1. Introduction
Most patients with PCa can be cured of their disease. However, if metastatic spread occurs, the lung is the second most common site [
1]. Lung metastases result in high morbidity and mortality. Patients suffer from cough, shortness of breath, and pleural effusion. Unraveling the processes of PCa progression and metastatic spread might lead to a better understanding and ultimately better treatment or even prevention of lung metastases. We recently performed a comprehensive transcriptomic analysis of a panel of 770 genes in PCa lung metastases. Carcinoembryonic antigen cell adhesion molecule 6 (
CEACAM6) was among the genes that were highly expressed in PCa lung metastases, and it was more than 8fold higher in lung metastases in comparison to bone metastases. CEACAM6 belongs to a large family of adhesion molecules which in turn belong to the immunoglobulin supergene family. Structurally, they have an extracellular domain and are anchored to the cell membrane [
2]. In normal tissues CEACAM6 is expressed on the surface of epithelial and myeloid cells [
3]. The biological function of CEACAMs is manifold, among others immune response, angiogenesis, and pathogen receptors [
4]. CEACAM6 has been implicated in cancer progression. For instance, CEACAM6 expression is an adverse factor of overall survival in colorectal cancer [
5]. Non-invasive atypical breast lesions with high CEACAM6 expression exhibit a greater risk of progressing to invasive breast cancer [
6]. CEACAM6 exerts its tumorigenic properties by enhancing adhesion and invasion [
