1. Introduction
Helicobacter pylori (
H. pylori) is among the most common bacterial infections in the world and one of the most common infectious agents linked to any malignancy (~ 89% of non-cardia gastric cancer [
1]). Eradication of
H. pylori is recommended to reduce the risk of gastric cancer and improve gastritis and gastric atrophy [
2].
Treatment for
H. pylori typically includes a proton-pump inhibitor and a combination of antibiotics which should be adjusted based on the resistance rates within the geographic region. Multiple factors affect treatment success, with compliance and antibiotic resistance as the two most commonly cited reasons for failure [
3]. More than three quarters of respondents in a national survey indicated compliance with eradication therapies was difficult due to gastrointestinal upset [
4]. Commonly used therapies are now estimated to fail in approximately 25% to 40% of patients, [
5,
6,
7] primarily due to increasing antibiotic resistance.
H. pylori antibiotic resistance rates vary greatly between each antibiotic and across geographic communities, and resistance rates are increasing [
8]. A systematic review of antibiotic resistance in the Asia-Pacific regions found overall resistance for clarithromycin around 17% (95% CI 15-18) and 44% (95% CI 39-48) for metronidazole, with clarithromycin resistance rates increasing over the 1990-2016 time period of the study [
9]. A review of articles published between 2007 and 2017 found clarithromycin rates in the Americas near 10% (95% CI 4-16%) with metronidazole rates of 23% (91% CI 2-44%) [
10]. The observed resistance prevalence rates were lower when the studies were restricted to only those among treatment-naïve patients, with resistance rates of 10% for clarithromycin and 23% for metronidazole [
10]. In Portugal, where overall clarithromycin resistance was 50%; higher rates were associated with failed eradication treatment [
11].
North American studies on the antibiotic resistance rates for
H. pylori remain scarce [
2]. A recent meta-analysis restricted to US only studies (N= 19 studies total) found high resistance to clarithromycin, 31.5% (95% CI 23.6%-40.6%; n=18 studies) and metronidazole, 42.1% (95% CI 27.3% - 58.6%; n=14 studies) [
12]. However, there was considerable heterogeneity across the studies used to estimate the pooled prevalence rates in these meta-analyses. As with a 2018 meta-analysis across the Americas, the resistance prevalence rates were lower when restricting to treatment-naïve patients: 16.7% for clarithromycin and 29.3% for metronidazole [
12].
Even less is known about resistance rates among rural and Indigenous populations. In an
H. pylori resistance surveillance study among Alaska Natives, metronidazole resistance was observed in 42% of cultured isolates followed by clarithromycin resistance in 29.8% of isolates [
13]. None of these prior studies included data from American Indian populations living in the lower 48 states. In this current cross-sectional study, we utilized biopsy samples from patients already undergoing an upper GI endoscopy at a clinic serving American Indians in northern Arizona where
H. pylori prevalence is near 60% [
14] and about 30% of gastric biopsy samples were
H. pylori positive [
15]. Microbial DNA was amplified in samples to assess
H. pylori antibiotic resistance-associated mutation rates for clarithromycin and metronidazole. Prevalence of mutations associated with infection was compared by age and sex for each antibiotic.
2. Materials and Methods
2.1. Ethical Considerations
Working with medical samples and with American Indian communities requires IRB oversight and approval from the communities within which we work. The protocol and consent documentation were first approved by the Northern Arizona University Institutional Review Board (IRB #8195001). We also received Chapter resolutions before seeking Navajo Nation Human Research Review Board (NNHRRB) protocol approval (NNR-16.263 approved 8/21/2021). Approval by the NNHRRB to submit the manuscript was received 1/7/2023. All biopsy samples and data were deidentified prior to transport to Northern Arizona University.
2.2. Sample Collection
Participants were recruited from the Winslow Indian Health Care Clinic (WIHCC) general surgery clinic during their visit to discuss esophagogastroduodenoscopy (EGD) for upper GI symptoms. WIHCC is located and serves eight-chapter communities in the southwestern Navajo Nation. During their medical visit, the H. pylori study was explained by the surgeon. Willing participants signed an informed consent form to have two additional biopsy samples collected and their medical records abstracted for histopathology findings, prior testing and treatment for H. pylori, and demographic characteristics. Inclusion criteria were that the individual be at least 18 years of age and a patient. Clinic staff completed medical record abstraction using a record abstraction form and individuals were given a participant identification number for the sample and the abstracted information to protect anonymity. During endoscopy, a biopsy from both the antrum and the fundus was collected, placed in 400μL RNAlater; and transported to the NAU laboratory for DNA isolation. All biopsy samples and data were deidentified prior to transport to Northern Arizona University.
