1. Introduction
In published guidelines, there is a pressing need for greater consensus regarding the minimum microbial threshold for diagnosing a urinary tract infection (UTI).[
1,
2,
3,
4,
5,
6,
7,
8,
9] The existing literature presents conflicting information on the commonly used diagnostic threshold of 100,000 CFU/mL with surprisingly scant and dated evidence to support it.[
1,
2,
3,
4,
5,
6] The confusion regarding a minimum threshold has led to uncertainty amongst clinicians, which can lead to increased use of empiric therapy or undertreatment of UTIs caused by lower microbial densities.[
10] Ensuring accurate diagnosis of symptomatic patients with complicated urinary tract infections (cUTIs) is crucial because these patients often possess one or more risk factors that can lead to treatment failure, adverse clinical outcomes, and/or severe complications.[
11,
12,
13] There is a gap in contemporary studies evaluating the correlation between rising microbial density and the presence or absence of a UTI.[
14]
Due to the limitations of SUC, which make it a flawed gold standard test for diagnosis, it is important to identify cases that have a very high likelihood of being true UTIs for any study evaluating diagnostic tests and thresholds.[
15,
16] This study defined true UTI cases as specimens from symptomatic patients that had a clinical diagnosis in a specialist setting for UTI, had a positive identification of known uropathogens by PCR or SUC, and had elevated urine biomarkers that have been documented to show inflammation of the urinary tract and have high specificity for UTI.
The biomarkers used in this study NGAL, IL-8, and IL-1 β are essential components of the constitutive immune response in the urinary tract and have previously demonstrated an ability to confirm the presence of cUTI.[
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31] The uroepithelium and resident innate immune cells in the bladder quickly protect against microbial threats by first identifying microbial patterns and triggering an immune response that produces antimicrobial peptides and pro-inflammatory cytokines.[
30,
31,
32,
33] One of our previous studies discovered that NGAL, IL-8, and IL-1β have sensitivities of 82.6%, 91.2%, and 69.8% with specificities of 90.8%, 76.8%, and 96.9%, respectively.[
34] A consensus of two or more of these biomarkers meeting the threshold of positivity gave a sensitivity of 84.0% and a specificity of 91.2%.[
34]
Using the biomarkers plus a UTI diagnosis from a specialty setting to identify UTI cases, we evaluated biomarker levels at different microbial densities using both M-PCR and SUC. The aim was to determine if there was a particular minimal microbial density threshold at which the biomarkers in these cases was significantly elevated, indicating that below this density a UTI was unlikely. There is common agreement that no absolute minimum density threshold should be set since some patients have been shown to have severe UTI even at very low densities of 10
2 CFU/mL.[
10] However, it would be useful to assess if there is a threshold over which a significant majority of UTIs are present in symptomatic patients and if that threshold matches the longstanding one of 100,000 CFU/mL or is lower.[
35] Evidence of a lower threshold would call into question the long-standing practice of considering lower densities as incidental findings, which has the potential to lead to underdiagnosis of patients symptomatic of a UTI.[
36]
2. Materials and Methods
Study design
This study compared urine biomarker results and microbial quantification in subjects symptomatic for UTI.
The subjects consisted of 583 patients presenting at urology clinics in 39 U.S. states with clinical presentations and ICD-10-CM diagnostic codes consistent with UTI between 01/17/2023 and 04/24/2023. Subjects’ de-identified urine samples were stored in a biorepository and evaluated at Pathnostics’ clinical laboratory. The Western Institutional Review Board exempted the study from review.
Specimen handling
All urine specimens in this study were collected into a sterile cup by the midstream clean-catch method. Specimens were split and transferred into two Vacutainers (Becton Dickinson, Franklin Lakes, NJ), one yellow top tube for the P-AST assays, and one gray top tube containing boric acid for the M-PCR, SUC, and biomarker testing. Upon receipt, each urine sample was separated into 1mL aliquots and placed in microcentrifuge tubes. Aliquots immediately underwent testing by M-PCR and SUC. Aliquots for biomarker (NGAL, IL-8, and IL-1β) testing were centrifuged at 13,200 rpm for 2 minutes. Aspiration of the supernatant was transferred to a clean tube, labeled, and frozen at -80oC +/- 10oC until ELISA testing.
Specimen testing
Enzyme-linked immunosorbent assay (ELISA)- ELISA kits purchased from R&D Systems/Bio-Techne (Minneapolis, MN), including human Lipocalin-2 / NGAL Quantikine ELISA Kit (Catalog number SLCN20), human IL-8 / CXCL8 Quantikine ELISA Kit (Catalog number S8000C), and human IL-1β / IL-1F2 Quantikine ELISA kit (Catalog number SLB50) were used. The assays measured the levels of NGAL [Range 0.2 - 500 ng/mL], IL-8 [Range 7.5 - 2,000 pg/mL], and IL-1β [Range 3.9 - 250 pg/mL] in the urine study specimens per the manufacturer’s instructions. An Infinite M Nano+ microplate reader (TECAN, Switzerland) measured absorbance at 450nm and 540nm, respectively. Frozen supernatants were thawed at room temperature before assaying.
Multiplex- Polymerase Chain Reaction (M-PCR) and Pooled Antibiotic Susceptibility Testing (P-AST)- The M-PCR/P-AST assay (Guidance® UTI, Pathnostics, Irvine, CA) provides susceptibility testing for 19 antibiotics, semi-quantitation of 27 pathogens, three bacterial groups, ESBL phenotype, and the identification of 32 antibiotic-resistance genes. This test is intended to be used for the diagnosis of complicated, persistent, recurrent UTIs, and for UTIs in elevated-risk patients. The assay was performed as previously described.[
37,
38,
39] First, the King Fisher/MagMAX™ automated DNA extraction instrument and the MagMAX™ DNA Multi-Sample Ultra Kit (Thermo Fisher, Carlsbad, CA) extracted the urine specimen microbial DNA according to the manufacturer’s instructions. The extracted DNA was used to identify and quantitate the specimen microbes. After combining a universal PCR master mix and extracted DNA, amplification was completed using TaqMan technology in a Life Technologies 12K Flex 112-format OpenArray System (Thermo Fisher Scientific, Wilmington, NC). The inhibition PCR control used was
Bacillus atrophaeus. Plasmids containing bacterial target DNA unique to each microbial species acted as positive controls. Duplicate specimen DNA samples were spotted on 112-format OpenArray chips (Thermo Fisher Scientific, Wilmington, NC). The Pathnostics data analysis tool (Pathnostics, Irvine, CA) sorted data, assessed data quality, summarized control sample data, identified positive assays, quantified bacterial load, and generated results. Results of the antibiotic resistance gene detection and the P-AST component of the test, which provides pooled phenotypic susceptibility results for 19 antibiotics, were not included in this analysis.