7]. Its surface location makes this protein an attractive target and indeed, antibody-conjugated and monoclonal therapies have been developed (reviewed in [
2]). So far, little evidence implicating CEACAM6 in PCa progression and metastases had been generated. Blumenthal et al. found CEACAM6 expression to be similar in PCa and normal tissues [
8]. Since
CEACAM6 emerged as a potential driver of metastatic spread in PCa in our own data from PCa lung metastases we performed functional validation experiments
in vitro.
2. Results
2.1. CEACAM6-Targeting siRNAs Effectively Silence the Expression of CEACAM6 in PC-3 Cells
To investigate the effect of CEACAM6 silencing on PCa cells (PC-3), the cells were transfected with CEACAM6-siRNA. As the results, CEACAM6-siRNAs gives a very high efficiency level knockdown of gene expression (
Figure 1-A). Based on the quantification analysis our siRNA experiment showed more than 85 % knockdown efficiency compared to the PC-3 cells with scrambled siRNA as control (
Figure 1-B).
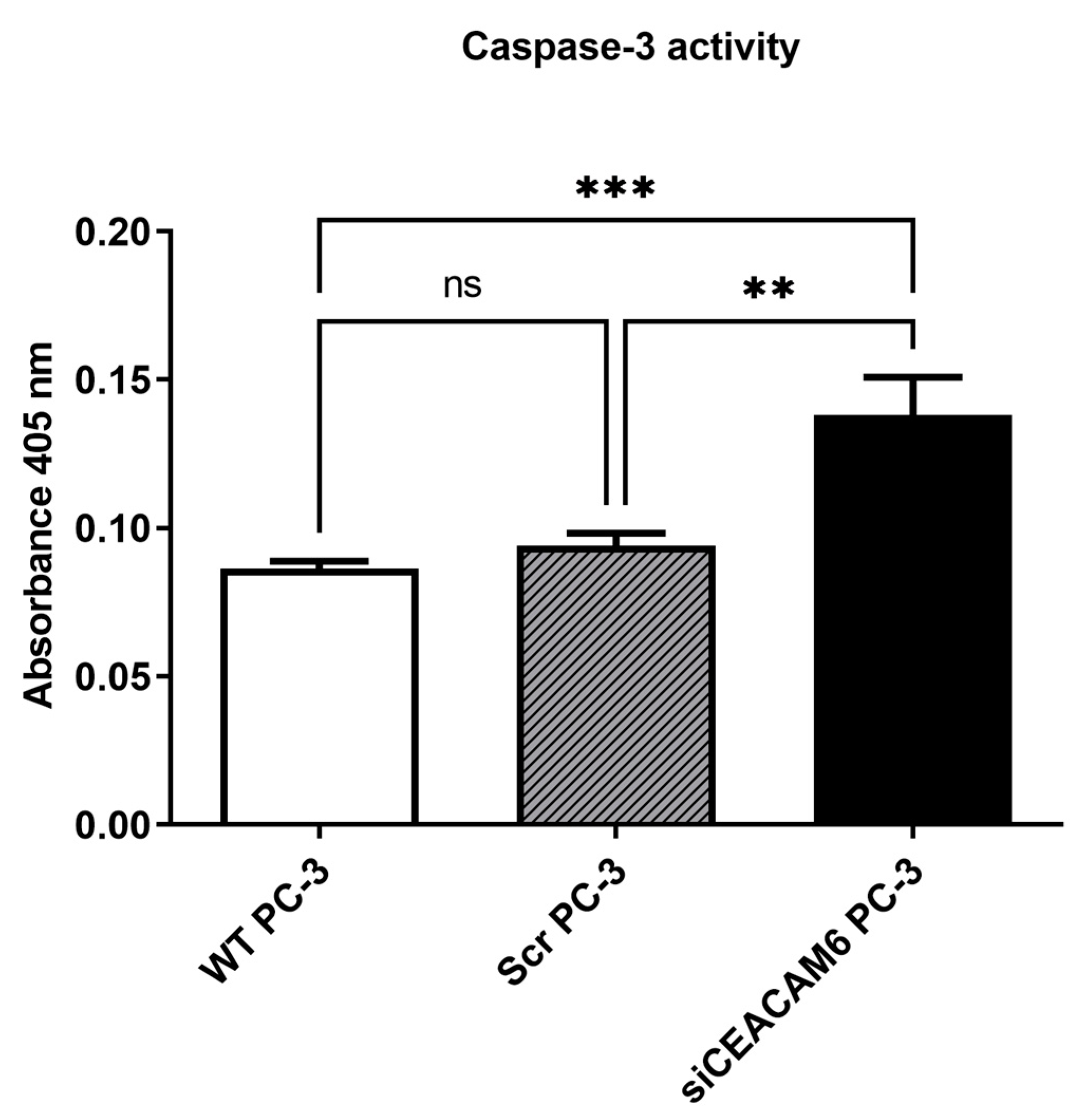
2.2. Knockdown of CEACAM6 Elevated Caspase-3 Mediated Apoptosis in PC-3 Cells.
It has been previously suggested that one of the primary apoptotic mediators in PC-3 cells is activation of caspase-3 [
9,
10]. To determine apoptotic properties in CEACAM6 knockdown PC-3 cells, we performed a caspase-3 assay 48 hours post transfection by using a colorimetric caspase-3 activity kit. We observed a significant elevation of caspase-3 activity in CEACAM6 knockdown cells compared with scrambled and WT PC-3 cells as controls (
Figure 2).
