1. Introduction
The multiclass classification problem is one of the most common and important tasks in modern machine and deep learning applications in various fields [
1,
2,
3,
4,
5]. One of such areas is medicine, where the solution of such problems allows improving the systems of diagnostics, rehabilitation, and the life quality support of patients. Currently, there are numerous works devoted to using of neural networks for solving classification problems in medicine.
For example, the classification of cancer on pathological images of cervical tissues using deep learning algorithms was considered by Pan Huang, et al. in [
6]. The authors used ResNet50 [
7], DenseNet12, Inception_v3 [
8] and VGGNet19 [
9] for an almost balanced dataset consisted of 468 RGB images, including 150 normal, 85 low-grade squamous intraepithelial lesions, and 104 high-grade squamous intraepithelial lesions. Swaraj and Verma in [
10] solved the COVID-19 classification problem on chest X-ray images. The dataset consisted of 15371 posterior-to-anterior chest x-ray images. The authors used LeNet [
11], VGG-16 [
12], ResNet-50, AlexNet [
13], Xception [
14] and Inception_v3 architectures as deep learning models for training. As a result, LeNet showed an accuracy of 74.75% on a validation dataset, AlexNet - 70.04%, ResNet-50 - 75.71%, VGG-16 - 87.34%, Inception_v3 - 84.29%, Xception - 67.76%. State of arts algorithms are also used for solving actual medical problems. For example, works [
15,
16] provide an overview of the application of transformers models for solving various medical problems related to the analysis of medical images. The types of problems that can be solved by transformers models, their architecture and metrics were considered. The paper [
17] states the approach of applying transformers to solving the problem of classifying medical images of the stage of chronic venous disease for self-diagnosis of a patient by using the image of the patient’s legs.
Deep learning algorithms are used to solve classification problems in other areas of science and technology too. Gangsar and Tiwari in [
18] studied a vibration and current monitoring for effective fault prediction in an induction motor by using multiclass support vector machine (MSVM) algorithms. Ceyhan et al. in [
19] shared the results of experiments aimed at classifying wheat varieties using deep learning methods, which made it possible to create a new approach for such classification, which can be used in industry, including on equipment with low memory/small computing power.
Despite the widespread use of deep learning methods, their training faces several challenges. So, one of the critical problems in multiclass classification tasks is the imbalance of the dataset. This is especially important when using contemporary pre-trained neural networks which were mentioned before, where, in fact, the last layers of the neural network are retrained. With an unbalanced dataset, the error of relating an object to the correct class increases, which cannot always be eliminated by changing the training parameters of a neural network.
There are a number of works devoted to various techniques for increasing the metrics of training models on an unbalanced dataset. A possible way to improve the quality of the classification is to change the number of objects in the dataset by data augmentation, data mining, etc. It is commonly said that data augmentation can substantially increase the values of training metrics. The authors of article [
20] propose the modified VGG16 model for classification pneumonia X-rays images. Presented IVGG13 model trained on augmented dataset produced good metrics with the F1-measure compared with the best convolutional neural networks for medical image recognition. At a time when COVID-19 was the most common in the world, Jain et al. [
21] presented an approach to form of a classifier neural network for COVID-19 diagnoses by X-ray images. This approach contained 4 phases including data augmentation. As it was shown in [
22], the strategy of training used with data augmentation techniques that avoid overfitting problems on ConvNets shows the best results.
At the same time, data mining usage can also have a positive impact on the performance of neural networks. In [
23] various approaches to medical data mining are considered, as well as the learning outcomes after applying each of the approaches. It also showed that all considered mining methods increase the accuracy of the neural networks. Bellazzi and Zupan [
24] stated that many data mining methods can be successfully applied to a variety of practical problems in clinical medicine and are an excellent means of increasing network performance for medical data processing tasks.
However, dataset imbalance affects the training results of each neural network differently, as does data preprocessing like data augmentation and data mining. It is also of interest to study which of the methods for reducing the imbalance of a dataset is most preferable for a particular neural network. In this paper, we studied the sensitivity to a dataset imbalance of the following contemporary neural networks: Xception, ViT-384 [
25], ViT-224, VGG19, ResNet34 [
26], ResNet50, ResNet101 [
27], Inception_v3, DenseNet201 [
28], DenseNet161 [
29], DeIT [
30]. Different imbalance reduction techniques will be used to determine this sensitivity.
The remainder of this paper is organized as follows. The data processing methods, data mining ways, as well as trained neural networks are described in
Section 2. The performance evaluation and research results are stated in
Section 3. Finally,
Section 4 discusses the study’s findings, results, and the main conclusion of the work.
2. Materials and Methods
2.1. Datasets
The study was carried out in three stages. The models’ parameters were the same for all stages.