Quantitative M-PCR used probes and primers to detect the following microbes (23 bacteria species, 4 yeast species, and 3 bacterial groups):
Acinetobacter baumannii (A. baumannii),
Actinotignum schaalii (A. schaalii),
Aerococcus urinae (A. urinae),
Alloscardovia omnicolens (A. omnicolens), Candida albicans (C. albicans),
Candida auris (C. auris),
Candida glabrata (C. glabrata),
Candida parapsilosis (C. parapsilosis),
Citrobacter freundii (C. freundii),
Citrobacter koseri (C. koseri),
Corynebacterium riegelii (C. riegelii), Enterococcus faecalis (E. faecalis),
Enterococcus faecium (E. faecium), Escherichia coli (E. coli),
Gardnerella vaginalis (G. vaginalis),
Klebsiella oxytoca (K. oxytoca),
Klebsiella pneumoniae (K. pneumoniae),
Morganella morganii (M. morganii),
Mycoplasma hominis (M. hominis),
Pantoea agglomerans (P. agglomerans),
Proteus mirabilis (P. mirabilis),
Providencia stuartii (P. stuartii),
Pseudomonas aeruginosa (P. aeruginosa),
Serratia marcescens (S. marcescens),
Staphylococcus aureus (S. aureus),
Streptococcus agalactiae (S. agalactiae),
Ureaplasma urealyticum (U. urealyticum), Coagulase Negative Staphylococci (CoNS) which includes
Staphylococcus epidermidis,
Staphylococcus haemolyticus, Staphylococcus lugdunenesis, and
Staphylococcus saprophyticus (S. saprophyticus); Enterobacter Group which includes
Klebsiella aerogenes (K. aerogenes) (formally known as Enterobacter aerogenes) and
Enterobacter cloacae (E. cloacae), and Viridans Group Streptococci (VGS) which includes
Streptococcus anginosus,
Streptococcus oralis, and
Streptococcus pasteuranus. Generated reports provided the name(s) of all yeasts detected at any level and all bacteria detected at a density range of <10,000, 10,000 - 49,999; 50,000 - 99,999; or > 100,000 in cells/mL. The cells/mL quantitation was previously shown to correlate 1:1 linearly with CFUs/mL as defined by SUC. [
40]
Standard Urine Culture (SUC)- The SUC method was performed as previously described.[
38,
41] Briefly, urine was vortexed, and a sample 1µL each was spread onto blood agar and colistin and nalidixic acid agar/MacConkey agar (CNA/MAC) plates using a sterile plastic loop. All plates were incubated for 24 hours at 35°C under aerobic conditions. Plates with < 10,000 CFUs/mL were reported as normal urogenital flora and plates with growth (≥ 10,000 CFU/mL) were used for colony counts (blood agar plates) and identification and quantitation of each morphologically distinct and separate colony (CNA/MAC plates). If ≥ 3 pathogens were present without a predominant species, results were reported as contaminated/mixed flora. Pathogen identification was confirmed with the VITEK 2 Compact System (bioMerieux, Durham, NC).[
38,
41]
Statistical analysis
Participant demographics and the ICD-10-CM code breakdown were described in the summary statistics table (e.g., mean and standard deviation (SD) for continuous variables, such as age, or count and percentage for categorical variables, such as sex and ICD-10-CM). The distribution of all the microorganisms listed above was provided with the number of each species detected and population percentages by M-PCR and by SUC. Summary statistics (n, median, mean) for the expression of the three biomarkers were provided for different subgroups: based on microbial density and detection method M-PCR or SUC. Each subgroup median value was compared to the ‘no microbes detected’ group median using the Wilcoxon test. Previously published thresholds for biomarker positivity were used as cutoffs for the analysis; NGAL ≥ 38 ng/mL, IL-8 ≥ 20.6 pg/mL, and IL-1β ≥ 12.4 pg/mL.[
42,
43] This study defined biomarker consensus as any two or all three of the biomarkers positive at or above the cutoff levels. Individual biomarker results were evaluated with summary statistics (n, median). Results were compared between different microbial density categories derived from both M-PCR and SUC. All hypothesis tests were 2-sided, and a p-value of < 0.05 was considered statistically significant. All data analyses were performed using R 4.2.2 (
https://www.r-project.org/).
3. Results
Patient demographics
We examined urine samples from a total of 583 individuals with a median age of 76.3 years (Range 60.0 – 99.7 years, Standard Deviation = 8.85). Of these, 68.3% (n = 398) were females, and 31.7% (n = 185) were males. Most symptomatic patient samples were submitted with an ICD-10-CM code (
https://www.icd10data.com) of N39.0 for Urinary tract infection, site not specified [81.8% (n = 534)], see
Table 1.
Some patients had more than one ICD-10-CM code associated with their case. The five most prevalent codes are listed individually and all remaining codes were grouped together as “other”.
Bacterial and yeast identification
Out of the 583 urine samples obtained from patients with UTI symptoms, M-PCR did not detect any microbes in 117 samples, whereas SUC did not detect any microbes in 193 samples. In samples where microorganisms were detected, M-PCR identified 883 microbes with densities at > 10,000 cells/mL, while SUC identified 496 samples with microorganisms at a threshold of > 10,000 CFU (
Supplementary Table S1).