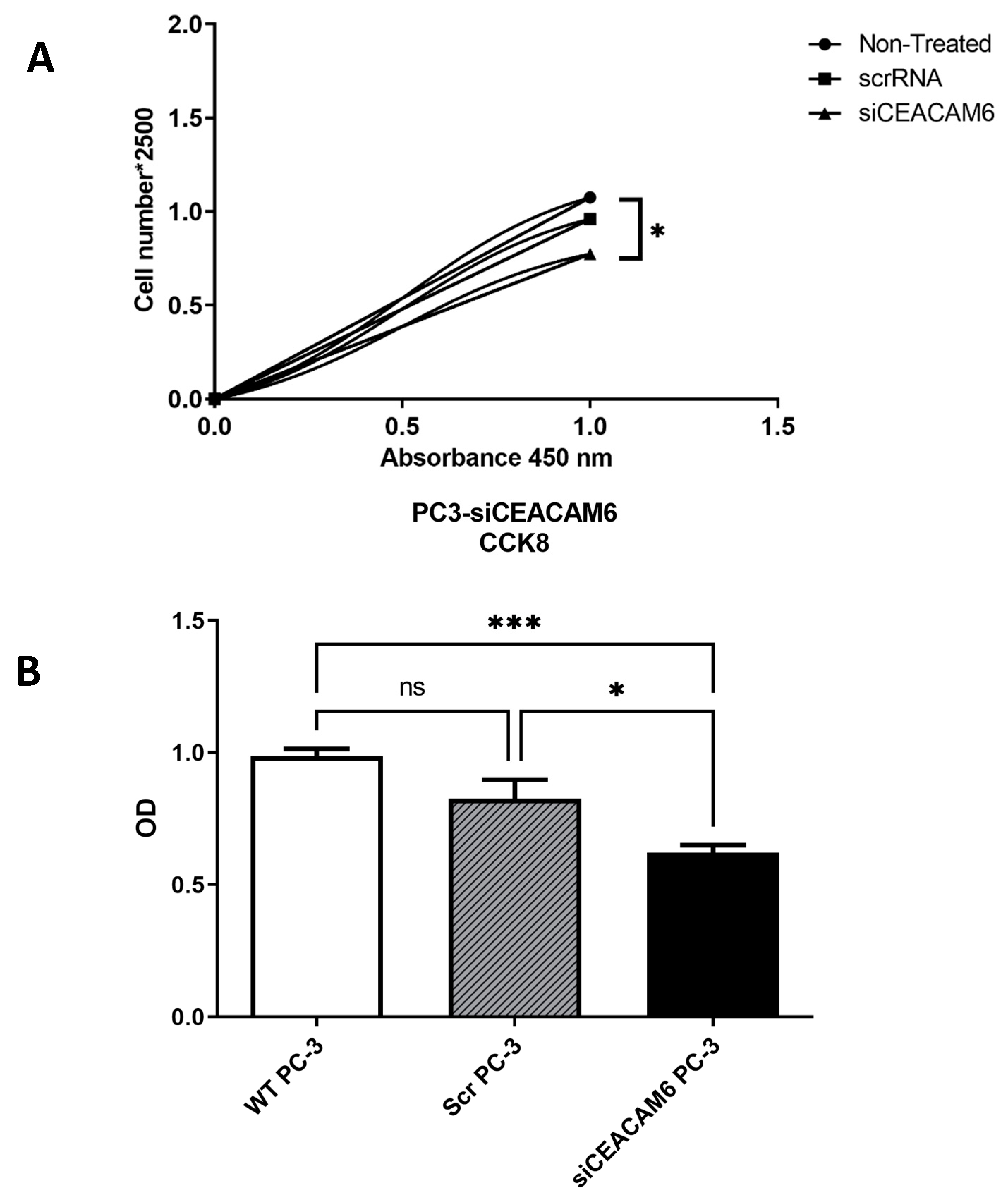
2.3. Silencing of CEACAM6 Reduced Cell Viability and Cell Count in PC-3 Cells.
Eventually establishing the efficacy of siRNA-CEACAM6 in PC-3 cells, we assessed functional alteration which induced by CEACAM6 knockdown in PC-3 cells. PC-3 cells transfected with CEACAM6-siRNA were assessed for cell viability and proliferation by using CCK-8 assay after 48 hours of transfection along with WT PC-3 cells with no treatment and PC-3 cells with scrRNA as controls. We observed a significant decrease of cell viability (
Figure 3-A) and cell counts (
Figure 3-B) in CEACAM6 knockdown PC-3 cells compared with scrambled and WT cells.
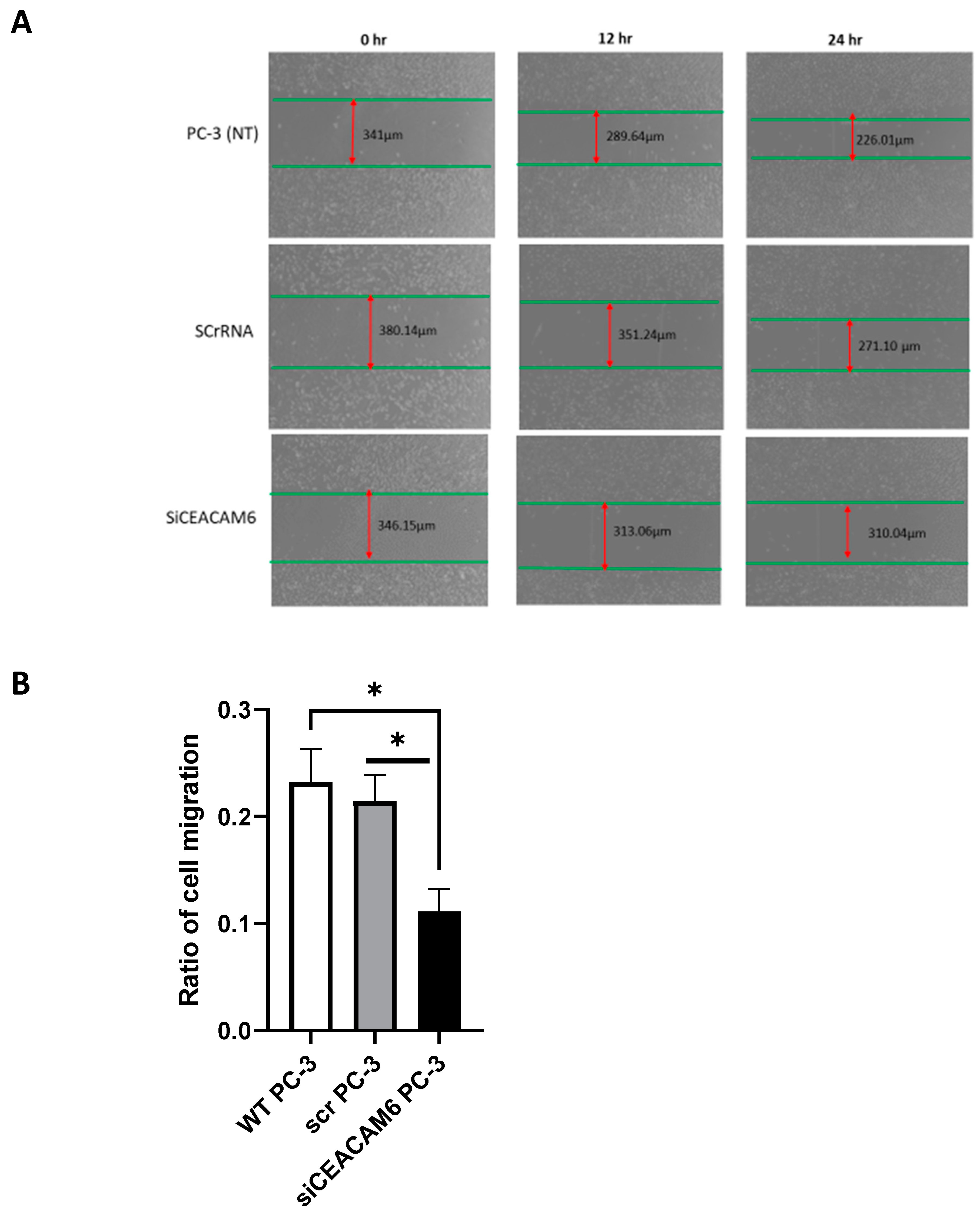
2.4. CEACAM6 Knockdown Significantly Diminished Cell Migration in PC-3 Cells.
Regarding the metastatic properties of PC-3 cells, we also studied the effect of CEACAM6 knockdown on the migratory functions of PC-3 cells. To distinguish migrated from proliferated cells, we minimized the proliferation ratio by using a serum-free growth medium for PC-3 cells during the scratch assay. Our results showed that, in the scratch test, the migration distance of PC-3 cell lines with CEACAM6 knockdown was significantly less shortened compared with that of the control group after 48 hours of post transfection incubation (
Figure 4-A, B).
3. Discussion
In a comparative screening approach, we identified elevated CEACAM6 in PCa lung metastases. We performed a functional analysis using siRNA to knockdown
CEACAM6 in PC-3 cells. We could show that all hallmarks of cancer cells i.e. cell viability, proliferation, apoptosis, and cell migration were influenced by