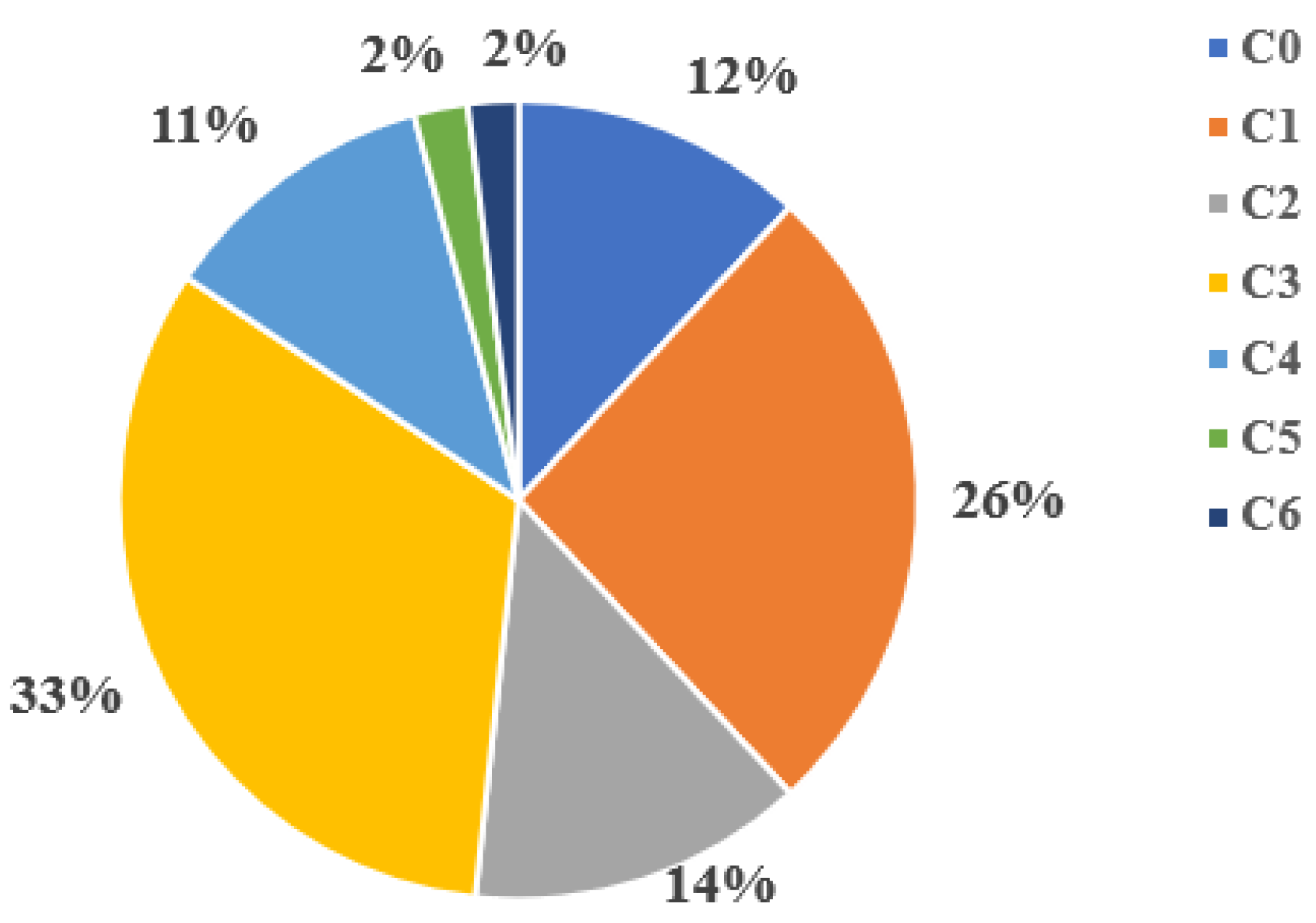
At the first stage, all the following neural networks Xception, ViT-384, ViT-224, VGG19, ResNet34, ResNet50, ResNet101, Inception_v3, DenseNet201, DenseNet161, DeIT were trained on the full dataset. The dataset was an unbalanced set of 20,971 leg images distributed across 7 classes (C0, C1, …, C6) according to the CEAP classification for chronic venous diseases [
17]. The distribution of images by classes was as follows: C0 - 12%; C1 - 26%; C2 - 14%; C3 - 33%; C4 - 11%; C5 and C6 classes - 2% each as shown in
Figure 1 and
Table 1.
As can be seen in
Figure 1, the initial dataset was significantly unbalanced. The number of classes C5 and C6 are extremely small compared to other classes, and class C3 is dominant.
