1. Introduction
In humans, a regular physical activity is considered one of the most valid tools able to reduce the risk of premature mortality and onset of many diseases, such as obesity, diabetes, cancer, metabolic and cardiovascular diseases [
1,
2]. Some of these diseases have been associated with DNA damage, including aberrant DNA methylation patterns, and telomere shortening [
2,
3]. Epidemiological studies have shown that subjects who exercise regularly have a lower risk of all-cause mortality with respect to sedentary subjects [
1,
4], with beneficial effects that are more pronounced in elderly people [
5]. Compared to inactive individuals, physically active adults exhibit improved cardiorespiratory fitness and muscle strength, cognitive function and favorable metabolic profile, as well as healthier body mass and composition [
1,
3].
At the cellular level, physical activity is able to determine nuclear changes, influencing gene expression and, in particular, inducing epigenetic modifications in terms of altered DNA methylation in muscle cells [
6]. These epigenetic changes induced by the physical activity were also found to have beneficial effects in cancer patients, inducing the increase of the tumor suppressor genes expression and decreased expression of oncogenes [
7]. In particular, it was observed that cancer cells exhibit an abnormal DNA methylation pattern, such as hypermethylation of tumor suppressor genes promoters and hypomethylation in promoter regions of oncogenes [
1]. Vice versa, physical activity seems to be able to reduce the consequences of this process and to determine increased levels of tumor suppressor genes expression and the inhibition of oncogenesis processes [
7]. Moreover, physical exercise acts as an inhibitor of the aging process, through the activation of telomerase, the preservation of telomere length [
2,
6] and the improvement of mitochondrial biogenesis and function [
8].
However, it is known that intense physical activity, combined with some lifestyles (e.g. smoking habits, alcohol intake, incorrect diet) or environmental factors (e.g. exposure to radiation, viruses and bacteria) is able to induce high concentration of free radicals, determining inflammation, increased levels of oxidative damage to cells and tissues and genomic damage [
9,
10]. On the contrary, regular but moderate exercise has been associated to lower levels of free radicals, as result of a favorable adaptive response by the organism, resulting in beneficial effects in relation at the onset and progression of various pathologies associated with oxidative stress [
11]. In particular, researches conducted on both humans and rodents, have shown that low levels of oxidative stress are able to promote adaptation and to activate cellular mechanisms of protection [
11,
12].
Based on these assumptions, we decided to evaluate, in a sample of amateur athletes practicing different disciplines, the level of genomic damage by means of the buccal micronuclei (MNi) assay, comparing obtained data with those from subjects who practice sports only occasionally and subjects who do not practice any physical activity. The aims of the present study was to evaluate whether physical activity affects background levels of genomic damage and whether the different sports disciplines differentially affect these levels.
MNi analysis in exfoliated buccal cells is a useful and non-invasive method to monitor genomic damage in humans, as consequence, for example, of an exposure to genotoxins, radiation, chemicals and environmental xenobiotics [
13]. MNi derive from whole chromosomes or chromosome fragments, which, during mitotic telophase, are not included in the daughter nuclei because they do not attach properly to the spindle during the segregation process in anaphase [
14]. These chromosomes or chromosome fragments are than enclosed by a nuclear membrane and, except for their smallest size, are morphologically similar to nuclei [
13].
Another nuclear abnormality observable with MNi test, known as Nuclear Bud (NBUD), has been associated with chromosomal instability events. Nuclear buds are the result of an abnormal gene amplification process, and are characterized by a similar morphology as MNi, with the exception that they are connected to the nucleus by a narrow or broad stem of nucleoplasmic material. The MNi assay also allows to record the presence of binucleated cells, the excess of which may represent the result of an imperfect cytodieresis mechanism [
14,
15,
16].
Finally, it is known that some gene polymorphisms are able to modulate the level of the genomic damage [
17,
18]. Therefore, we also decided to evaluate the association of some polymorphisms of genes, encoding enzymes belonging to the different damage repair systems and metabolic genes, with the MNi frequency observed in the studied samples. In particular, we analyzed the following polymorphisms of phases I and II metabolic genes and damage repair genes, that are the most studied gene polymorphisms associated to genomic damage [
17]: Cytochrome P450 1A1 (
CYP1A1) exon 7 (A>G, rs1048943), the Glutathione S-transferases
GSTT1(positive>null, rs1601993659),
GSTM1(positive>null, rs1183423000),
XRCC1 194 (C>T, rs1799782) and
XPC exon 15 (A>C, rs2228001).
2. Materials and Methods
2.1. Population sample
The study population included 125 athletes (belonging to the following disciplines: Martial Arts n=35, Basket n= 29, Volley n=40 and Sprint n=22) and 81 control subjects (50 sedentary controls and 31 sports controls, i.e. subjects who practice sports occasionally, and no more than two times a week).
It is well known that cigarette smoke, alcohol consumption, drugs and X-rays could alter level of genomic damage [
19,
20,
21,
22]. For this reason, we excluded from the sample smokers and individuals who reported alcohol consumption, treatment for acute infections and/or chronic non-infectious diseases, history of cancer and exposure to diagnostic X-rays, for at least one year prior the analysis.
Subjects received detailed information about aims and experimental procedures of the study and gave their informed consent. Selected volunteers were anonymously identified by a numeric code. The study was approved by the University of Turin ethics committee (protocol number 0609375, 10-28-2021) and was performed in agreement with the ethical standards laid down in the 2013 Declaration of Helsinki.