CEACAM6 silencing resulting in a less aggressive phenotype. Our findings are in concordance with published literature, where CEACAM6 was investigated in other cancer cell lines [
7]. The role of members of the CEACAM family in PCa progression has not been fully established. This is the first functional analysis that provides evidence, that indeed CEACAM6 promotes PCa aggressiveness. Therefore,
CEACAM6 might pose an attractive therapeutic target. Being a surface antigen, CEACAM6 should be amenable to antibody-based therapy. Recently, a fully-human anti-CEACAM5 antibody and Chimeric Antigen Receptor T-cells have been developed for the treatment of CEACAM5-positive neuroendocrine PCa [
11]. Similar immunoconjugates are being tested for other entities [
12]. CEACAM6 is also involved in suppressing T-cell mediated immune response. A CEACAM6 inhibitory antibody was able to restore T-cell function
in-vitro [
13]. In PCa, Sipuleucel-T is approved as immunotherapy for PCa. Hagihara et al. could show that neoadjuvant treatment with Sipuleucel-T in localized PCa induces gene expression of CEACAM6 and other immunoinhibitory genes. This suggests that treatment with Sipuleucel-T might be counteracted by CEACAM6. Combination treatment of Sipuleucel-T and anti-CEACAM6 antibodies could be a feasible option. In conclusion,
CEACAM6 promotes PCa aggressiveness and poses a very interesting therapeutic target. Both the immunoinhibitory properties of CEACAM6 as well as its tumour-promoting role should be further explored clinically.