Biomarker Detections
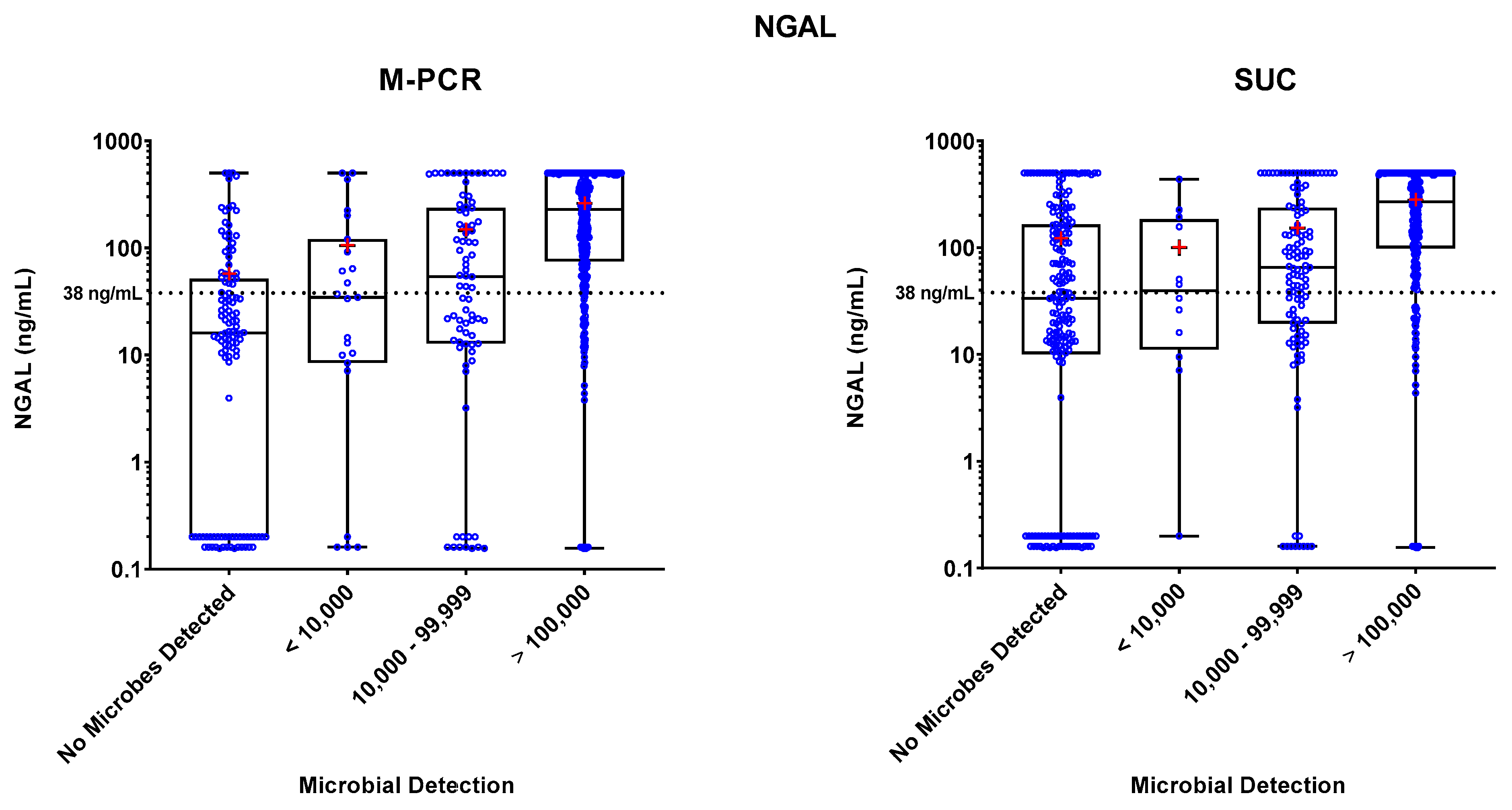
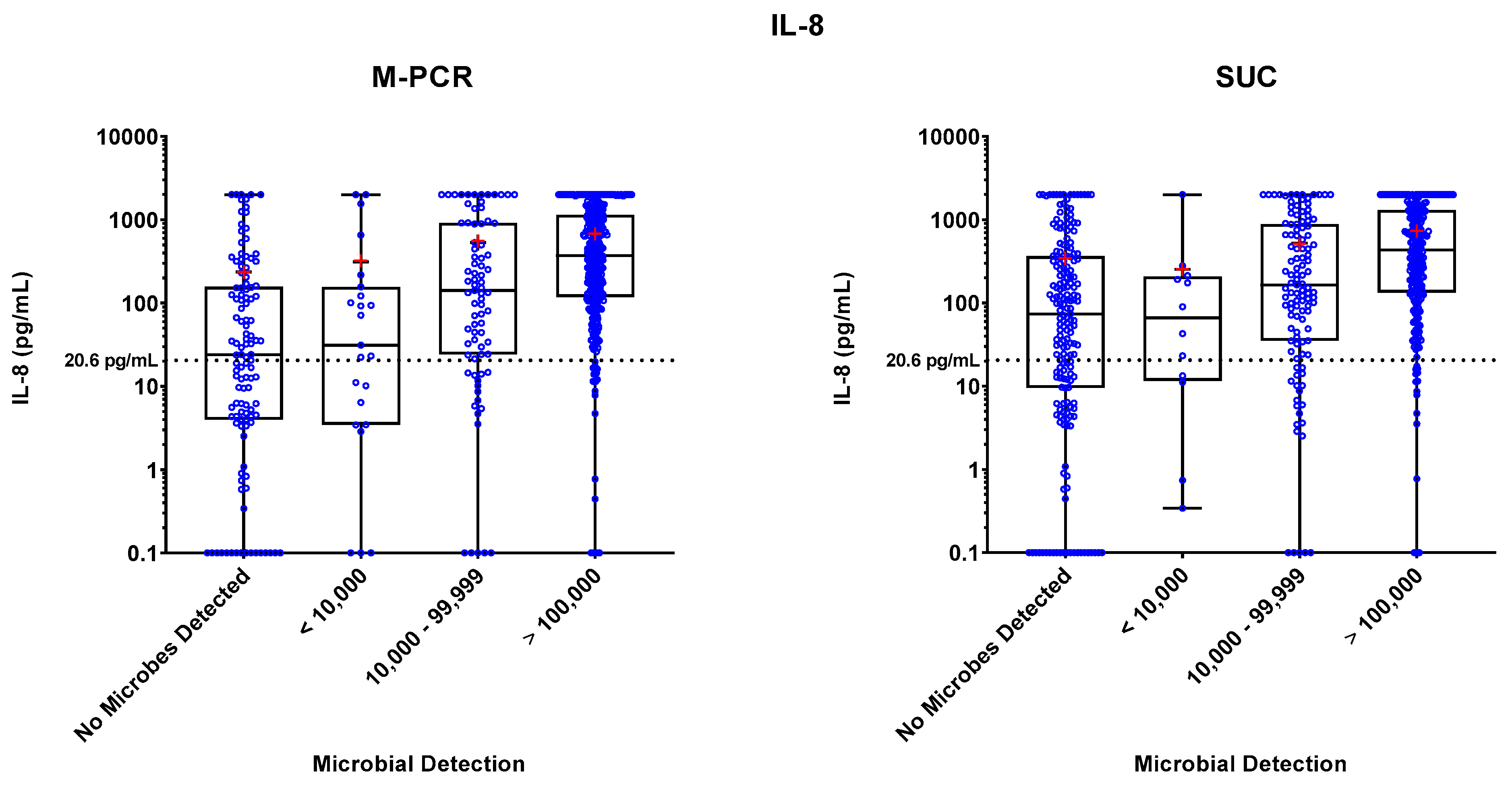
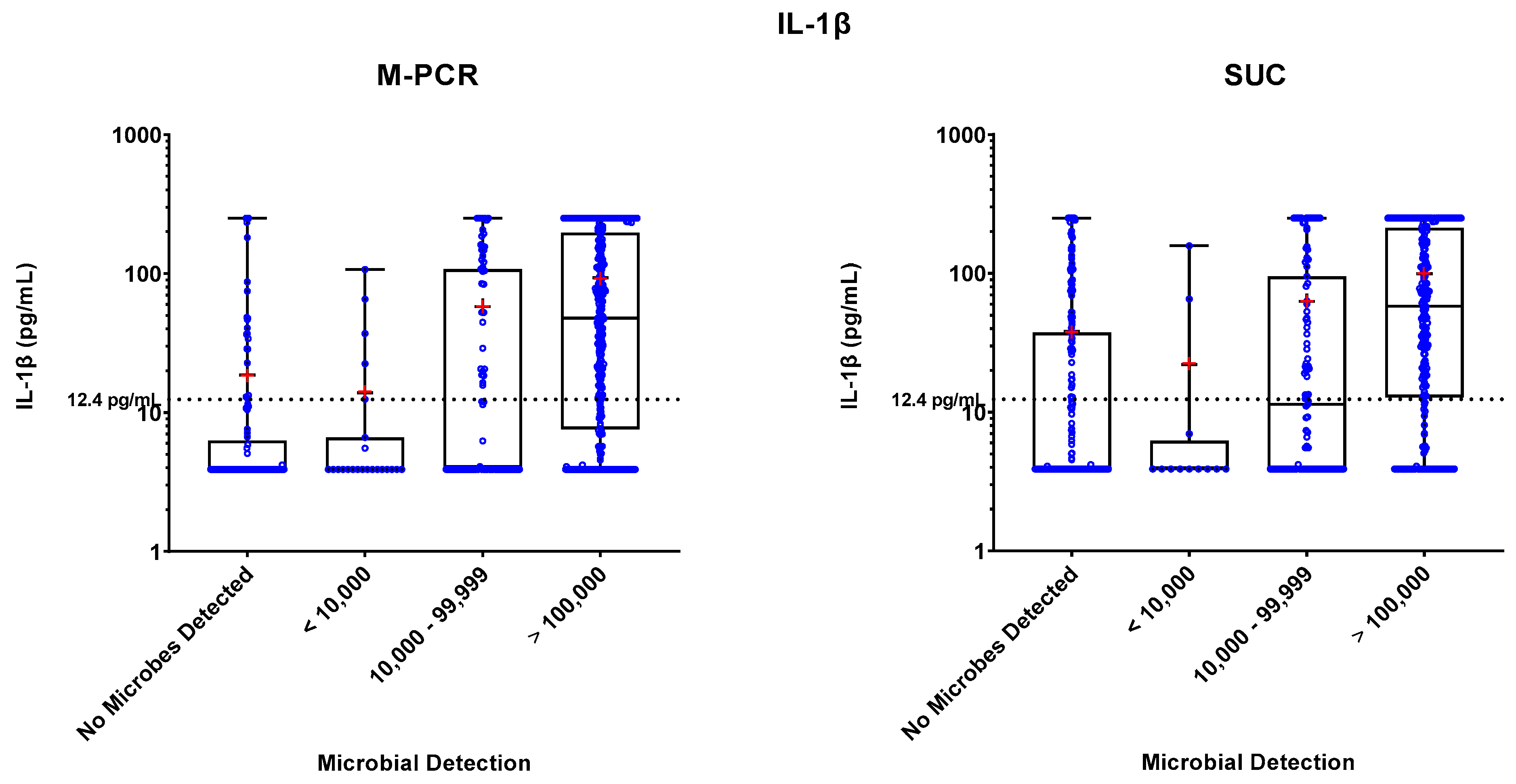
Specimen levels of NGAL, IL-8, and IL-1β biomarkers were measured by ELISA. These levels were plotted against four ranges of microbial densities detected by M-PCR and SUC: no microorganisms detected, < 10,000, 10,000 to 99,999, and > 100,000 cells or CFU per mL. Levels of each biomarker versus density ranges are shown in
Figure 1,
Figure 2, and
Figure 3. The "no microorganisms detected" group was used for statistical comparisons, employing the Wilcoxon test to evaluate the differences between the biomarker level medians from each of the microbe density ranges.
Table 2,
Table 3, and
Table 4 present the comprehensive summary of comparisons between each of the three biomarkers and density categories from both PCR and SUC.
In both the M-PCR and SUC methodologies, there was a consistent rise in the median levels of the three biomarkers as the microbial density increased. Compared to the group with no microbes detected, biomarkers showed significantly higher median levels starting at the 10,000 to 99,999 cells/mL category for M-PCR (p = 0.0002, p < 0.0001, p < 0.0001) and SUC (p = 0.0104, p = 0.0014, p = 0.0064) for NGAL, IL-8, and IL-1β, respectively. The > 100,000 cells/mL category also displayed a significantly higher median level of each biomarker for both M-PCR (p < 0.0001 for all) and SUC (p < 0.0001 for all). There was no significant difference in the median levels between specimens with no microbes detected and specimens with microbes detected at < 10,000 cells/mL for M-PCR (p = 0.2076, p = 0.7018, p = 0.86) or CFU/mL for SUC (p = 0.8056, p = 0.7767, p = 0.2083) for NGAL, IL-8, or IL-1β, respectively.
Biomarker Patterns: Microbial Density and Technique Differences
As observed in
Table 2,
Table 3 and
Table 4 and
Figure 1,
Figure 2 and
Figure 3, a consistent correlation exists between increased microbial density and elevated biomarker levels, regardless of the microbial detection method used. There was a significantly higher number of SUC specimens with negative results (n = 193) compared to M-PCR (n = 117).