2.3. Sequencing
For the 23S ribosomal RNA (23S) and the oxygen-insensitive NADPH nitroreductase (rdxA) regions were sequenced for clarithromycin and metronidazole resistance, respectively (Table 1). First the glmM, which encodes for a phosphoglucosamine mutase, was used to confirm that samples were PCR-positive for H. pylori.
2.4. Primers
Custom primers were developed to detect the resistance associated mutations (Table 1). Clarithromycin-resistant mutations were defined as A2142G, A2143G, or T2182C of the 23S ribosomal RNA [19-22]. Mutations associated with metronidazole were defined as R16H, R16C, R16P, H25R, H53R, H53A, D59N, L62V, A68T, A68V, A68S, A68N, G98S, G163V, G163D, V204I, and A206T [
23]. Real time PCR was used to amplify these sequences using the primers listed below. Sequencing of the amplified
H. pylori DNA was performed by Genewiz (Azenta Life Sciences, South Plainfield, NJ). The analytic method was validated using a commercially available resistance testing kit, the Viasure
H. pylori-Clarithromycin resistance kit (Jant Pharmaceutical Corporation, Encino CA). Testing with this kit showed 100% sensitivity and specificity for the kits’ target mutations (A2142G, A2143G; see supplemental material 1). There is no commercial kit available to determine metronidazole resistance as point mutations seem to change depending on the population and country.
2.5. Analysis
The analysis was restricted to individuals with no history of H. pylori infection as recorded in the medical record, and thus assumed to be their primary infection. In addition, only samples for which the region of the pathogen, 23S ribosomal RNA for clarithromycin and rdxA for metronidazole, was successfully amplified are included in the analysis. Analyses were performed in triplicate for each sample.