4. Materials and Methods
4.1. Cell Culture
For this work, we used the human PCa cell lines PC3 which were purchased from ATCC (Manassas, VA) and maintained according to provider's instructions. The cells were incubated at 37°C with 5% CO2 in DMEM medium. After reaching confluency of roughly 80% (2-3 days of incubation), the cells were washed with PBS two times and then detached from the culture dishes with the help of Accutase® and trypsin. The trypsinization was stopped by adding the DMEM medium to the PC3 cells. The accruing suspension was centrifugated at 800 U/min and 20°C for 10 minutes, then the supernatant liquor was aspirated, and the pellet was resuspended in the respective medium. The suspension was split into 3 parts and allocated into bigger culture bottles. After another two days of growing, at a confluency of 60-70%, the cells were washed with PBS and detached again, and cell pellets were prepared as described above. The pellets were directly used for the following experiments or frozen at -80°C for long-term storage or later usage. Mycoplasma-test was performed for detection of possible contamination and turned out to be negative. To measure the final cell concentrations of both cell lines, PierceTM BCA Protein Assay Kit (Thermo ScientificTM) was used according to the manufacturer’s users guide from January 30th 2020.
4.2. siRNA Transfection
Gene silencing was performed in PC3 cells using siRNAs targeting
CEACAM6 and scrambled RNA as a non-targeting control (
Table 1). PC3 cells seeded in 6-well-plates (45-50,000 cells/well) were transfected in serum-free medium with 100 nM small-interfering RNA (siRNA) diluted in Opti-MEM
TM medium, using Lipofectamine® 2000 reagent (2-4 µl/well). After 5-6 hours the medium was replaced with DMEM medium.
4.3. Protein Isolation and Western Blot Analysis
Cells were detached from the culture bottles with Accutase® and centrifugated at 1200 rpm for 10 minutes. The supernatant was aspirated, and the cells were resuspended in warm PBS and again centrifugated at the same conditions as before. For protein isolation, a stock solution of 98% RIPA-buffer, 1% of each protease-inhibitor and 1% of phosphatase-inhibitor was used. The cells were washed with cold PBS and resuspended in RIPA-lysis buffer. This mixture incubated for 30 minutes stored on ice and then was centrifugated at 13000 rpm at 4°C for 30 minutes. The supernatant contains the lysed proteins.
For the protein assay PierceTM BCA Protein Assay Kit was utilized. For that purpose, 9 µl RIPA-buffer were put in each well of a 96-well plate and 1 µl protein-extract was added to each well. Additionally, some wells were filled with the samples of the standard range to calculate the concentration of protein and estimate the inaccuracies due to manual pipetting. The working solution was added and the plate was put to incubate at 37°c for 30 minutes. The protein concentration of the samples was calculated based on the absorbance-measurement of the TECAN Spark®.
For western blot analysis, an amount of 30 µg of total protein was used for each line. After separating the isolated protein in 10 to 12% SDS polyacrylamide gel, they were transferred to a polyvinylidene fluoride membrane of 0.45 µm by electric current. The reaction was blocked with TBS-T buffer containing 6% non-fat dry milk. One of the following different antibodies was added to each membrane: anti-CEACAM6 (dilution 1:1000; Life Technology, Thermo ScientificTM), anti-SLPI1 (dilution 1:1000; Life Technology, Thermo ScientificTM) and anti-FIGF (dilution 1:1000, Life Technology, Thermo ScientificTM). The blocked membranes were left for incubation for the night. After incubation, the abundant antibody-mixture was washed off the membranes using TBS-T buffer in 3 cycles of 10 minutes each. Subsequently, anti-mouse IgG (Goat-anti-rabbit HRP, dilution 1:1000, Life Technology, Thermo ScientificTM) was added to the washed membranes as a secondary antibody and the membranes were left for another hour of incubation at room temperature. After incubation, the membranes were washed for 10 minutes 5 ties in a row. For showing the resulting specific immune-reactive signals, enhanced chemiluminescence was used the ECL-substrate. Afterwards the membranes were stripped and the housekeeping gene anti-beta-actin was added (1:5000; Life Technology, Thermo ScientificTM) to the membranes as a positive control. The membranes were treated just like described above.