2.2. Buccal MNi assay
Buccal MNi assay was performed according to the following protocol: exfoliated buccal mucosa cells were collected, with a toothbrush, by gently scraping the mucosa of the inner lining of one or both cheeks and/or the inner side of the lower lip and palate. The toothbrush tip was, than, immersed in a fixative solution consisting of methanol/acetic acid 3:1, shaken for at least 1 min and stored at 4 °C before the analysis. Successively, cells were gathered by centrifugation, the supernatant was discarded, and the pellet was dissolved in a minimal amount of fixative, which was seeded on the slides to detect MNi by conventional staining with 5% Giemsa (pH 6.8) prepared in Sörensen buffer. Microscopic analysis was performed at 1000X magnification on a light microscope. According to the established criteria, MNi, NBUDs, and binucleated cells were scored in 1,000 cells with well-preserved cytoplasm per subject [
16].
2.3. DNA extraction and Genotyping
In order to extract DNA, a second toothbrush was used to collect exfoliated buccal mucosa cells by scraping the mucosa of the inner lining of one or both cheeks. DNA extraction was performed by using a salting-out procedure: samples were centrifuged at 14,000 rpm for 1 min and the pellet, constituted by buccal mucosa cells, was resuspended in a solution containing 340 µL lysis buffer (10 mM Tris pH 7.6, 10 mM EDTA and 50 mM NaCl), 30 µL of SDS 10% and 30 µL of proteinase K. After incubation at 55 °C for 1 h, 200 µL of saturated sodium acetate was added to the solution. The samples were vigorously shaken and centrifuged at 14,000 rpm for 5 min. Subsequently, 0.6 mL of isopropanol for DNA precipitation were added to the supernatant and, after centrifugation at 14,000 rpm for 1 min, 0.5 mL of 70% ethanol was added to remove salt from the pellet. After subsequent centrifugation, the pellet was dried at room temperature for at least 60 min and the pellet was resuspended in 50 µL of ultrapure distilled water.
PCR-based genotyping was performed for the following gene polymorphisms: CYP1A1 (rs1048943, A>G), GSTM1 (rs 1183423000, presence/absence), GSTT1 (rs1601993659, presence/absence), XRCC1 (rs1799782, C>T), and XPC (rs2228001, A>C). Primer sequences, annealing temperatures, PCR methodologies and expected PCR product sizes are reported in Supplementary Material 1.
PCR reactions were performed in a 25 μL volume containing about 10 ng DNA (template), with a final concentration of 1X Reaction Buffer, 1.5 mM of MgCl2, 5% of DMSO, 250 µM of dNTPs, 0.5 μM of each primer, and 1 U/sample of Taq DNA polymerase (Fischer, U.S.). Cycles were set as follows: 35 cycles, 1 min at 95°C, 1 min at 60-65 °C, 1 min at 72°C, and a final extension step 10 min at 72 °C. Amplification products were detected by ethidium bromide staining after 2.5% agarose gel electrophoresis.
2.4. Statistical Analysis
All statistical analyses were performed using the SPSS software statistical program (version 28.0, SPSS Inc., Chicago, USA). Counts of micronuclei and other abnormalities are presented as the mean frequency (± standard deviation) in a sample of 1,000 cells/subject. For data not normally distributed, we used the Mann-Whitney test to compare two sample groups whereas, for data normally distributed, the comparison between groups was performed by ANOVA. The correlation between gene polymorphisms and the genomic damage markers was performed using the Sperman’ Rho correlation. All P-values were two-tailed, and the level of statistical significance was set at P<0.05 for all tests.
3. Results
Results of the Shapiro-Wilk test showed that all the analyzed categories, with the exception of the weight (P=0.388), were not normally distributed (P<0.001).
The demographic features of the studied samples are reported in
Table 1. A total of 206 subjects were recruited, 148 males (mean age±S.D. 21.858±3.720; mean weight±S.D. 74.418±10.081) and 58 females (mean age±S.D. 21.052±2.781; mean weight±S.D. 60.716±9.942). As expected, significant differences were found between sexes in terms of means weight (
P<0.001, Mann-Whitney test).
The athlete sample was represented by 125 subjects (mean age ± S.D. 21.528±4.063; mean weight ± S.D. 72.654±12.103), 89 males (mean age ± S.D. 21.697±4.342; mean weight ± S.D. 76.526±10.496) and 36 females (mean age ± S.D. 21.111±3.293; mean weight ± S.D. 63.083±10.470). Among athletes, 35 were martial arts athletes (mean age ± S.D. 22.500±4.460; mean weight ± S.D. 70.941±9.739); 29 were basketball players (mean age ± S.D. 20.207±3.668; mean weight ±S.D. 81.652±9.992); 40 were volleyball players (mean age ± S.D. 21.300±4.201; mean weight ± S.D. 72.223±12.722) and 22 were athletic sprinters (mean age ± S.D. 22.182±3.319; mean weight ± S.D. 64.227±9.532).The basketball athletes showed a significantly higher weight with respect to martial art and sprinter athletes (P<0.001 for both) as well as to volleyball athletes (P = 0.002).
The control sample was represented by 81 subjects (mean age ± S.D. 21.790±2.375; mean weight ± S.D. 67.327±10.512), 59 males (mean age ± S.D. 22.102±2.524; mean weight ± S.D. 71.237±8.563) and 22 females (mean age ± S.D. 20.955±1.704; mean weight ± S.D. 56.841±7.763).