Sample concordance was calculated as the number of pairs that were concordant (including those with no mutations) from all successfully amplified pairs. For each antibiotic, mutations associated with antibiotic resistance were compared by age (t-test) and sex (chi square). Treatment is qualitatively described for patients for whom we had treatment information.
2.6. Statistical Analysis
Statistical analyses were performed using StataBE v17 (StataCorp LLC, College Station, TX).
3. Results
3.1. Recruitment and Inclusion
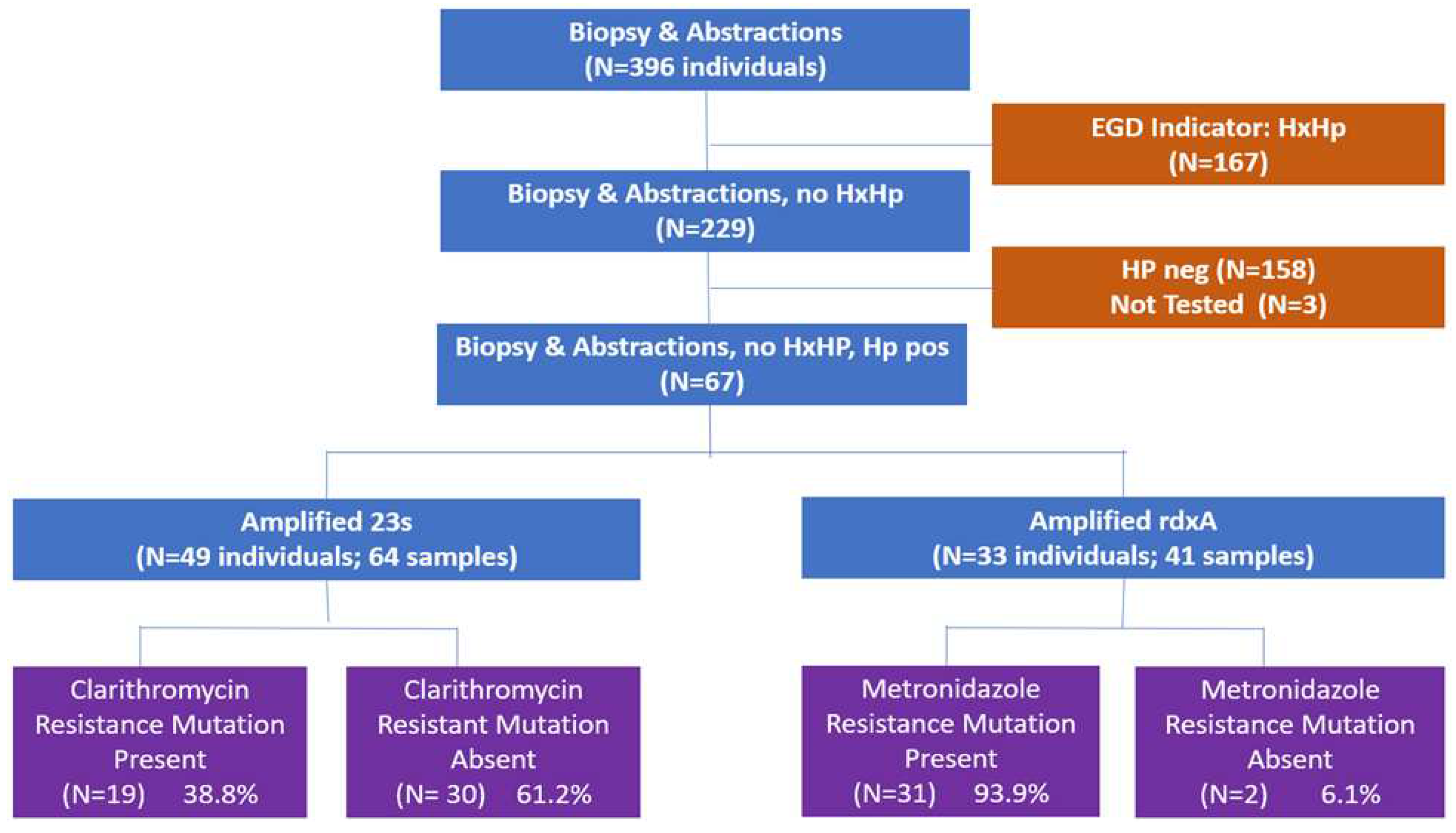
Individuals were recruited into the study during their EGD appointment. The project and the process were discussed. After consenting to the project, two additional biopsy samples were taken from both the antrum and fundus for 396 individuals and medical records were abstracted. Based on the information abstracted from the medical records, patients for which a history of
H. pylori infection was noted were excluded (N=167). After removing participants from this study because their samples were negative for
H. pylori (N=158) or could not be tested (N=3), there were 67 participants with
H. pylori-positive biopsy samples. Restricting the analysis to those samples which were
H. pylori positive with no prior history of
H. pylori, this process yielded 49 individuals (64 samples) for clarithromycin and 33 individuals (41 samples) for metronidazole (
Figure 1).
3.2. Antibiotic Clarithromycin
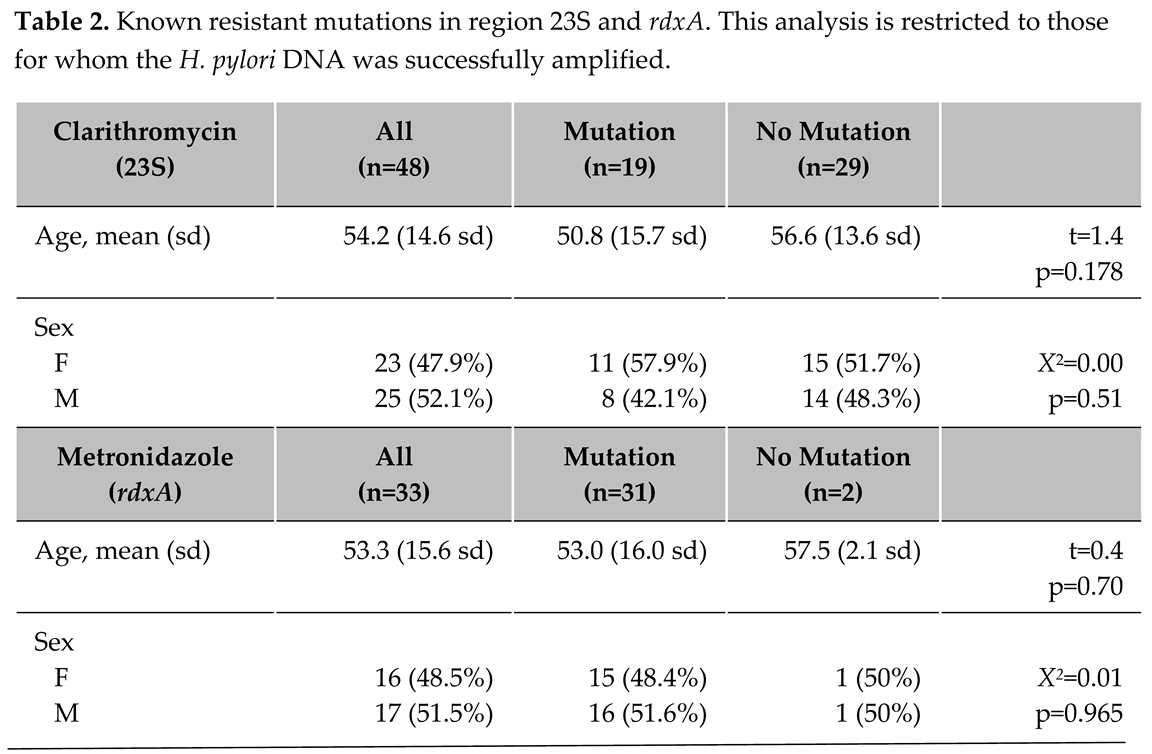
Clarithromycin resistance, defined as the presence of either T2182C, A2142G or A2143G mutations, was present in 38.8% (N=19) of the participants for whom 23S ribosomal RNA was successfully amplified. T2182C (N=18) was the most observed mutation, followed by A2142G (N=1) and A2143G (N=1). There were no statistically significant differences between those with and without clarithromycin-resistant mutations regarding mean age (p=0.18) or sex (p=0.51; Table 2). If the resistance definition was restricted to the more conservative A2142G or A2143G, the resistance prevalence was 4% (N=2 of 49 individuals).