At the second stage, small datasets were used. Cutting off the dataset by the smallest class is a good approach to avoid unbalanced, when the original dataset is big enough. To form small but balanced datasets, the initial dataset was cut off by the smallest class (C6). The excluded images were used as a validation dataset. To avoid randomness in the result obtained, three datasets were formed, where classes C0-C5 contained, if possible, different images. So, we constructed three datasets – “small_ds_1”, “small_ds_2”, “small_ds_3”. The distribution of images by classes is shown in
Table 2.
At the third stage, the dataset was formed as follows. Each class was cropped to the number of images equal to the size of class C6 + 15% of it. Then the missing images in classes C5 and C6 were augmented. Such approach can help generate new samples from existing samples using standard image conversion algorithms. It is based on deep learning concepts where a slightly modified original image is considered as new image.
But a lot of artificial data can be detrimental to the ability of a machine learning model to generalize. So, misapply of this approach can lead to overfitting. That's why we augmented the dataset not more than 15%. The distribution of images by classes for “aug_ds” is shown in
Table 3.
We used the following image transformation algorithms for data augmentation:
Shift Scale Rotate. Randomly apply affine transforms: translate, scale, and rotate images.
Random Crop. Crop random part of the images with fixing size.
Horizontal Flip. Flip an image 180 degrees to get a mirror image.
RGB Shift. Randomly shift values for each Red-Green-Blue channel of an image.
Random Brightness Contrast. Randomly change brightness and contrast of an image.
2.2. Deep Learning Neural Networks
We trained the following 11 different contemporary architectures of deep learning neural networks on the obtained datasets: ResNet34. ResNet50, ResNet101, VIT-base-patch16-224, VIT-base-patch16-384, DeIT-base-patch16-224, DenseNet161, DenseNet201, VGG19, Inception, Xception. Our goal was to formulate the best approach to train these networks on an initially unbalanced dataset.
ResNet (Residual Neural Network) models are the most popular models in computer vision. ResNet is a deep learning model in which the weight layers learn residual functions with reference to the layer inputs. The fundamental difference between ResNet and classical convolutional neural networks is shortcut connections. Thanks to them, ResNet can contain a lot of layers while bypassing the vanishing gradient problem, which helps to catch more features from input images.
Visual image transformer (VIT) and Data efficient image transformer (DeIT) are the newest architecture in computer vision at this moment. They are based on transformer architecture, which was created for natural language processing tasks. The main idea of these architectures is to use not pixels as layers, but fixed-size image pieces called tokens (or patches). These tokens processing like texts tokens on NLP, using attention layers for getting image embeddings. DeiT can be considered as VIT model with an additional distillation token.
DenseNet is modified ResNet. DenseNet establishes a dense connection between all the front and back layers. Another important feature of DenseNet is the implementation of feature reuse via feature bonding on a channel. These features allow DenseNet to achieve higher performance than ResNet with fewer parameters and computational overhead.
VGG architecture is one of the first convolution neural networks models. Its main idea is to learn by parts. This is achieved by adding new layers to the old frozen layers and training model.
The ideas behind Inception were first created by Google in their GoogLeNet model. This model was based on inception blocks, which parallel apply on previous layer different convolution filters and then concatenate results. The authors of Inception changed the kernel size and number of convolutional filters, which made it possible to reduce the number of blocks and size of model.
Xception is an upgrade of Inception based on xception blocks. Unlike the inception block, it applies a 1x1 convolution to the previous layer, splits it into channels, and applies its own convolution filter with the same kernel size to each. This mechanism allows the network to be even more compact and give better results.
The training parameters for all models are shown in
Table 4. The rest parameters had default values.
2.3. Metrics
The following metrics were used for the evaluation of the quality of classification models for the imbalanced dataset:
where TruePositive—the number of images which were classified correctly; FalseNegative—the number of images that should have been assigned to this class but were not; FalsePositive—the number of images that were erroneously assigned to this class; TrueNegative—the number of images correctly marked as not being in this class.
Obviously, and
Note, that the accuracy could be misleading for imbalanced datasets. But since we formed balanced datasets, we can use this metric for the quality evaluation.
4. Discussion
As can be seen in
Table 5Table 6Table 7Table 8Table 9Table 10Table 11Table 12Table 13Table 14 and
Table 5, training on an unbalanced dataset leads to the fact that the model can poorly see the small classes, or not see at all. ResNet models are very sensitive to class imbalance. So, the ResNet34 model does not see the C5 class at all when training on the initial dataset (TPR = 0.00), but TPR≈0.46 after training on small datasets (small_ds_1, small_ds_2, small_ds_3 and aug_ds). For ResNet50, we can see an increase of TPR for class C5 from 0.29 (initial ds) to ≈0.58(small ds) and 0.60 (aug_ds). For the C6 class, the TPR increases from 0.56 (initial ds) to 0.77 (aug_ds). TPR for class C5 for ResNet101 increases from 0.15 (initial ds) to 0.67(aug_ds).