A total of 206,000 buccal cells were observed.
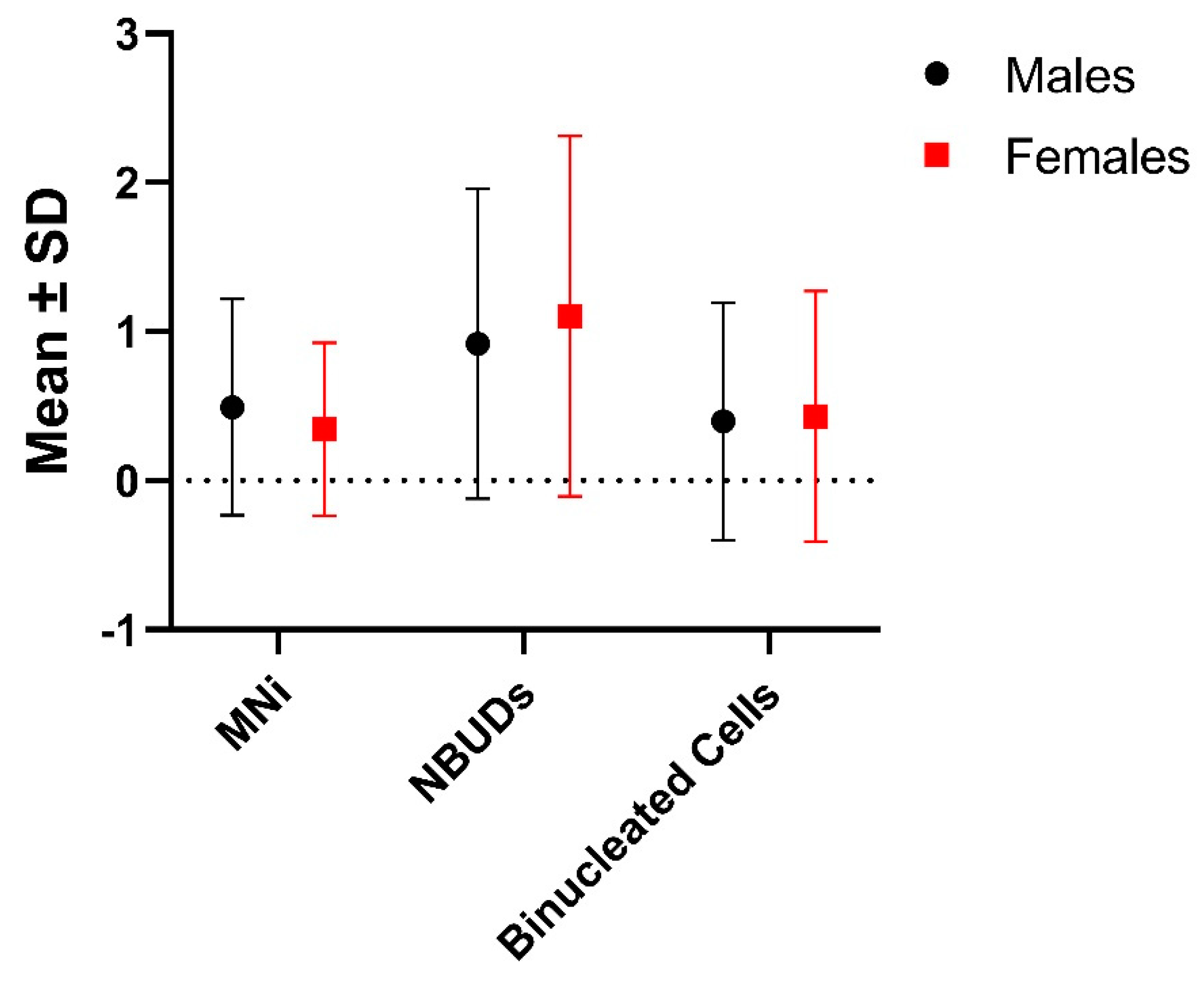
No significant differences were found between sexes in the frequency of MNi, NBUDs and other nuclear aberrations, as well as in terms of binucleated cells (
Table 2,
Figure 1).