Among the 13 individuals for whom both the antrum and fundal biopsy samples successfully amplified, nine had no clarithromycin-associated mutations, and three had the same mutations in both samples. The one discordant pair exhibited the T2182C mutation only in the antrum sample. Sensitivity analysis was performed using the Viasure clarithromycin resistance kit which confirmed presence of A2142G and A2143G (
Supplementary Table S1;
Supplementary Figure S1).
3.3. Antibiotic Metronidazole
Of the mutations associated with metronidazole resistance, we observed R16H (N=4 individuals), R16C (N=2), H53R (N=17), D59N (N=31), L62V (N=22), and G98S (N=15). Among the 33 individuals with no history of H. pylori infection, where the rdxA was successfully amplified, 93.9% (N=31) showed at least one of these mutations. There was no observed difference in mean age or sex differences; however, the H. pylori DNA in only two individuals did not express any of the mutations of interest (Table 2).
Five of the seven individuals with mutations known to be resistant to metronidazole who had both biopsy samples successfully amplified in the rdxA gene had identical mutations in both the antrum and fundus biopsy pairs. Of the two discordant samples, one exhibited metronidazole-associated mutations only in the antrum sample, while the other shared one matching mutation.
3.4. Antibiotic Treatment
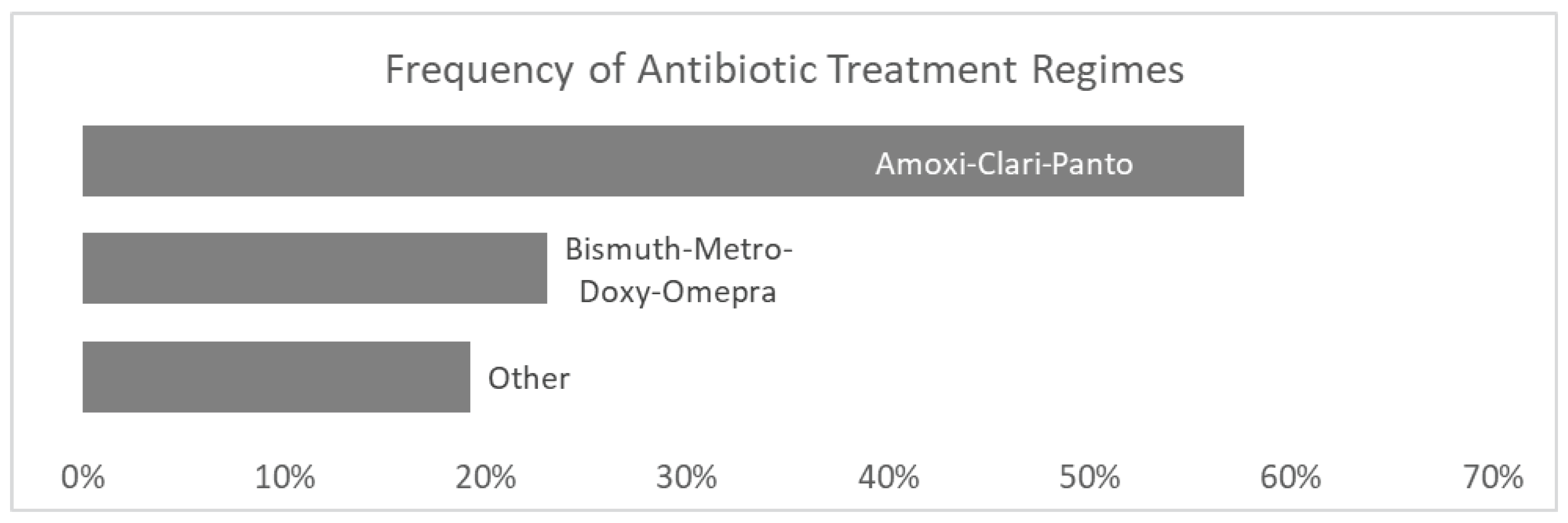
Among the 67 individuals positive for
H. pylori who met the inclusion criteria for this study, the prescribed treatment was known for 26 individuals (
Figure 2). Triple therapy consisting of amoxicillin, clarithromycin and pantoprazole was the most prescribed treatment regimen (57.7% of known treatments). Bismuth with metronidazole, doxycycline and omeprazole was the next most prescribed treatment (23.1%).