Biomarker levels in the “no microorganisms detected” group were significantly higher for NGAL (p = 0.002), IL-8 (p = 0.009), and IL-1β (p = 0.001) when microbial detection was assessed by the SUC method compared to M-PCR. (
Figure 4). There were also a higher number of specimens identified with microbial densities in the > 100,000 cells/mL category when tested by M-PCR (n = 364) compared to SUC (n = 263). Several specimens from the “no microorganisms detected” by SUC group had high biomarker levels and high microbial densities when tested by M-PCR. Out of the 193 samples that were negative based upon SUC, 60 (31%) showed high levels of both microbes > 10,000 by M-PCR and NGAL (above threshold), 70 (36.2%) had elevated microbial levels > 10,000 by M-PCR along with high levels of IL-8 (above threshold), and 47 (44.4%) had microbial levels > 10,000 by M-PCR combined with biomarker levels that surpassed the threshold for IL-1β (above threshold).
Biomarker Consensus by Microbial Density
The assessment of each specimen for biomarker consensus (defined as two or more biomarkers above the established cutoffs) was conducted and organized based on microbial density and detection method (
Figure 5). Specimens having microbial densities ≥ 10,000 cells/mL detected by either M-PCR or SUC exhibited more than 50% biomarker consensus positivity with the percent of positivity increasing as microbial density rose. SUC negative cases, where no microbes were detected (n = 193), had biomarker consensus positive results in 46% of cases, which was much higher than M-PCR negative cases (n = 117) with 29% consensus positive.
4. Discussion
In this study, we employed the current standard-of-care SUC method, and the culture-independent M-PCR assay to identify and quantify microbes from patients with presumptive UTIs. Previous studies have demonstrated a 1:1 correlation for microbial quantitation in the linear range between SUC and M-PCR, allowing for direct comparisons between culture-dependent and culture-independent methods across various microbial densities.[
40] Additionally, we assessed the immune response by measuring infection-associated biomarkers (NGAL, IL-8, and IL-1β) within the same urine specimens. This unique approach facilitated meaningful comparisons between SUC and M-PCR and provided direct associations between the presence and density of microorganisms against the immune response in clinically relevant specimens.
Traditionally, a microbial density of ≥ 100,000 cells/mL has been deemed diagnostically significant; however, recent clinical reviews and guidelines have proposed lower thresholds that are still clinically relevant.[
1,
2,
3,
4,
5,
6] In our study, we observed that lower microbial densities (≥ 10,000 cells/mL) detected by M-PCR and SUC in symptomatic subjects showed a notable increase in infection-associated biomarker levels. Additionally, for these subjects suspected of having a UTI, a cell density of ≥ 10,000 cells/mL strongly correlated with biomarker consensus in M-PCR and SUC techniques. These findings suggest that a microbial detection threshold of 10,000 cells/mL could be an indicative criterion for diagnosing a UTI.
Using a threshold of >100,000 cells/mL as a criterion for initiating antimicrobial therapy carries significant clinical implications. One consequence is the potential for undertreatment of certain UTI patients, allowing microbes to proliferate, infiltrate host cells, develop biofilms, or acquire antibiotic resistance before treatment is administered.[
44,
45] Delaying treatment can lead to a progression of clinical severity, potentially necessitating higher antibiotic doses for extended durations or increasing the likelihood of complications, including recurrent infections and bacteremia.[
46]
Previous studies consistently show that M-PCR outperforms SUC in identifying and quantifying more causative uropathogens.[
37,
38,
39] This is significant in that the SUC method favors the growth of gram-negative microbes like
E. coli but are less effective for non-
E. coli microorganisms and fastidious microbes, which are increasingly recognized as common uropathogens.[
41,
47,
48,
49,
50,
51,
52] Additionally, SUC is less likely to identify polymicrobial infections.[
37,
38,
39]
Building on these findings, our study also observed more specimens where no microorganisms were detected in the SUC group (n = 193) compared to the M-PCR group (n = 117). Interestingly, the specimens with no microorganisms detected by SUC exhibited significantly higher levels of NGAL, IL-8, and IL-1β than those with no microbe detected by M-PCR. Furthermore, SUC specimens in which no organisms were detected demonstrated significantly higher (46%) consensus scores (two or more biomarkers above the established thresholds) when compared to negative M-PCR cases (29%).
The significantly higher biomarker levels in SUC-negative cases compared to M-PCR-negative cases have important implications. Firstly, the difference in biomarker levels underscores the potential variation in effectiveness between SUC and M-PCR as testing methods for detecting potential pathogens, suggesting that M-PCR may have higher sensitivity and specificity for detecting microbes that are causing a UTI, thereby enhancing diagnostic accuracy for symptomatic cases of UTI. Furthermore, SUC cases displaying low or no microbial densities alongside high biomarker levels suggest a potential failure in detecting the organisms causing the UTI leading to these elevated levels. These findings challenge the sensitivity of SUC for the identification of uropathogens, raising questions about false negatives in culture-based testing. Consequently, the increase in biomarker levels between SUC-negative cases highlights the need to carefully consider the testing method employed when interpreting results and making treatment decisions, as SUC-negative cases may warrant closer monitoring.
In analyzing urine samples from patients symptomatic of a UTI, a few specimens exhibited outlier data points characterized by lower inflammatory biomarker levels despite high microbial densities. These outliers with low inflammatory biomarker levels, even in symptomatic cases, could reflect scenarios where immune responses are compromised due to medications or underlying health conditions or instances of a resolving UTI.[
53,
54,
55,
56,
57] Furthermore, it is worth noting that different microbial strains may vary in their ability to elicit an immune response, and the combined presence of multiple strains can also influence the observed biomarker levels. Future work could explore the thresholds specific to individual or combinations of microbes and their pathogenic densities in detecting urinary tract infections.
This study has several strengths. Firstly, a large number of samples were concurrently tested for levels of three different biomarkers and microbial detection using two different semiquantitative microbial detection assays. This comprehensive approach allowed us to semi-quantitatively measure microbial density while simultaneously assessing the optimal density for diagnosing a urinary tract infection (UTI) and determining the accuracy of a negative diagnostic test result. The inclusion of multiple biomarkers and assays enhances the robustness of the findings and strengthens the overall conclusions.
However, there are some limitations to consider. One weakness of the study is that we were unable to measure the biomarker levels based on specific symptoms experienced by the patients. Understanding how the biomarkers correlate with different symptoms could provide valuable insights into the diagnostic and prognostic capabilities of the assays. Additionally, the number of cases in our study was not sufficient to measure the biomarker levels across densities for specific microbial species. Further investigations with a larger sample size would enable a more comprehensive analysis of the relationship between microbial densities, biomarker levels, and specific microbial species.
Future studies could build upon these findings by assessing the biomarker levels across different species or combinations of species, allowing for a more targeted approach to diagnosing and managing UTIs. Additionally, exploring the association between biomarker levels and specific symptoms of UTI would provide valuable clinical information for personalized treatment strategies. These potential avenues of research can further enhance our understanding of urinary tract infections and contribute to improved diagnostic accuracy and patient care.