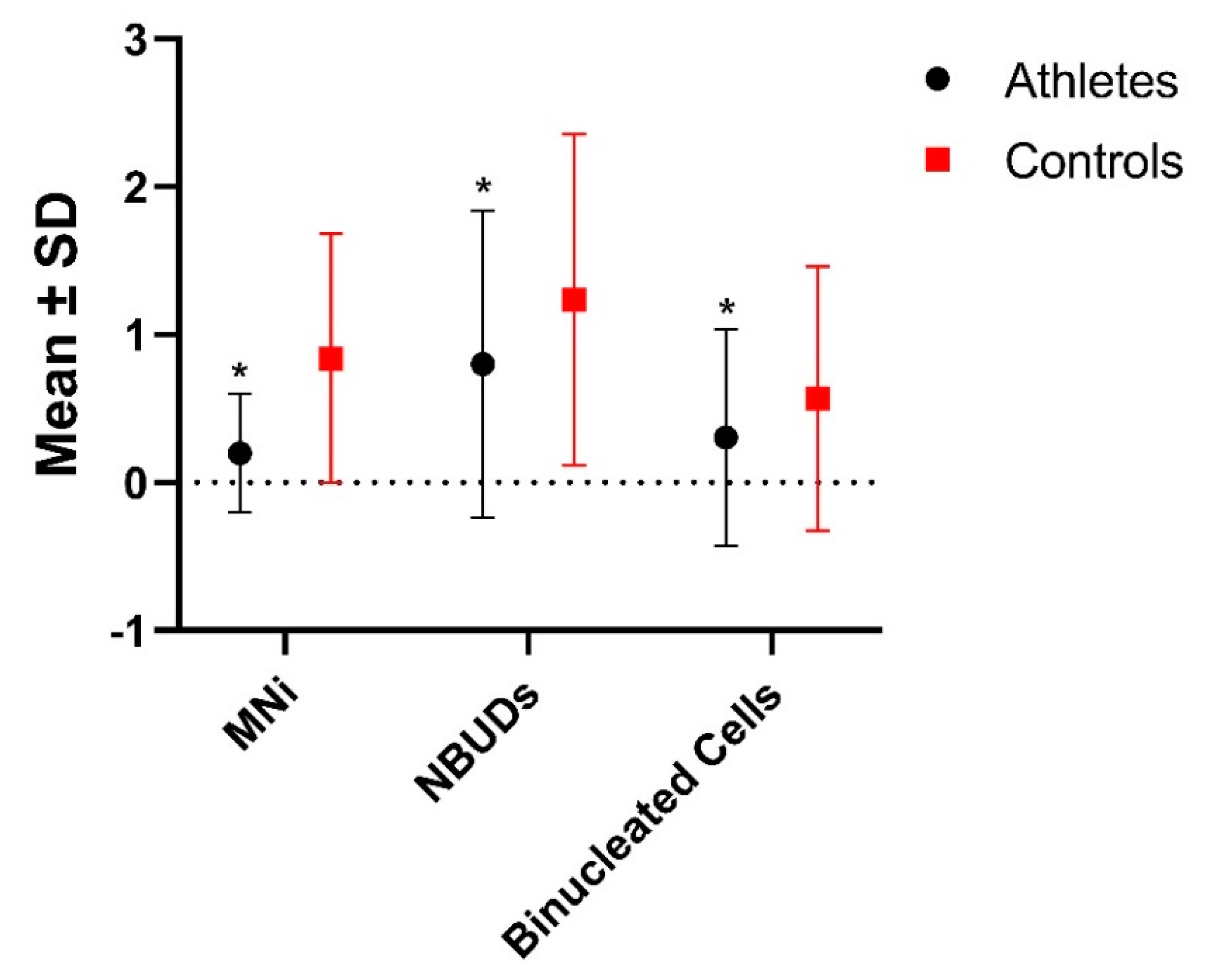
In
Table 3 and
Figure 2, results about the statistic evaluation of the differences in the level of genomic and nuclear damage are showed. In
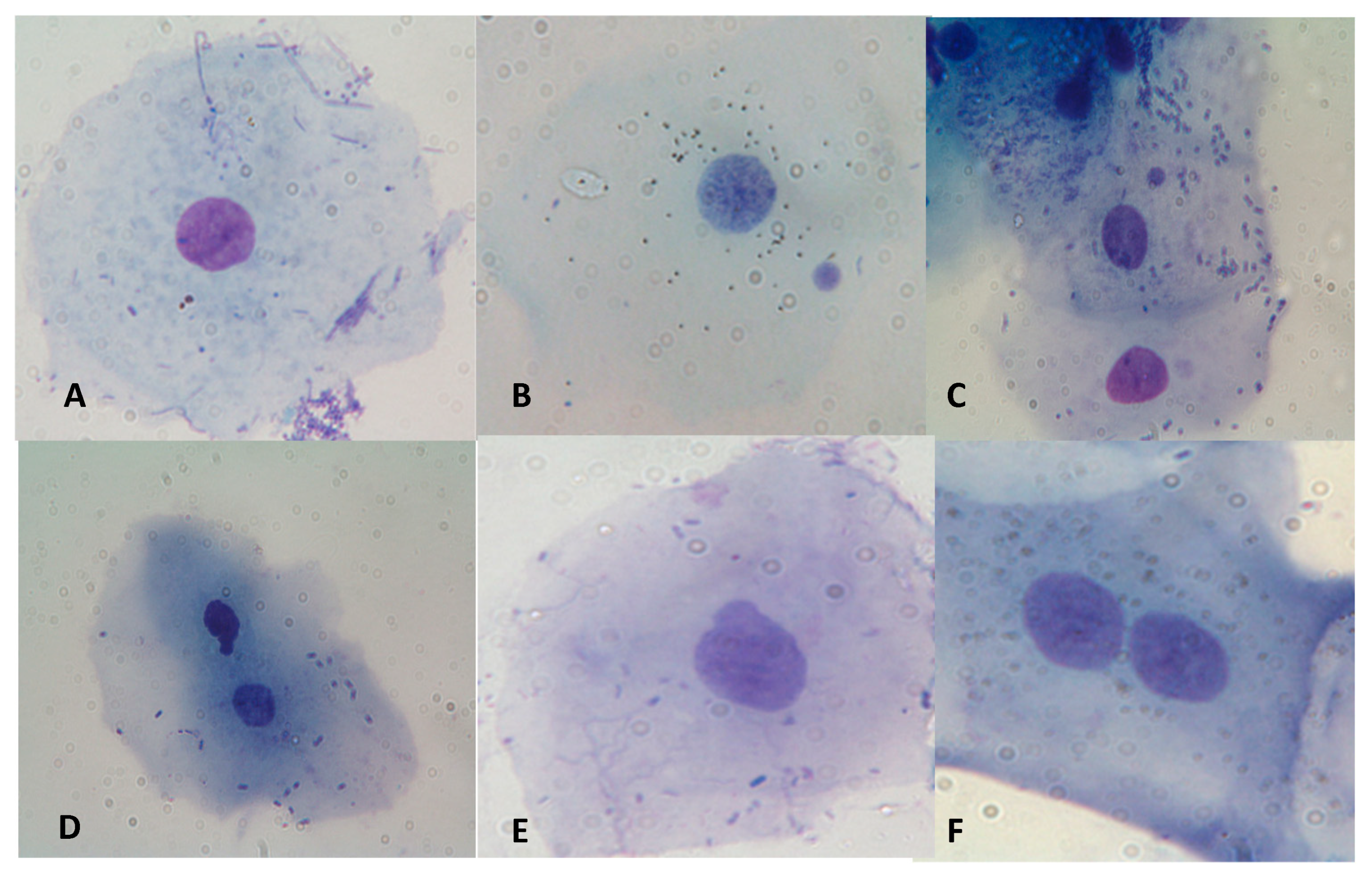
Figure 3 some examples of cells with MNi, NBUDs, and Binucleated cell are showed. Athletes showed significantly lower values of MNi (P<0.001), NBUDs (P = 0.002) and Binucleated Cells (P<0.001) (
Table 3) with respect to control subjects.