4.4. RNA Isolation, cDNA Synthesis and qPCR
For RNA isolation the RNeasy Mini Kit (Qiagen®) was used according to the manufacturer’s protocol. For quantification of the RNA amount, NanoDropTM-Spectrophotometer (Thermo ScientificTM) was applied. The synthesis of cDNA with help of reverse transcription, was performed using the SuperScriptTM IV First-Strand Synthesis System (Thermo ScientificTM) according to the manufacturer’s SuperScript IV cDNA synthesis protocol with the application of a random hexamer primer. For reverse transcription-control, the reverse transcriptase IV was substituted with water.
For qPCR itself, the LightCycler® 480 SYBR Green I Master Kit (Roche Molecular Systems Inc.) was utilized according to the manufacturer’s recommendations with a few small alterations concerning the volumes. The measurement was performed in a 96-well plate with a volume of 20 µl for each well, thereof 1 µl cDNA (which equates to 50 ng cDNA) and 7 µl distilled water to get to a total volume of 20 µl. The other components were used in the amounts recommended by the manufacturer (10 µl of MasterMix and 2 µl Primers). The evaluation was performed using the LightCycler® 480 Instrument (Roche Molecular Systems Inc.). The primer sequences are shown in
Table 2.
4.5. Proliferation Assay: CCK8-Method
A cell proliferation assay was performed using the cell counting Kit-8 (CCK-8) from GlpBio Technology LLC. PC3 cells were seeded into a 96-well plate at a density of 103-104 cells/well within 100 µl DMEM medium and incubated at 37°C and 5% CO2 for 24 hours. Afterwards 10 µl of the CCK8 working solution were added to the wells and incubated under the same conditions again for 6 hours. Then, 10 µl of CCK8 were added to each well and the plate was incubated for 3 hours one more time. Before measurement, the content of the wells was carefully mixed to make sure the color is distributed equally all over the well. Via microplate reader the absorbance of the mixture in each well was measured at 450 nm wavelength.
4.6. Apoptosis Assay: Caspase-3 Method
For cell apoptosis assay, the Caspase-3/CPP32 Colorimetric Assay Kit (BioVision Life Science SourceTM) was used according to the manufacturer’s protocol. For the assay, apoptosis was induced in PC-3 cells and pellets of about 1x106 cells were suspended in the cell lysis buffer. This mixture was positioned on ice to incubate for 10 minutes. After incubation the mixture was centrifugated and the supernatant was isolated for further usage. The extract was mixed with Cell Lysis Buffer, 2X Reaction Buffer and 4 mM DEVD-pNA substrate and then left for incubation for about 1 hour at 37°C. Afterwards the samples were analysed by use of a spectrophotometer.
4.7. Cell Migration (Scratch Wound) Assay
For the cell migration assay, we used the well-known classical method. Briefly, the PC3 cells were seeded into a 6-well plate at 1 x 105 cells per well (two to three replicates per group). When the cells were 70–80% confluent, the transfection was performed, and after 6 hours, the medium was removed and replaced with serum-free medium. Following starvation for 24 hours, the monolayer cells were scratched (at least three even scratches) using a pipette tip and washed three times with phosphate-buffered saline (PBS) to remove the detached and dead cells. Images were taken at 0 h, 12 h, and 24 h under a light microscope to measure the post-wounding widths of the scratches. At the end the rate of cell migration was calculated as following:
Cell migration rate = (scratch width at 0 h - scratch width at 24/12 h) / scratch width at 0 h.