5. Conclusions
In conclusion, our study shows that symptomatic subjects with UTI exhibit a significant immune response at a microbial density threshold of > 10,000 cells/mL, regardless of the detection method used. This suggests that a lower diagnostic microbial density threshold is clinically appropriate for UTI diagnosis and management, applicable to both microbial identification and quantitation techniques used here. Additionally, our study emphasizes the need for closer monitoring when SUC yields a negative result, given the possibility of false negatives and their clinical implications.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Supplementary Table S1: Bacterial Identification and Frequency by M-PCR and SUC in UTI symptomatic patients.
Author Contributions
Conceptualization, Mohit Mathur and David Baunoch; Data curation, Richard Festa; Formal analysis, Jimin Wang and Yan Jiang; Funding acquisition, David Baunoch; Investigation, Richard Festa and David Baunoch; Methodology, Natalie Luke and David Baunoch; Project administration, Richard Festa; Resources, David Baunoch; Software, Richard Festa, Jimin Wang and Yan Jiang; Supervision, Natalie Luke and Richard Festa; Validation, Richard Festa; Visualization, Emery Haley and Jimin Wang; Writing – original draft, Laura Parnell; Writing – review & editing, Natalie Luke, Mohit Mathur, Emery Haley, Lori Anderson and David Baunoch. All authors have read and agreed to the published version of the manuscript. .
Funding
This research was funded by Pathnostics.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical review and approval were waived for the symptomatic cohort in this study due to the use of deidentified samples from a biobank repository.
Informed Consent Statement
Patient consent was waived for the symptomatic cohort in this study due to the use of deidentified samples from a biobank repository.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy concerns.
Acknowledgments
The authors would like to acknowledge Michael Percaccio, Jesus Magallon, Mariana Remedios-Chan4, Alain Rosas, Marzieh Akhlaghpour, Max Murphy, Jasmine Nguyen, Kelli Eugenio, Tim Cho, and Annie Ngo for their contributions toward generating the data presented in this manuscript.
Conflicts of Interest
NL, MM, RF, EH, LA, and DB are employees of Pathnostics, and JW, YJ, and LP are paid consultants of Pathnostics.
References
- Rubin, R.H.; Shapiro, E.D.; Andriole, V.T.; Davis, R.J.; Stamm, W.E. Evaluation of New Anti-Infective Drugs for the Treatment of Urinary Tract Infection. Clin Infect Dis 1992, 15, S216–S227. [Google Scholar] [CrossRef]
- Hovelius, B.; Mårdh, P.-A.; Bygren, P. Urinary Tract Infections Caused by Staphylococcus Saprophyticus: Recurrences and Complications. J Urology 1979, 122, 645–647. [Google Scholar] [CrossRef]
- McNulty, C. PHE/NHS Diagnosis of Urinary Tract Infections. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/927195/UTI_diagnostic_flowchart_NICE-October_2020-FINAL.pdfov.uk) (accessed on 15 February 2023).
- Kouri, T.; Fogazzi, G.; Gant, V.; Hallander, H.; Hofmann, W.; Guder, W.G. European Urinalysis Guidelines. Scand J Clin Laboratory Investigation 2000, 60, 1–96. [Google Scholar] [CrossRef]
- Roberts, F.J. Quantitative Urine Culture in Patients with Urinary Tract Infection and Bacteremia. Am J Clin Pathol 1986, 85, 616–618. [Google Scholar] [CrossRef] [PubMed]
- Kunin Urinary Tract Infections: Detection, Prevention, and Management; Kunin, M., Ed.; Lea & Febiger: : Philadelphia, 1997.
- Choe, H.; Lee, S.; Yang, S.S.; Hamasuna, R.; Yamamoto, S.; Cho, Y.; Matsumoto, T. STI, the C. for D. of the U.G. for U. and Summary of the UAA-AAUS Guidelines for Urinary Tract Infections. Int J Urol 2018, 25, 175–185. [Google Scholar] [CrossRef]
- Cueto, M. de; Aliaga, L.; Alós, J.-I.; Canut, A.; Los-Arcos, I.; Martínez, J.A.; Mensa, J.; Pintado, V.; Rodriguez-Pardo, D.; Yuste, J.R.; et al. Executive Summary of the Diagnosis and Treatment of Urinary Tract Infection: Guidelines of the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC). Enfermedades Infecciosas Y Microbiol Clínica 2017, 35, 314–320. [Google Scholar] [CrossRef]
- Naber, K.G.; Bergman, B.; Bishop, M.C.; Bjerklund-Johansen, T.E.; Botto, H.; Lobel, B.; Cruz, F.J.; Selvaggi, F.P.; (EAU), U.T.I. (UTI) W.G. of the H.C.O. (HCO) of the E.A. of U. EAU Guidelines for the Management of Urinary and Male Genital Tract Infections1. Eur Urol 2001, 40, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Polage, C.R.; Bard, J.D.; May, L.; Lee, F.M.; Fabre, V.; Hayden, M.K.; Doernberg, S.D.B.; Haake, D.A.; Trautner, B.W.; et al. Envisioning Future UTI Diagnostics. Clin Infect Dis 2021. [CrossRef]
- Sabih Complicated Urinary Tract Infections; [Internet], S., Ed.; StatPearls Publishing: Treasure Island (FL), 2023.
