13.26 | 1.54 | 0.010 |
| Pre-pregnancy body composition | |||||
| Pre-pregnancy fat mass (%) | 29.32 | 4.03 | 26.80 | 4.85 | 0.262 |
| Pre-pregnancy fat mass (kg) | 18.75 | 4.19 | 15.82 | 4.40 | 0.167 |
| Pre-pregnancy fat-free body mass (Kg) | 44.17 | 4.60 | 42.26 | 3.85 | 0.188 |
| Pre-pregnancy muscle mass (kg) | 41.94 | 4.35 | 40.04 | 3.58 | 0.135 |
| Fetuses | |||||
| T2 Fetal weight (g) | 629.30 | 204.02 | 598.87 | 186.40 | 0.875 |
| T3 Fetal weight (g) | 2172.82 | 353.43 | 2132.18 | 457.47 | 0.514 |
| T2 SCFT (mm) | 2.84 | 0.52 | 2.95 | 0.56 | 0.721 |
| T3 SCFT (mm) | 4.13 | 0.76 | 4.07 | 1.07 | 0.521 |
| T2 Total thigh tissue (cm3) | 5.23 | 1,78 | 5.12 | 1.53 | 0.945 |
| T3 Total thigh tissue (cm3) | 13.37 | 2.97 | 13.53 | 3.25 | 0.955 |
| T2 Thigh muscle mass (cm3) | 2.97 | 1.04 | 2.90 | 0.93 | 0.729 |
| T3 Thigh muscle mass (cm3) | 7.69 | 1.72 | 7.54 | 1.80 | 0.643 |
| T2 Subcutaneous thigh fat (cm3) | 2.26 | 0.84 | 2.27 | 0.76 | 0.991 |
| T3 Subcutaneous thigh fat (cm3) | 5.68 | 1.64 | 6.03 | 1.61 | 0.225 |
| T2 Total arm tissue (cm3) | 3.05 | 0.93 | 2.85 | 0.83 | 0.358 |
| T3 Total arm tissue (cm3) | 7.01 | 1.55 | 7.07 | 1.87 | 0.860 |
| T2 Arm muscle mass (cm3) | 1.57 | 0.50 | 1.46 | 0.46 | 0.224 |
| T3 Arm muscle mass (cm3) | 3.45 | 0.82 | 3.52 | 0.98 | 0.683 |
| T2 Subcutaneous arm fat (cm3) | 1.46 | 0.52 | 1.46 | 0.63 | 0.991 |
| T3 Subcutaneous arm fat (cm3) | 3.55 | 0.93 | 3.56 | 1.02 | 0.928 |
| Neonates | |||||
| T5 Weight (g) | 3354.87 | 298.47 | 3068.50 | 386.57 | 0.027 |
| T5 Length (cm) | 50.03 | 1.78 | 48.80 | 2.33 | 0.182 |
| T5 Fat-free mass percentage (%) | 90.39 | 3.98 | 91.57 | 5.65 | 0.120 |
| T5 Fat mass percentage (%) | 9.61 | 3.98 | 8.43 | 5.65 | 0.120 |
| T5 Fat-free mass (kg) | 3.08 | 0.19 | 2.76 | 0.27 | 0.218 |
| T5 Fat mass (kg) | 0.34 | 0.13 | 0.26 | 0.21 | 0.039 |
| Terms name | Binom Raw P-Value | Binom Fold Enrichment |
| Transient neonatal diabetes mellitus | 0.0010 | 1041.1 |
| Neonatal insulin-dependent diabetes mellitus | 0.0015 | 656.2 |
| Severe failure to thrive | 0.0037 | 269.7 |
| Insulin-resistant diabetes mellitus | 0.0185 | 53.7 |
| Insulin resistance | 0.0236 | 42.0 |
| Breast carcinoma | 0.0255 | 38.7 |
| Neoplasm of the breast | 0.0272 | 36.4 |
| Hyperglycemia | 0.0275 | 35.9 |
| Dehydration | 0.0379 | 25.9 |
| T2 Total thigh tissue | Β | r² | p | 95% CI |
| DMR 2 | 9.172 | 0.853 | 0.014 | 2.340; 16.005 |
| Gestational weight gain | -0.010 | 0.843 | -0.127; 0.106 | |
| Pre-pregnancy BMI | -0.780 | 0.005 | -1.249; -0.310 | |
| Maternal age | -0.048 | 0.400 | -0.172; 0.076 | |
| Sex | 1.026 | 0.092 | -0.205; 2.257 | |
| Gestational age | 0.833 | 0.002 | 0.411; 1.255 | |
| DMR 6 | 21.516 | 0.039 | 1.407; 41.625 | |
| Gestational weight gain | -0.072 | 0.322 | -0.228; 0.084 | |
| Pre-pregnancy BMI | -0.786 | 0.820 | 0.008 | -1.316; -0.256 |
| Maternal age | -0.044 | 0.489 | -0.181; 0.093 | |
| Sex | 1.313 | 0.070 | -0.131; 2.757 | |
| Gestational age | 0.939 | 0.002 | 0.443; 1.434 | |
| T3 Total thigh tissue | Β | r² | p | 95% CI |
| DMR 2 | 8.265 | 0.715 | 0.018 | 1.790; 14.740 |
| Gestational weight gain | 0.045 | 0.371 | -0.064; 0.154 | |
| Pre-pregnancy BMI | -0.393 | 0.080 | -0.844; 0.058 | |
| Maternal age | 0.036 | 0.487 | -0.077; 0.150 | |
| Sex | 0.679 | 0.127 | -0.235; 1.593 | |