4. Discussion
To our knowledge, this is the first contemporary survey of H. pylori resistance among American Indians in the lower 48 states. In this sample of American Indians from northern Arizona undergoing EGD, we found high rates of mutations associated with antibiotic resistance (38.6% of patients exhibited mutations associated with clarithromycin and 95.6% had mutations associated with metronidazole. Our analysis indicates concerning rates of resistance although the clinical phenotype for some of these mutations needs to be further addressed using samples where full culture and sensitivity is possible. The high prevalence of H. pylori in the population and the high frequency of resistance associated mutations suggest that incorporating susceptibility testing is important when determining optimal treatment regimen.
The presence of the T2182C clarithromycin resistance mutation in this population is high compared with other populations [
16]. Agudo et al. [
25] found this mutation in only 5.9% of 118 patients in Madrid, while finding higher A to G 2142/3 point mutations, 88.1% and 85.3%, respectively [
17]. The T2182C mutation seems to be predominant in Asian countries where the infection is also high and it may indicate the long-term association between
H. pylori and our population [
16,
18] and further investigation of this mutation is warranted. While the A-to-G mutation at the 2142 or 2143 position or the A-to-C mutation at the 2142 region of the 23S rRNA gene is well established, the clarithromycin resistance role of the T2182C mutation has been questioned [
19]. Specifically, while the mutation may be observed among individuals with clarithromycin resistance, culturing yields mixed sensitive and resistant colonies [
20]. Others have shown that the association between established mutations is highly variable across populations and with mixed results even in culturing [
17,
18]. This is why whole genome sequencing has been proposed to identify multiple antibiotic resistance markers [
21] and improve therapy outcomes.
Our findings are closer to the 93.2% metronidazole resistance rates observed among a high-risk Colombian population, though none of their samples were resistant to clarithromycin (N=59) based on in vitro experiments and PCR amplification [
22]. Other studies in high-risk populations have found higher clarithromycin resistance, for example 31.2% in a study in Santiago Chile where
H. pylori positive rates were 48.7% [
23]. While studying the pattern of antibiotic resistance in Mexican mestizo for 20 years, it was found that while metronidazole resistance decreased from 75% to 51%, resistance for clarithromycin increased from 10% to 32% [
24]. Our results have provided a limited insight into antibiotic resistance in northern Arizona, and it may help explain some of the reasons for treatment failure, while emphasizing the need for further studies.
Limitations. These results are representative of patients undergoing EGD at a general surgery clinic in northern Arizona. We restricted the analysis to those individuals who did not have a history of H. pylori infection in their medical record. While this limited the sample size, it provided an estimate of antibiotic resistance in treatment-naïve patients. It is possible that this reflects higher use of macrolides during adolescence than in older patients.
Like most
H. pylori antibiotic resistance studies [
12], the resistance estimates were generated by molecular sequencing for the presence of resistance associated genes rather than by culture which is used to determine minimum inhibitory concentrations. This is a limitation as it measures the existence of resistance mutations in the
H. pylori DNA rather than clinical evidence of resistance. Metronidazole has shown a general lack of reproducibility in
in vitro testing. Neither Etest nor agar dilution is recommended as a reliable means for assessing
H. pylori resistance to metronidazole [
25]. In meta-analyses, stratifying by diagnostic method did not show significant differences in the estimated resistance rates [
9,
10].
These findings regarding antibiotic resistance are clinically relevant to the choices around treatment for
H. pylori. At the time of this study, we had treatment information for only 39% of the individuals who met the inclusion criteria. This low percentage may be because of the timing of record abstraction or may be indicative of differences in the sensitivity of clinical diagnostic tests. DNA extraction is more sensitive than both histopathology and the rapid CLO tests performed in clinics, which may mean patients go undetected and untreated using clinical diagnostics [
26]. However, given the observed resistance mutations in this sample and that more than half are treatments included clarithromycin (57.7% of known treatments) and another quarter (23.1%) of treatments include Metronidazole is cause for pause. Current guidelines recommend bismuth and non-bismuth quadruple, concomitant (PPI, amoxicillin, clarithromycin and a nitroimidazole) as a first line of treatment in areas with clarithromycin resistance over 15% [
8]. Others have reported that clarithromycin should be avoided in countries where resistance prevalence is greater than 25% [
9].
Depending on the definition for clarithromycin resistance, the observed resistance was either well above both guideline thresholds, that is 38.6% when including T2182C in the definition of resistance, or well below them, 4% when restricting to just A2142G or A2143G mutations. Perhaps a recommended way to address this issue is by reporting the association of mutations at A2143G and at A2142G with the T2182C mutation [
27].