- Gupta, K.; Hooton, T.M.; Naber, K.G.; Wullt, B.; Colgan, R.; Miller, L.G.; Moran, G.J.; Nicolle, L.E.; Raz, R.; Schaeffer, A.J.; et al. International Clinical Practice Guidelines for the Treatment of Acute Uncomplicated Cystitis and Pyelonephritis in Women: A 2010 Update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 2011, 52, e103–e120. [Google Scholar] [CrossRef]
- Wagenlehner, F.M.E.; Johansen, T.E.B.; Cai, T.; Koves, B.; Kranz, J.; Pilatz, A.; Tandogdu, Z. Epidemiology, Definition and Treatment of Complicated Urinary Tract Infections. Nat Rev Urol 2020, 17, 586–600. [Google Scholar] [CrossRef]
- Hernández-Hernández, D.; Padilla-Fernández, B.; Ortega-González, M.Y.; Castro-Díaz, D.M. Recurrent Urinary Tract Infections and Asymptomatic Bacteriuria in Adults. Curr Bladder Dysfunct Reports 2021, 1–12. [Google Scholar] [CrossRef]
- Price, T.K.; Dune, T.; Hilt, E.E.; Thomas-White, K.J.; Kliethermes, S.; Brincat, C.; Brubaker, L.; Wolfe, A.J.; Mueller, E.R.; Schreckenberger, P.C. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. Journal of Clinical Microbiology 2016, 54, 1216–1222. [Google Scholar] [CrossRef]
- Price, T.K.; Hilt, E.E.; Dune, T.J.; Mueller, E.R.; Wolfe, A.J.; Brubaker, L. Urine Trouble: Should We Think Differently about UTI? Int Urogynecol J 2018, 29, 205–210. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Smith, S.N.; Mody, L.; Mobley, H.L.T. Urine Cytokine and Chemokine Levels Predict Urinary Tract Infection Severity Independent of Uropathogen, Urine Bacterial Burden, Host Genetics, and Host Age. Infect Immun 2018, 86. [Google Scholar] [CrossRef]
- Edwards, G.; Seeley, A.; Carter, A.; Smith, M.P.; Cross, E.L.; Hughes, K.; Bruel, A.V. den; Llewelyn, M.J.; Verbakel, J.Y.; Hayward, G. What Is the Diagnostic Accuracy of Novel Urine Biomarkers for Urinary Tract Infection? Biomark Insights 2023, 18, 11772719221144460. [Google Scholar] [CrossRef]
- Gadalla, A.A.H.; Friberg, I.M.; Kift-Morgan, A.; Zhang, J.; Eberl, M.; Topley, N.; Weeks, I.; Cuff, S.; Wootton, M.; Gal, M.; et al. Identification of Clinical and Urine Biomarkers for Uncomplicated Urinary Tract Infection Using Machine Learning Algorithms. Sci Rep-uk 2019, 9, 19694. [Google Scholar] [CrossRef]
- Horváth, J.; Wullt, B.; Naber, K.G.; Köves, B. Biomarkers in Urinary Tract Infections – Which Ones Are Suitable for Diagnostics and Follow-Up? Gms Infect Dis 8, Doc24. [CrossRef]
- Hosman, I.S.; Roić, A.C.; Lamot, L. A Systematic Review of the (Un)Known Host Immune Response Biomarkers for Predicting Recurrence of Urinary Tract Infection. Frontiers Medicine 2022, 9, 931717. [Google Scholar] [CrossRef]
- Martino, F.K.; Novara, G. Asymptomatic Bacteriuria or Urinary Tract Infection? New and Old Biomarkers. Int J Transl Medicine 2022, 2, 52–65. [Google Scholar] [CrossRef]
- Masajtis-Zagajewska, A.; Nowicki, M. New Markers of Urinary Tract Infection. Clin Chim Acta 2017, 471, 286–291. [Google Scholar] [CrossRef]
- Nanda, N.; Juthani-Mehta, M. Novel Biomarkers for the Diagnosis of Urinary Tract Infection–-A Systematic Review. Biomark Insights 2009, 4, BMIS3155. [Google Scholar] [CrossRef]
- Otto, G.; Burdick, M.; Strieter, R.; Godaly, G. Chemokine Response to Febrile Urinary Tract Infection. Kidney Int 2005, 68, 62–70. [Google Scholar] [CrossRef]
- Rodhe, N.; Löfgren, S.; primary …, S.-J. of Cytokines in Urine in Elderly Subjects with Acute Cystitis and Asymptomatic Bacteriuria. 2009. [CrossRef]
- Shaikh, N.; Martin, J.M.; Hoberman, A.; Skae, M.; Milkovich, L.; McElheny, C.; Hickey, R.W.; Gabriel, L.V.; Kearney, D.H.; Majd, M.; et al. Biomarkers That Differentiate False Positive Urinalyses from True Urinary Tract Infection. Pediatr Nephrol 2020, 35, 321–329. [Google Scholar] [CrossRef]
- Yilmaz, A.; Sevketoglu, E.; Gedikbasi, A.; Karyagar, S.; Kiyak, A.; Mulazimoglu, M.; Aydogan, G.; Ozpacaci, T.; Hatipoglu, S. Early Prediction of Urinary Tract Infection with Urinary Neutrophil Gelatinase Associated Lipocalin. Pediatr Nephrol 2009, 24, 2387. [Google Scholar] [CrossRef]
- Yim, H.E.; Yim, H.; Bae, E.S.; Woo, S.U.; Yoo, K.H. Predictive Value of Urinary and Serum Biomarkers in Young Children with Febrile Urinary Tract Infections. Pediatr Nephrol 2014, 29, 2181–2189. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Yang, J.; Xie, X.; Chen, H. The Immune Responses to Different Uropathogens Call Individual Interventions for Bladder Infection. Front Immunol 2022, 13, 953354. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.N.; Miao, Y. The Nature of Immune Responses to Urinary Tract Infections. Nat Rev Immunol 2015, 15, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Pradeu, T.; Masters, S.L.; Mogensen, T.H. Constitutive Immune Mechanisms: Mediators of Host Defence and Immune Regulation. Nat Rev Immunol 2021, 21, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Mariano, L.L.; Ingersoll, M.A. The Immune Response to Infection in the Bladder. Nat Rev Urol 2020, 17, 439–458. [Google Scholar] [CrossRef]
- Akhlaghpour, M.; Haley, E.; Parnell, L.; Luke, N.; Mathur, M.; Festa, R.; Percaccio, M.; Magallon, J.; Remedios-Chan, M.; Rosas, A.; et al. 2023. Urine Biomarkers Individually and as a Consensus Model Show High Sensitivity and Specificity for Detecting UTIs. Diagnostics. Unpublished manuscript, last modified June 27. Submitted. 27 June.