| Gestational age | 0.472 | 0.155 | -0215; 1.160 | |
| T2 Thigh muscle mass | Β | r² | p | 95% CI |
| DMR 2 | 5.314 | 0.814 | 0.021 | 1.006; 9.622 |
| Gestational weight gain | -0.026 | 0.440 | -0.100; 0.047 | |
| Pre-pregnancy BMI | -0.416 | 0.011 | -0.712; -0.120 | |
| Maternal age | -0.012 | 0.739 | -0.090; 0.066 | |
| Sex | 0.773 | 0.051 | -0.003; 1.549 | |
| Gestational age | 0.431 | 0.005 | 0.165; 0.697 | |
| T3 Thigh muscle mass | Β | r² | p | 95% CI |
| DMR 2 | 6.373 | 0.687 | 0.032 | 0.694; 12.052 |
| Gestational weight gain | 0.067 | 0.147 | -0,029; 0.162 | |
| Pre-pregnancy BMI | -0.177 | 0.339 | -0,572; 0.219 | |
| Maternal age | 0.036 | 0.429 | -0,063; 0.136 | |
| Sex | 0.442 | 0.244 | -0.360; 1.243 | |
| Gestational age | 0.358 | 0.213 | -0.246; 0.961 | |
| T2 Subcutaneous thigh fat | Β | r² | p | 95% CI |
| DMR 2 | 3.858 | 0.846 | 0.029 | 0.506; 7.211 |
| Gestational weight gain | 0.016 | 0.549 | -0.041; 0.073 | |
| Pre-pregnancy BMI | -0.364 | 0.006 | -0.594; -0.133 | |
| Maternal age | -0.037 | 0.207 | -0.097; 0.024 | |
| Sex | 0.254 | 0.367 | -0.350; 0.858 | |
| Gestational age | 0.402 | 0.002 | 0.195; 0.609 | |
| DMR 6 | 10.933 | 0.017 | 2.494; 19.372 | |
| Gestational weight gain | -0.019 | 0.532 | -0.084; 0.047 | |
| Pre-pregnancy BMI | -0.385 | 0.862 | 0.004 | -0.607; -1.162 |
| Maternal age | -0.035 | 0.202 | -0.093; 0.023 | |
| Sex | 0.424 | 0.148 | -0.182; 1.030 | |
| Gestational age | 0.463 | 0.001 | 0.255; 0.671 | |
| T3 Subcutaneous thigh fat | Β | r² | p | 95% CI |
| DMR 10 | 7.604 | 0.596 | 0.033 | 0.763; 14.445 |
| Gestational weight gain | -0.034 | 0.267 | -0.100; 0.031 | |
| Pre-pregnancy BMI | -0.247 | 0.064 | -0.511; 0.018 | |
| Maternal age | 0.015 | 0.619 | -0.052; 0.083 | |
| Sex | 0.257 | 0.311 | -0.284; 0.797 | |
| Gestational age | -0.004 | 0.982 | -0.401; 0.393 | |
| T3 Total arm tissue | Β | r² | p | 95% CI |
| DMR 6 | -25.640 | 0.804 | 0.002 | -39.368; -11.911 |
| Gestational weight gain | 0.115 | 0.039 | 0.007; 0.222 | |
| Pre-pregnancy BMI | 0.410 | 0.043 | 0.016; 0.805 | |
| Maternal age | 0.038 | 0.400 | -0.059; 0.134 | |
| Sex | -0.269 | 0.460 | -1.059; 0.521 | |
| Gestational age | 0.311 | 0.257 | -0.271; 0.893 | |
| T3 Subcutaneous arm fat | Β | r² | p | 95% CI |
| DMR 6 | -17.433 | 0.667 | 0.010 | -29.597; -5.270 |
| Gestational weight gain | 0.078 | 0.097 | -0.017; 0.172 | |
| Pre-pregnancy BMI | 0.233 | 0.165 | -0.116; 0.583 | |
| Maternal age | 0.014 | 0.716 | -0.071; 0.099 | |
| Sex | -0.339 | 0.302 | -1.039; 0.361 | |
| Gestational age | 0.175 | 0.461 | -0,340; 0.691 | |
| T5 Fat mass percentage | Β | r² | p | 95% CI |
| DMR 2 | -20.299 | 0.761 | 0.039 | -39.362; -1.236 |
| Gestational weight gain | 0.500 | 0.013 | 0.135; 0.865 | |
| Pre-pregnancy BMI | 0.651 | 0.275 | -0.615; 1.916 | |
| Maternal age | 0.071 | 0.530 | -0.175; 0.318 | |
| Sex | -5.207 | 0.002 | -7.968; -2.446 | |
| Gestational age | -0.653 | 0.196 | -1.710; 0.403 | |
| T5 Fat mass | Β | r² | p | 95% CI |
| DMR 2 | -0.719 | 0.780 | 0.040 | -0.395; -0.042 |
| Gestational weight gain | 0.019 | 0.009 | 0.006; 0.032 | |
| Pre-pregnancy BMI | 0.026 | 0.216 | -0.018; 0.071 | |
| Maternal age | 0.003 | 0.391 | -0.005; 0.012 | |
| Sex | -0.180 | 0.002 | -0.279; 0.082 | |
| Gestational age | -0.016 | 0.374 | -0.053; 0.022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