While a screen-and-treat strategy is clearly recommended in high-risk communities, the implication of antibiotic resistance means that a screen-and-treat policy for communities with an intermediate to low risk of gastric cancer is only weakly supported [
8]. 2017). A Norwegian cohort showed an increased frequency of metronidazole (69.5%) and clarithromycin (38.5%) resistance among those who failed treatment [
28]. As susceptibility testing becomes increasingly available, it can guide initial therapies where empiric therapy success rates are low and should be considered for patients with a history of prior treatment failure [
29].
5. Conclusions
Given the risk of treatment failure with antibiotic resistance, there is a need to consider the resistance profile of the patient population during treatment. Little is known about resistance rates among rural indigenous populations in northern Arizona, where H. pylori prevalence is near 60%, and about one-third of gastric biopsy samples are H. pylori positive. The resistance rates we observed in this population of patients undergoing endoscopy are similar to that reported in other high-risk populations: 38.8% for clarithromycin and 93.9% for metronidazole.
Supplementary Materials
The following supporting information can be downloaded at:
Preprints.org, Supplemental Figure S1: Sample run for clarithromycin positive
H. pylori detected in the FAM channel. Arrow pointing to a suspicious sample. Also, the wild type phenotype was detected in the HEX channel which confirmed lack of clarithromycin resistance in this sample. The clarithromycin-resistant
H. pylori commercial kit from Viasure measures resistance to clarithromycin by targeting two known mutations; Supplemental Table S1: Testing of clarithromycin-resistant
H. pylori using the commercial kit from Viasure that measures resistance to clarithromycin by targeting two known mutations, A2142G and A2143G.
Author Contributions
F.P.M., H.E.B. and R.B.H. conceived and supervised the study; R.D. and L.P. recruited and consented participants at WIHCC; F.P.M., C.M.A.S. and A.R.G. conducted the DNA isolation, and characterization of antibiotic resistance; H.E.B. and F.P.M. completed the data analyses and led the writing of main manuscript. Specifically, conceptualization, F.P.M. and H.E.B.; methodology, F.P.M., C.M.A.S, A.R.G. and H.E.B.; investigation, F.P.M., C.M.A.S, A.R.G. and R.D.; resources, F.P.M. and R.B.H.; data curation, H.E.B., R.D. and L.P.; writing—original draft preparation, F.P.M. and H.E.B.; writing—review and editing, F.P.M., H.E.B., R.D. and R.B.H.; funding acquisition, F.P.M., H.E.B. and R.B.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Cancer Institute of the National Institutes of Health under the awards for the Partnership of Native American Cancer Prevention U54CA143924 (UACC) and U54CA143925 (NAU). Molecular resistance testing was supported by a grant from RedHill Biopharma.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
This work was funded in part by RedHill Biopharma. While the representatives provided their opinions, they did not influence the analysis or results. The authors confirm no other conflict of interest associated with this manuscript.
References
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr [Internet]. 2016 [cited 2023 Jun 21];7(2):418–9. [CrossRef]
- Chey, W. D., G. I. Leontiadis, C. W. Howden, and S. F. Moss. 2017. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 112: 212–238. [CrossRef]
- Shah, S. C., P. G. Iyer, and S. F. Moss. 2021. AGA Clinical Practice Update on the Management of Refractory Helicobacter pylori Infection: Expert Review. Gastroenterology. 160: 1831–1841. [CrossRef]
- Bailey, K. S., H. E. Brown, V. Lekic, K. Pradeep, J. L. Merchant, and R. B. Harris. 2023. Helicobacter pylori treatment knowledge, access and barriers: A cross-sectional study. Helicobacter. 28: e12954. [CrossRef]