- Tullus, K. Low Urinary Bacterial Counts: Do They Count? Pediatric Nephrology 2016, 31, 171–174. [Google Scholar] [CrossRef]
- Scott, V.; Haake, D.A.; Churchill, B.M.; Justice, S.S.; Kim, J.-H. Intracellular Bacterial Communities: A Potential Etiology for Chronic Lower Urinary Tract Symptoms. Urology 2015, 86, 425–431. [Google Scholar] [CrossRef]
- Baunoch, D.; Luke, N.; Wang, D.; Vollstedt, A.; Zhao, X.; Ko, D.S.C.; Huang, S.; Cacdac, P.; Sirls, L.T. Concordance Between Antibiotic Resistance Genes and Susceptibility in Symptomatic Urinary Tract Infections. Infect Drug Resist 2021, 14, 3275–3286. [Google Scholar] [CrossRef] [PubMed]
- Wojno, K.J.; Baunoch, D.; Luke, N.; Opel, M.; Korman, H.; Kelly, C.; Jafri, S.M.A.; Keating, P.; Hazelton, D.; Hindu, S.; et al. Multiplex PCR Based Urinary Tract Infection (UTI) Analysis Compared to Traditional Urine Culture in Identifying Significant Pathogens in Symptomatic Patients. Urology 2020, 136, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Vollstedt; D, B.; KJ, W.; N, L.; K, C.; Belkoff, L.; Milbank, A.; N, S.; R, H.; N, G.; et al. Multisite Prospective Comparison of Multiplex Polymerase Chain Reaction Testing with Urine Culture for Diagnosis of Urinary Tract Infections in Symptomatic Patients. Journal of Surgical Urology 2020. [CrossRef]
- Festa, R.A.; Opel, M.; Mathur, M.; Luke, N.; Parnell, L.K.S.; Wang, D.; Zhao, X.; Magallon, J.; Percaccio, M.; Baunoch, D. 2023. Quantitative Multiplex PCR in Copies Ml–1 Linearly Correlates with Standard Urine Culture in Colonies Ml−1 for Urinary Tract Infection (UTI) Pathogens. Letters in Applied Microbiology. Unpublished manuscript, last modified June 9. Submitted.
- Price, T.K.; Dune, T.; Hilt, E.E.; Thomas-White, K.J.; Kliethermes, S.; Brincat, C.; Brubaker, L.; Wolfe, A.J.; Mueller, E.R.; Schreckenberger, P.C. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol 2016, 54, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Clinic, M. Neutrophil Gelatinase-Associated Lipocalin (NGAL) 2008.
- Yuan, Q.; Huang, R.; Tang, L.; Yuan, L.; Gao, L.; Liu, Y.; Cao, Y. Screening Biomarkers and Constructing a Predictive Model for Symptomatic Urinary Tract Infection and Asymptomatic Bacteriuria in Patients Undergoing Cutaneous Ureterostomy: A Metagenomic Next-Generation Sequencing Study. Dis Markers 2022, 2022, 7056517. [Google Scholar] [CrossRef]
- Mysorekar, I.; National, H.S. of the Mechanisms of Uropathogenic Escherichia Coli Persistence and Eradication from the Urinary Tract. 2006. [Google Scholar] [CrossRef]
- Holá, V.; Opazo-Capurro, A.; Scavone, P. Editorial: The Biofilm Lifestyle of Uropathogens. Front Cell Infect Mi 2021, 11, 763415. [Google Scholar] [CrossRef]
- Yang, X.; Chen, H.; Zheng, Y.; Qu, S.; Wang, H.; Yi, F. Disease Burden and Long-Term Trends of Urinary Tract Infections: A Worldwide Report. Frontiers Public Heal 2022, 10, 888205. [Google Scholar] [CrossRef] [PubMed]
- Price, T.K.; Hilt, E.E.; Dune, T.J.; Mueller, E.R.; Wolfe, A.J.; Brubaker, L. Urine Trouble: Should We Think Differently about UTI? Int Urogynecol J 2018, 29, 205–210. [Google Scholar] [CrossRef]
- Harding, C.; Rantell, A.; Cardozo, L.; Jacobson, S.K.; Anding, R.; Kirschner-Hermanns, R.; Greenwell, T.; Swamy, S.; Malde, S.; Abrams, P. How Can We Improve Investigation, Prevention and Treatment for Recurrent Urinary Tract Infections – ICI-RS 2018. Neurourol. Urodyn. 2019, 38, S90–S97. [Google Scholar] [CrossRef]
- Szlachta-McGinn, A.; Douglass, K.M.; Chung, U.Y.R.; Jackson, N.J.; Nickel, J.C.; Ackerman, A.L. Molecular Diagnostic Methods Versus Conventional Urine Culture for Diagnosis and Treatment of Urinary Tract Infection: A Systematic Review and Meta-Analysis. European Urology Open Sci 2022, 44, 113–124. [Google Scholar] [CrossRef]
- Kline, K.A.; Lewis, A.L. Gram-Positive Uropathogens, Polymicrobial Urinary Tract Infection, and the Emerging Microbiota of the Urinary Tract. Microbiol Spectr 2016, 4. [Google Scholar] [CrossRef]
- Lotte, R.; Lotte, L.; Ruimy, R. Actinotignum Schaalii (Formerly Actinobaculum Schaalii): A Newly Recognized Pathogen—Review of the Literature. Clin Microbiol Infec 2016, 22, 28–36. [Google Scholar] [CrossRef]
- Kaido, M.; Yasuda, M.; Komeda, H.; Okano, M.; Ito, Y.; Ohashi, H.; Ohta, H.; Akai, Y. Prediction of Presence of Fastidious Bacteria by the Fully Automated Urine Particle Analyzer UF-1000i in the Case of Ineffective Antimicrobial Therapy for Urinary Tract Infection. J Infect Chemother 2023, 29, 443–452. [Google Scholar] [CrossRef]
- Hooton, T.M.; Roberts, P.L.; Stapleton, A.E. Asymptomatic Bacteriuria and Pyuria in Premenopausal Women. Clin Infect Dis 2020, 72, 1332–1338. [Google Scholar] [CrossRef]
- Trautner, B.W. Urinary Tract Infection as a Continuum—Implications for Diagnostic and Antibiotic Stewardship. Clin Infect Dis 2020, 72, 1339–1341. [Google Scholar] [CrossRef]
- Taha, A.S.; Grant, V.; Kelly, R.W. Urinalysis for Interleukin-8 in the Non-Invasive Diagnosis of Acute and Chronic Inflammatory Diseases. Postgrad Med J 2003, 79, 159. [Google Scholar] [CrossRef]
- Devarajan, P. Neutrophil Gelatinase-associated Lipocalin (NGAL): A New Marker of Kidney Disease. Scand J Clin Laboratory Investigation 2008, 68, 89–94. [Google Scholar] [CrossRef]
- Sharif-Askari, F.S.; Sharif-Askari, N.S.; Guella, A.; Alabdullah, A.; Sheleh, H.B.A.; AlRawi, A.M.H.; Haddad, E.S.; Hamid, Q.; Halwani, R.; Hamoudi, R. Blood Neutrophil-to-Lymphocyte Ratio and Urine IL-8 Levels Predict the Type of Bacterial Urinary Tract Infection in Type 2 Diabetes Mellitus Patients. Infect Drug Resist 2020, 13, 1961–1970. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).