4.8. Statistical Analyses
Two samples were compared by utilization of student’s two-tailed t-test. Comparisons of three or more samples were performed by usage of the 1-way-ANOVA with Tukey post hoc test. As a cutoff for statistical significance a p-value of p<0.05 was chosen. The data is declared as the mean of each value within the range of one standard deviation. Different software was applied for analyzing and illustration, such as QubitTM 2.0 IQ Analyzer, nanoString nSolver® analysis software v4.0 and Prism® 6 (GraphPad Software Inc. San Diego, USA).
5. Conclusions
In conclusion, CEACAM6 promotes PCa aggressiveness and poses a very interesting therapeutic target. Both the immunoinhibitory properties of CEACAM6 as well as its tumour-promoting role should be further explored clinically.
Author Contributions
VS: AS, AO, SP, JK, KW planned the study. AS, VS, DK, KW analyzed and interpreted the data. JSF, KW, AO, VS, AS performed experiments. VS and AS wrote the manuscript and prepared all figures. All authors read and approved the final manuscript.
Funding
This research was funded by Deutsche Forschungsgemeinschaft (DFG) Schwerpunktprogramm µBone SPP2084 (to VS and SP).
Institutional Review Board Statement
This study was approved by the Ethics Committee of the University of Luebeck (project code 18-053, date of approval: March 2nd, 2018, date of amendment: June 17th, 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and analyzed during this study can be made available upon reasonable request form the corresponding author.
Acknowledgments
The abstract of this manuscript was presented in 13. SYMPOSIUM Urologische Forschung der Deutschen Gesellschaft für Urologie (DGU) on November 2022. This work was funded by Deutsche Forschungsgemeinschaft (DFG) Schwerpunktprogramm µBone SPP2084 (to VS and SP). Duan Kang was supported by China Scholarship Council at the University of Luebeck (No. 202008440263). We thank Eva Dreyer for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results”.
References
- Bubendorf, L.; Schopfer, A.; Wagner, U.; et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol 2000, 31, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, N.; Arabzadeh, A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer metastasis reviews 2013, 32, 643–671. [Google Scholar] [CrossRef] [PubMed]
- Scholzel, S.; Zimmermann, W.; Schwarzkopf, G.; et al. Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am J Pathol 2000, 156, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Rizeq, B.; Zakaria, Z.; Ouhtit, A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci 2018, 109, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Jantscheff, P.; Terracciano, L.; Lowy, A.; et al. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic significance. J Clin Oncol 2003, 21, 3638–3646. [Google Scholar] [CrossRef] [PubMed]
- Poola, I.; Shokrani, B.; Bhatnagar, R.; et al. Expression of carcinoembryonic antigen cell adhesion molecule 6 oncoprotein in atypical ductal hyperplastic tissues is associated with the development of invasive breast cancer. Clin Cancer Res 2006, 12, 4773–4783. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, R.D.; Hansen, H.J.; Goldenberg, D.M. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen). Cancer Res 2005, 65, 8809–8817. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, R.D.; Leon, E.; Hansen, H.J.; et al. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer 2007, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.R.; Garner, D.S.; Williams, S.D.; et al. Comparison of immunohistochemistry for activated caspase-3 and cleaved cytokeratin 18 with the TUNEL method for quantification of apoptosis in histological sections of PC-3 subcutaneous xenografts. The Journal of pathology 2003, 199, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Slee, E.A.; Adrain, C.; Martin, S.J. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. The Journal of biological chemistry 2001, 276, 7320–7326. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.S.; Kim, Y.J.; Vergara, S.; et al. A highly-specific fully-human antibody and CAR-T cells targeting CD66e/CEACAM5 are cytotoxic for CD66e-expressing cancer cells in vitro and in vivo. Cancer letters 2022, 525, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Piha-Paul, S.; Simon, G.; Belani, C.P.; et al. A Phase 1, Open-Label, Dose-Escalation Study of L-DOS47 in Combination With Pemetrexed Plus Carboplatin in Patients With Stage IV Recurrent or Metastatic Nonsquamous NSCLC. JTO Clin Res Rep 2022, 3, 100408. [Google Scholar] [CrossRef] [PubMed]
- Pinkert, J.; Boehm, H.H.; Trautwein, M.; et al. T cell-mediated elimination of cancer cells by blocking CEACAM6-CEACAM1 interaction. Oncoimmunology 2022, 11, 2008110. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).