- Malfertheiner, P., F. Megraud, C. A. O’Morain, J. P. Gisbert, E. J. Kuipers, A. T. Axon, F. Bazzoli, A. Gasbarrini, J. Atherton, D. Y. Graham, R. Hunt, P. Moayyedi, T. Rokkas, M. Rugge, M. Selgrad, S. Suerbaum, K. Sugano, E. M. El-Omar, L. P. Andersen, J. Atherton, A. Axon, E. El-Omar, P. Hungin, F. Di, G. Gasbarrini, G. Gensini, A. Pilotto, M. Rugge, R. Stockbru¨gger, D. Vaira, A. Burette, X. Calvet, J. Gisbert, A. Lanas, F. Chan, L. Coelho, J. C. Delchier, F. Megraud, W. Fischbach, M. Kist, S. Koletzko, P. Malfertheiner, M. Vieth, K. M. Fock, K. L. Goh, D. Y. Graham, N. Vakil, R. Hunt, V. A. Isakov, E. J. Kuipers, L. Kupcinskas(Kaunas), T. Rokkas, S. Ladas, M. Leja, J. C. Machado, V. Mahachai, K. McColl, Y. Niv, C. O’Morain, A. Ristimaki, and K. Sugano. 2012. Management of Helicobacter pylori infection—the Maastricht IV/ Florence Consensus Report. Gut. 61: 646–664. [CrossRef]
- O’Connor, A., and C. O’Moráin. 2013. Helicobacter pylori infection in Europe: current perspectives. Expert Rev. Gastroenterol. Hepatol. 7: 541–548. [CrossRef]
- Venerito, M., T. Krieger, T. Ecker, G. Leandro, and P. Malfertheiner. 2013. Meta-Analysis of Bismuth Quadruple Therapy versus Clarithromycin Triple Therapy for Empiric Primary Treatment of Helicobacter pylori Infection. Digestion. 88: 33–45. [CrossRef]
- Malfertheiner, P., F. Megraud, C. O’Morain, J. Gisbert, E. Kuipers, A. Axon, F. Bazzoli, A. Gasbarrini, J. Atherton, D. Graham, R. Hunt, P. Moayyedi, T. Rokkas, M. Rugge, M. Selgrad, S. Sauerbaum, K. Sugano, and E. El-Omar. 2017. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 66: 6–30. [CrossRef]
- Kuo, Y. T., J. M. Liou, E. M. El-Omar, J. Y. Wu, A. H. R. Leow, K. L. Goh, R. Das, H. Lu, J. T. Lin, Y. K. Tu, Y. Yamaoka, and M. S. Wu. 2017. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: a systematic review and meta-analysis. Lancet. Gastroenterol. Hepatol. 2: 707–715. [CrossRef]
- Savoldi, A., E. Carrara, D. Y. Graham, M. Conti, and E. Tacconelli. 2018. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology. 155: 1372-1382.e17. [CrossRef]
- Almeida, N., J. M. Romãozinho, M. M. Donato, C. Luxo, O. Cardoso, M. A. Cipriano, C. Marinho, A. Fernandes, C. Calhau, and C. Sofia. 2014. Helicobacter pylori antimicrobial resistance rates in the central region of Portugal. Clin. Microbiol. Infect. 20: 1127–1133. [CrossRef]
- Ho, J. J. C., M. Navarro, K. Sawyer, Y. Elfanagely, and S. F. Moss. 2022. Helicobacter pylori Antibiotic Resistance in the United States Between 2011 and 2021: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 117: 1221-1230. [CrossRef]
- Mosites, E., D. Bruden, J. Morris, A. Reasonover, K. Rudolph, D. Hurlburt, T. Hennessy, B. McMahon, and M. Bruce. 2018. Antimicrobial resistance among Helicobacter pylori isolates in Alaska, 2000–2016. J. Glob. Antimicrob. Resist. 15: 148–153. [CrossRef]
- Harris, R. B., H. E. Brown, R. L. Begay, P. R. Sanderson, C. Chief, F. P. Monroy, and E. Oren. 2022. Helicobacter pylori Prevalence and Risk Factors in Three Rural Indigenous Communities of Northern Arizona. Int. J. Environ. Res. Public Heal. 2022, Vol. 19, Page 797. 19: 797. [CrossRef]
- Monroy, F. P., H. E. Brown, P. R. Sanderson, G. Jarrin, M. Mbegbu, S. Kyman, and R. B. Harris. 2022. Helicobacter pylori in Native Americans in Northern Arizona. Diseases. (Basel, Switzerland). 10: 19. [CrossRef]
- Abadi, A. T. B., A. M. Mobarez, M. J. M. Bonten, J. A. Wagenaar, and J. G. Kusters. 2014. Clinical relevance of the cagA, tnpA and tnpB genes in Helicobacter pylori. BMC Gastroenterol. 14: 1–5. [CrossRef]
- Sun, L., S. Talarico, L. Yao, L. He, S. Self, Y. You, H. Zhang, Y. Zhang, Y. Guo, G. Liu, N. R. Salama, and J. Zhang. 2018. Droplet Digital PCR-Based Detection of Clarithromycin Resistance in Helicobacter pylori Isolates Reveals Frequent Heteroresistance. J. Clin. Microbiol. 56. [CrossRef]
- Jeong, J. Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatino, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Y. Wong, Shiu Kum Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H. K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182: 5082–5090. [CrossRef]
- Kim, K. S., J. O. Kang, C. S. Eun, D. S. Han, and T. Y. Choi. 2002. Mutations in the 23S rRNA gene of Helicobacter pylori associated with clarithromycin resistance. J. Korean Med. Sci. 17: 599. [CrossRef]
- Kim, J. M., J. S. Kim, N. Kim, Y.-J. Kim, I. Y. Kim, Y. J. Chee, C.-H. Lee, and H. C. Jung. 2008. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J. Microbiol. Biotechnol. 18: 1584–1589. PMID: 18852516.