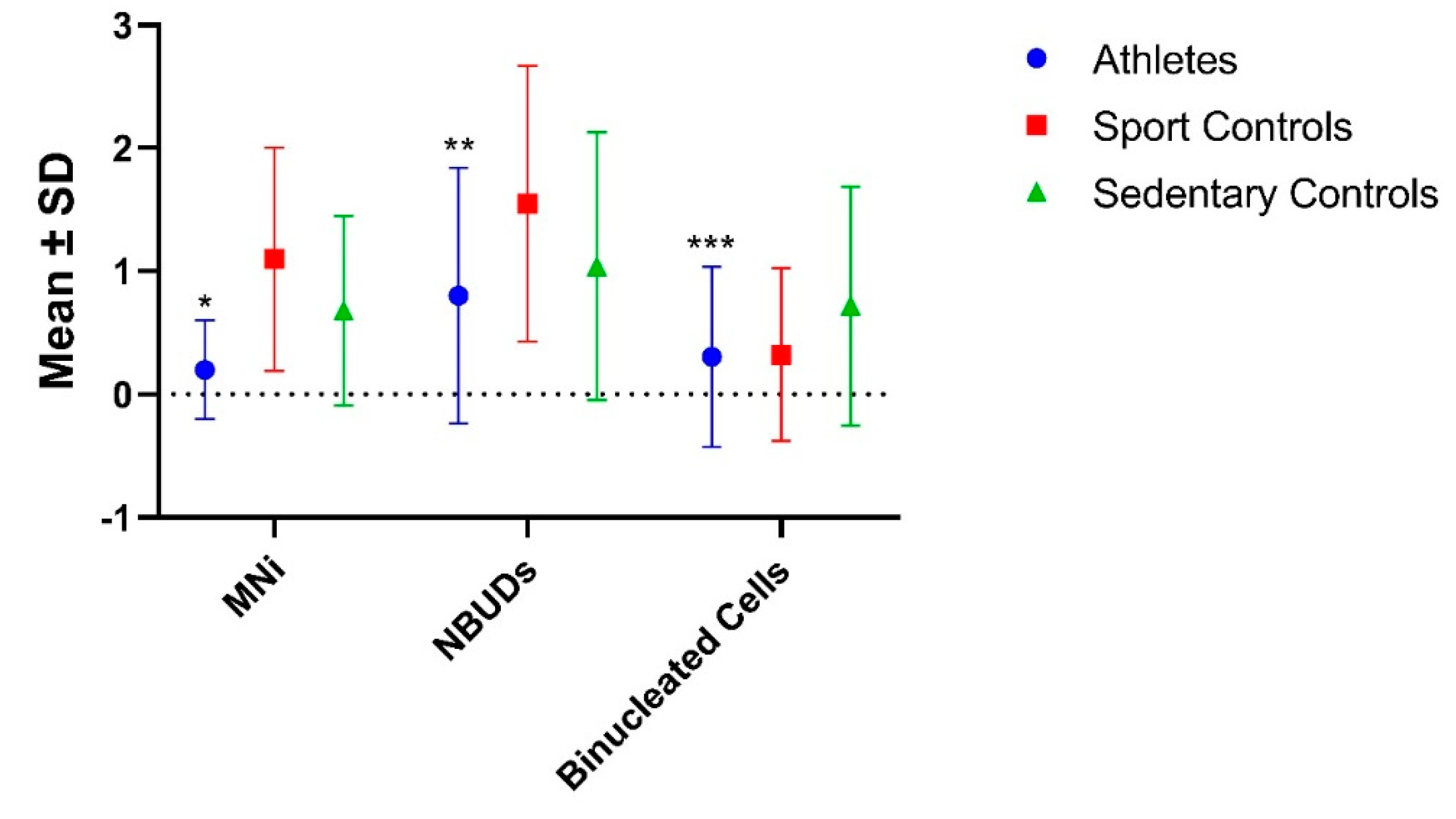
In order to evaluate the existence of possible differences within the control group, the latter was subdivide into "Sedentary Controls", which include subjects who do not practice any sport activity, and "Sport Controls", a category which include subjects who only practice sport occasionally (
Table 4 and
Figure 4).With regard to genomic and nuclear damage, the group of athletes showed significantly lower MNi than both the Sports and Sedentary Controls (P<0.001 for both), whereas for NBUDs, the significance was observed only with respect to the Sport Controls (P<0.001). No significant differences were found in terms of Binucleated cells. Interestingly, the group of Sportive Controls showed significantly higher values of MNi and NBUDs with respect to Sedentary Controls (P = 0.040 and P<0.001 for MNi and NBUDs, respectively).
In
Table 5 the group of athletes was subdivided on the basis of the different sport activity and the differences in the frequencies of MNi, NBUDs and Binucleated Cells were statistically evaluated. Sprinters and Martial Arts athletes showed a significantly higher frequency of MNi (P = 0.010) and binucleated cells (P<0.001), respectively, with respect to the other athletes categories, whereas the volleyball athletes showed a significantly higher NBUDs frequency with respect to the Basketball athletes (P = 0.030).
Finally, in order the analyse the possible influence of some metabolic and DNA-repair genes on the level of genomic damage, a correlation analyses between some phase I, phase II and DNA-repair genes and the analysed genomic damage markers was performed. No significant differences were found between the analysed gene polymorphisms and the genome damage markers.
4. Discussion
It is known that intense physical activity is associated with the production of high levels of free radicals, which can deplete the non-enzymatic antioxidant system, inducing impaired cellular function, apoptosis, necrosis and genomic damage [
10,
23].
On the other hand, the production of free radicals induced by moderate and constant physical exercise is considered one of the most powerful natural stimuli capable of improving the expression of antioxidant enzymes. In fact, trained subjects showed a higher number of mitochondria, with consequent lower levels of respiratory activity, oxidative stress, and chronic inflammation than untrained subjects [
4]. In general, moderate and regular physical activity was found to preserve genomic integrity and tissue function and to reduce the occurrence of age-related chronic diseases [
3].
In the present study, we evaluated the possible influence of moderate and constant physical activity on the levels of genomic damage, also comparing the frequencies of analyzed genomic markers between the different groups of athletes.
Athletes showed a significantly lower frequency of MNi than controls (
Table 3), demonstrating that regular physical exercise is able to reduce the level of genomic damage. In fact, it is known the existence of an adaptive response induced by ROS following regular long-term training, results in an over-expression of antioxidant enzymatic systems, such as superoxide dismutase, catalase and glutathione peroxidase. These endogen antioxidant enzymes act synergistically with non-enzymatic antioxidants, i.e. vitamins C and E, to counterbalance the negative effects of oxidative stress induced by physical exercise [
24].
However, it is known that the duration and intensity of physical exercise also appear to differently influence the level of oxidative stress [
25]. In particular, it has been observed that, while moderate and regular physical exercise induces the upregulation of antioxidant and oxidative damage repair systems [
24], acute and vigorous physical activity triggers a massive production of free radicals, with depletion in antioxidant defenses and increase in oxidative damage to proteins and DNA [
26,
27,
28].
These findings are congruent with our results, in fact when we compared the group of total athletes with both the sedentary and sport controls, we observed that the group of sport controls, i.e. subjects who perform sport activities occasionally and often intensely, showed significantly higher MNi values than both sedentary controls and athlete groups (
Table 3). It could means that, with respect to subject who practice sport in a regular and moderate manner, as in the case of amateur athletes, subjects who practice sport occasionally could represent subjects more prone to genomic damage and, consequently, with an increased risk for some diseases.
It should be noted that the group of sedentary controls did not show significant differences, in terms of frequencies of MNi and NBUDs, compared to the group of athletes (
Table 4). This found could also be explained with the fact that the number of "background" MNi observable in unexposed young subjects is very low, falling within a range from 0 to 12 MNi observed for 1000 analysed cells [
29] and, therefore, it is unlikely that significant differences will emerge in a comparative analysis.
The next step of the present work was to separate the group of athletes on the basis of the different disciplines (
Table 5). Sprinters, subjects practicing an anaerobic activity, showed a significantly higher frequency of MNi with respect to the other athletes. Although it is known that aerobic and resistance training was considered the leading cause of oxidative stress, it has been observed that free radicals can also be produced through other pathways, which are not necessarily related to oxygen demand. In fact, several studies have shown that even anaerobic exercise (high intensity training) can produce similar levels of oxidative stress, as can aerobic physical activity [
26].
Contrary to what has been observed in some published papers [
29,
30], sex was not found to influence the frequency of genomic damage (
Table 2).
Finally, it is known in literature that
GSTM1 and
GSTT1 gene polymorphisms can affect the levels of exercise-induced oxidative stress [
31,
32], and that some polymorphisms of damage repair genes can code for enzymes that show greater or lesser efficiency in repairing DNA damage and, in this sense, can affect the level of MNi [
17]. In our sample, the gene polymorphisms analyzed do not seem to influence the frequencies of MNi and NBUDs, further confirming that the observed differential genomic damage is probably due to the intensity and type of the sport activity. However, in interpreting these data, it is necessary to consider the fact that the response to the sport activity is multifactorial, being influenced by various factors including physiology, metabolism and genetics. From this point of view, it is unlikely that the genetic component can be explained by a variation in the DNA sequence affecting a few genes, while it is more probable that several gene loci, each with a small but significant contribution, could be responsible for this component [
24].