- Khan, R., S. Nahar, J. Sultana, M. M. Ahmad, and M. Rahman. 2004. T2182C mutation in 23S rRNA is associated with clarithromycin resistance in Helicobacter pylori isolates obtained in Bangladesh. Antimicrob. Agents Chemother. 48: 3567–3569. [CrossRef]
- Park, C. G., S. Kim, E. J. Lee, H. S. Jeon, and S. Han. 2018. Clinical relevance of point mutations in the 23S rRNA gene in Helicobacter pylori eradication: A prospective, observational study. Medicine (Baltimore). 97. [CrossRef]
- Mannion, A., J. A. Dzink-Fox, Z. Shen, M. Blanca Piazuelo, K. T. Wilson, P. Correa, R. M. Peek, M. Constanza Camargo, and J. G. Fox. 2021. Helicobacter pylori Antimicrobial Resistance and Gene Variants in High- and Low-Gastric-Cancer-Risk Populations. J Clin. Microbiol. 59. [CrossRef]
- Jung, D. H., J. H. Kim, S. J. Jeong, S. Y. Park, I. M. Kang, K. H. Lee, and Y. G. Song. 2018. Peptide Nucleic Acid Probe-Based Analysis as a New Detection Method for Clarithromycin Resistance in Helicobacter pylori. Gut Liver. 12: 641. [CrossRef]
- Agudo, S., G. Pérez-Pérez, T. Alarcón, and M. López-Brea. 2010. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J. Clin. Microbiol. 48: 3703–3707. [CrossRef]
- Hashemi, S. J., A. F. Sheikh, H. Goodarzi, M. J. Yadyad, S. S. Seyedian, S. Aslani, and M. A. Assarzadegan. 2019. Genetic basis for metronidazole and clarithromycin resistance in Helicobacter pylori strains isolated from patients with gastroduodenal disorders. Infect. Drug Resist. 12: 535–543. [CrossRef]
- Burucoa, C., C. Landron, M. Garnier, J. L. Fauchère, R. Khan, and M. Rahman. 2005. T2182C mutation is not associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 49: 868-870. [CrossRef]
- Hulten, K. G., R. M. Genta, I. N. Kalfus, Y. Zhou, H. Zhang, and D. Y. Graham. 2021. Comparison of Culture with Antibiogram to Next-Generation Sequencing Using Bacterial Isolates and Formalin-Fixed, Paraffin-Embedded Gastric Biopsies. Gastroenterology. 161: 1433-1442.e2. [CrossRef]
- Gonzalez-Hormazabal, P., M. Musleh, S. Escandar, H. Valladares, E. Lanzarini, V. G. Castro, L. Jara, and Z. Berger. 2018. Prevalence of clarithromycin resistance in Helicobacter pylori in Santiago, Chile, estimated by real-time PCR directly from gastric mucosa. BMC Gastroenterol. 18. [CrossRef]
- Camorlinga-Ponce, M., A. Gómez-Delgado, E. Aguilar-Zamora, R. C. Torres, S. Giono-Cerezo, A. Escobar-Ogaz, and J. Torres. 2021. Phenotypic and Genotypic Antibiotic Resistance Patterns in Helicobacter pylori Strains from Ethnically Diverse Population in México. Front. Cell. Infect. Microbiol. 10. [CrossRef]
- Osato, M. S., R. Reddy, S. G. Reddy, R. L. Penland, and D. Y. Graham. 2001. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int. J. Antimicrob. Agents. 17: 39–44. [CrossRef]
- Weiss, J., T.-K. Tsang, X. Meng, H. Zhang, E. Kilner, E. Wang, and W. Watkin. 2008. Detection of Helicobacter pylori Gastritis by PCR Correlation with Inflammation Scores and Immunohistochemical and CLOtest Findings. Am J Clin Pathol. 129: 89–96. [CrossRef]
- Gehlot, V., S. Mahant, A. K. Mukhopadhyay, K. Das, J. Alam, P. Ghosh, and R. Das. 2016. Low prevalence of clarithromycin-resistant Helicobacter pylori isolates with A2143G point mutation in the 23S rRNA gene in North India. J. Glob. Antimicrob. Resist. 6: 39–43. [CrossRef]
- Nestegard, O., B. Moayeri, F. A. Halvorsen, T. Tønnesen, S. W. Sørbye, E. Paulssen, K. M. Johnsen, R. Goll, J. R. Florholmen, and K. K. Melby. 2022. Helicobacter pylori resistance to antibiotics before and after treatment: Incidence of eradication failure. PLoS One. 17: e0265322. [CrossRef]
- Graham, D. Y., and S. F. Moss. 2022. Antimicrobial Susceptibility Testing for Helicobacter pylori Is Now Widely Available: When, How, Why. Am. J. Gastroenterol. 117: 524–528. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).