5. Conclusions
With the present study, we showed, in a sample of amateur athletes, that regular physical exercise is able to reduce the level of genomic damage measured in terms of MNi and NBUDs frequencies. This result may probably be attributable to an over-regulation of the endogen antioxidant systems induced by moderate sport practice, as widely reported in the literature [
11,
12].
Another significant result we obtained is that sport controls showed significantly higher MNi values than both the sedentary controls and athletes, demonstrating the fact that sport activity practiced occasionally may result in an increase of the genomic damage.
Finally, a limitation of the present study lies in the fact that some categories of athletes are under-represented, in particular that of endurance athletes. However, we would like to point out that we sampled subjects "clean" of any confounding factors that may affect the levels of genomic damagte, such as smoking, alcohol and radiation, characteristics that make subjects not easy to find.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Supplementary Material 1 – Gene Polymorphisms, Reference Sequence (rs) numbers, Primers, annealing temperatures and genotypig methodologies used.
Author Contributions
Conceptualization, A.S.; methodology, A.S.; laboratory procedure, A.S., A.N., A.A.N., M.S., Am.Sel.; Microscope cell observation: A.S., A.N., A.A.N., M.S., Am. Sel.; Genotyping: A.S., A.N., A.A.N., M.S., Am.Sel.; software analysis: A.S., A.N.; writing—original draft preparation, A.S.; writing—review and editing, A.S. and A.N.; project administration, A.S.. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by Italian Minister of University and Public Education (grant “ex 60%”).
Institutional Review Board Statement
The study was approved by the University of Turin ethics committee (protocol number 0609375, 10-28-2021).
Informed Consent Statement
All study participants gave their informed consent.
Data Availability Statement
The analytical data will be made available to interested parties upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grazioli, E.; Dimauro, I.; Mercatelli, N.; Wang, G.; Pitsiladis, Y.; Di Luigi, L.; Caporossi, D. Physical activity in the prevention of human diseases: role of epigenetic modifications. BMC Genomics 2017, 18, 111–131. [Google Scholar] [CrossRef] [PubMed]
- Sellami, M.; Bragazzi, N.; Prince, MS.; Denham, J.; Elrayess, M. Regular, Intense Exercise Training as a Healthy Aging Lifestyle Strategy: Preventing DNA Damage, Telomere Shortening and Adverse DNA Methylation Changes Over a Lifetime. Frontiers in Genetics 2021, 12, 652497. [Google Scholar] [CrossRef]
- Semeraro, M.D.; Smith, C.; Kaiser, M.; Levinger, I.; Duque, G.; Gruber, H.J.; Herrmann, M. . Physical activity, a modulator of aging through effects on telomere biology. Aging 2020, 12, 13803–13823. [Google Scholar] [CrossRef] [PubMed]
- Powers, S. K.; Deminice, R.; Ozdemir, M.; Yoshihara, T. Exercise-induced oxidative stress: Friend or foe? J. Sport Health Sci. 2020, 9, 415–425. [Google Scholar] [CrossRef]
- Rebelo-Marques, A.; De Sousa Lages, A.; Andrade, R.; Ribeiro, C.F.; Mota-Pinto, A.; Carrilho, F.; Espregueira-Mendes, J. The Benefits of Physical Exercise. Front Endocrinol (Lausanne). Aging Hallmarks 2018, 9, 258. [Google Scholar]
- Rezapour, S.; Shiravand, M.; Mardani, M. Epigenetic changes due to physical activity. Biotechnol. Appl. Biochem. 2018, 65, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.F.; Cardarelli, R.; Caroll, J.; Zhang, S.; Fulda, K.G.; Gonzalez, K.; Vishwanatha, J.K.; Morabia, A.; Santella, R.M. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics 2011, 6, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Larsen, F.J.; Schiffer, T.A.; Ørtenblad, N.; Zinner, C.; Morales-Alamo, D.; Willis, S.J.; Calbet, J.A.; Holmberg, H.C.; Boushel, R. High-intensity sprint training inhibits mitochondrial respiration through aconitase inactivation. FASEB Journal 2016, 30, 417–427. [Google Scholar] [CrossRef]
- Nikolaidis, M.G.; Margaritelis, N.V.; Matsakas, A. Quantitative Redox Biology of Exercise. Int. J sports med 2020, 41, 633–645. [Google Scholar] [CrossRef]
- Kruk, J.; Aboul, E.B.H.; Duchnik, E. Exercise-induced oxidative stress and melatonin supplementation: current evidence. J. Physiol. Sci. 2021, 71, 27. [Google Scholar] [CrossRef]
- Brancaccio, M.; Menniti, C.; Cesaro, A.; Fimiani, F.; Moscarella, E.; Caiazza, M.; Gragnano, F.; Ranieri, A.; D’Alicandro, G.; Tinto, N.; Mazzacara, C.; Lombardo, B.; Pero, R.; Limongelli, G.; Frisso, G.; Calabrò, P.; Scudiero, O. Dietary Thiols: A Potential Supporting Strategy against Oxidative Stress in Heart Failure and Muscular Damage during Sports Activity. Int. J. Environ Res Pub. Health 2020, 17, 9424. [Google Scholar] [CrossRef] [PubMed]
- Hollander, J.; Fiebig, R.; Gore, M.; Ookawara, T.; Ohno, H.; Ji, L.L. Superoxide dismutase gene expression is activated by a single bout of exercise in rat skeletal muscle. Eur. J. Physiol. 2001, 442, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Harvey, S.; Gruner, T.; Fenech, M. The buccal cytome and micronucleus frequency is substantially altered in Down's syndrome and normal ageing compared to young healthy controls. Mutat. Res. 2008, 638, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Kirsch-Volders, M.; Natarajan, A.T.; Surralles, J.; Crott, J.W.; Parry, J.; Norppa, H.; Eastmond, D.A.; Tucker, J.D.; Thomas, P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagen 2011, 26, 125–132. [Google Scholar] [CrossRef]
- Bolognesi, C.; Knasmueller, S.; Nersesyan, A.; Thomas, P.; Fenech, M. The HUMNxl scoring criteria for different cell types and nuclear anomalies in the buccal micronucleus cytome assay –An update and expanded photogallery. Mutat. Res. Rev. Mutat. Res. 2013, 753, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Holland, N.; Bolognesi, C.; Kirsch-Volders, M.; Bonassi, S.; Zeiger, E.; Knasmueller, S.; Fenech, M. Buccal micronucleus cytome assay. Nat. Prot. 2009, 4, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Ghelli, F.; Cocchi, E.; Buglisi, M.; Squillacioti, G.; Bellisario, V.; Bono, R.; Santovito, A. The role of phase I, phase II, and DNA-repair gene polymorphisms in the damage induced by formaldehyde in pathologists. Scient. Rep. 2021, 11, 10507. [Google Scholar] [CrossRef]
- Santovito, A.; Delsoglio, M.; Manitta, E.; Picco, G.; Meschiati, G.; Chiarizio, M.; Gendusa, C.; Cervella, P. Association of GSTT1 null, XPD 751 CC and XPC 939 CC genotypes with increased levels of genomic damage among hospital pathologists. Biomarkers 2017, 22, 557–565. [Google Scholar] [CrossRef]
- Santovito, A.; Cervella, P.; Delpero, M. Chromosomal damage in peripheral blood lymphocytes from nurses occupationally exposed to chemicals. Hum Exp Toxicol. 2014, 33, 897–903. [Google Scholar] [CrossRef]
- Santovito, A.; Cervella, P.; Delpero, M. Increased frequency of chromosomal aberrations and sister chromatid exchanges in peripheral lymphocytes of radiology technicians chronically exposed to low levels of ionizing radiations. Environ. Toxicol. Pharmacol. 2014, 37, 396–403. [Google Scholar] [CrossRef]
- Santovito, A.; Caravella, P.; Delpero, M. Evaluation of Genomic Damage in Peripheral Lymphocytes from Occupationally Exposed Anesthetists: Assessment of the Effects of Age, Sex, and GSTT1 Gene Polymorphism. J. Biochem. Mol. Toxicol. 2015, 29, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Santovito, A.; Cervella, P.; Delpero, M. . Evidence of genotoxicity in lymphocytes of non-smoking alcoholics. Mol. Biol. Rep. 2015, 42, 53–9. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, M.A.; Filaire, E.; McCall, A.; Fabre, C. Radical Oxygen Species, Exercise and Aging: An Update. Sports Med. 2015, 45, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Vecchio, M.; Currò, M.; Trimarchi, F.; Naccari, S.; Caccamo, D.; Riccardo, L.; Barreca, D.; Di Mauro, D. The Oxidative Stress Response in Elite Water Polo Players: Effects of Genetic Background. BioMed Res. Int. 2017, 7019694. [Google Scholar] [CrossRef]
- Bloomer, R.J. Effect of exercise on oxidative stress biomarkers. Adv. Clin. Chem 2008, 46, 1–50. [Google Scholar]
- Bloomer, R. J.; Goldfarb, A.H. Anaerobic exercise and oxidative stress: a review. Can. J. Appl. Physiol. 2004, 29, 245–263. [Google Scholar] [CrossRef]
- Peake, J.; Suzuki, K. Neutrophil activation, antioxidant supplements and exercise-induced oxidative stress. Exerc. Immunol. Rev. 2004, 10, 129–141. [Google Scholar]
- Koyama, K. Exercise-induced oxidative stress: A tool for “hormesis” and “adaptive response”. J. Physic. Fit. Sports Med. 2014, 3, 115–120. [Google Scholar] [CrossRef]
- Santovito, A.; Gendusa, C. Micronuclei frequency in peripheral blood lymphocytes of healthy subjects living in Turin (North-Italy): contribution of body mass index, age and sex. Ann. Hum. Biol. 2020, 47, 48–54. [Google Scholar] [CrossRef]
- Gajski, G.; Gerić, M.; Oreščanin, V.; Garaj-Vrhovac, V. Cytokinesis-block micronucleus cytome assay parameters in peripheral blood lymphocytes of the general population: contribution of age, sex, seasonal variations and lifestyle factors. Ecotoxicol Environ Saf. 2018, 148, 561–570. [Google Scholar] [CrossRef]
- Cotton, S.C.; Sharp, L.; Little, J.; Brockton, N. , Glutathione S-transferase polymorphisms and colorectal cancer: a HuGE review. Am. J. Epidemiol. 2000, 151, 7–32. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Lee. S.Y.; Ki, C.S.; Jong-Won, K.. GSTM1, GSTT1 and GSTP1 polymorphisms in the Korean population. J. Korean Med. Sci. 2005, 20, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).