For VIT models we can improve TPR by 0.5 for small classes if we will use a balanced dataset. For example, VIT-base-patch16-224: TPR = 0.12(initial ds) and TPR = 0.61 (aug_ds). DeIT models are more stabilized for the training on an unbalanced dataset. For them, the improvement in metrics is no more than 0.2.
DenseNet proved to be very sensitive to training on an unbalanced dataset. For them, training on a balanced dataset is preferable, then class recognition metrics will be significantly improved (by 0.6 or more).
A similar conclusion can be drawn for Inception_v3, Xception, and VGG. The training these models on balanced dataset allow improve metrics by 0.2 for some classes.
It should be noted that there is no deterioration in the recognition of classes that contained a large number of objects in the initial data set. It can also be seen from the obtained results that the metrics on the “small_ds_1”, “small_ds_2”, “small_ds_3” datasets are different and obviously depend on those objects that remained in these datasets after the initial dataset was cut off.
The best and the most stable metrics of the neural networks under consideration were obtained on “aug_ds” dataset, which was formed from the initial dataset, when the initial dataset was cropped by the number of images in C6 class + 15%. Then the missing images in all classes were augmented.
Thus, it seems appropriate for unbalanced datasets to form balanced datasets on their basis according to the described methodology, and train neural network models using the resulting balanced dataset.
Author Contributions
Conceptualization, M.B.; methodology, M.B.; software, S.O. and I.U.; validation, S.O., I.U. and A.S.; formal analysis, S.O.; investigation, I.U.; resources, M.B.; data curation, A.S.; writing—original draft preparation, I.U. and S.O.; writing—review and editing, M.B.; visualization, S.O.; supervision, M.B. and A.S.; project administration, M.B.; funding acquisition, M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huang, G.-B.; Zhou, H. Extreme Learning Machine for Regression and Multiclass Classification. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics) 2012, 42, 513–529. [Google Scholar]
- Har-Peled, S.; Roth, D.; Zimak, D. Constraint classification: A new approach to multiclass classification. Algorithmic Learning Theory: 13th International Conference 2002, 365–379. [Google Scholar]
- Li, T.; Zhang, C.; Ogihara, M. A comparative study of feature selection and multiclass classification methods for tissue classification based on gene expression. Bioinformatics 2004, 20, 2429–2437. [Google Scholar]
- Suri, M.; Parmar, V.; Sassine, G.; Alibart, F. OXRAM based ELM architecture for multi-class classification applications. International Joint Conference on Neural Networks (IJCNN) 2015, 1–8. [Google Scholar]
- Jobi-Taiwo, A. A.; Cudney, E. A. Mahalanobis-Taguchi system for multiclass classification of steel plates fault. International Journal of Quality Engineering and Technology 2015, 5, pp. 25–39. [Google Scholar]
- Huang, P.; Tan, X.; Chen, C.; Lv, X.; Li, Y. AF-SENet: Classification of Cancer in Cervical Tissue Pathological Images Based on Fusing Deep Convolution Features. Sensors 2020, 21, 122. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, S. S.; Hiremath, J.; Kulkarni, V. V.; Harshit, B. C.; Kumar, S.; Hiremath, M. S. Facial Expression Recognition Using Transfer Learning with ResNet50. 2023, 281–300. [Google Scholar] [CrossRef]
- Huang, J.; Gong, W.-J.; Chen, H. Menfish Classification Based on Inception_V3 Convolutional Neural Network. 2019, 677(5), 052099–052099. [Google Scholar] [CrossRef]
- Fajri, D. M. N.; Mahmudy, W. F.; Yulianti, T. Detection of Disease and Pest of Kenaf Plant Based on Image Recognition with VGGNet19. Knowledge Engineering and Data Science 2021, 4(1), 55. [Google Scholar] [CrossRef]
- Swaraj, A.; Verma, K. Classification of COVID-19 on Chest X-Ray Images Using Deep Learning Model with Histogram Equalization and Lungs Segmentation. 2021. [Google Scholar] [CrossRef]
- Mahmoud, S.; Gaber, M. ; Gamal Farouk; Arabi Keshk. Heart Disease Prediction Using Modified Version of LeNet-5 Model. 2022, 14(6), 1–12. [Google Scholar] [CrossRef]
- Juan Augusto, Campos-Leal; Yee-Rendon, A.; Vega-Lopez, I. F. Juan Augusto Campos-Leal; Yee-Rendon, A.; Vega-Lopez, I. F. Simplifying VGG-16 for Plant Species Identification. 2022, 20(11), 2330–2338. [Google Scholar] [CrossRef]
- Josephine, Atchaya; J., Anitha; Asha Gnana, Priya; J. Jacinth, Poornima; Hemanth, J. Josephine Atchaya; J. Anitha; Asha Gnana Priya; J. Jacinth Poornima; Hemanth, J. Multilevel Classification of Satellite Images Using Pretrained AlexNet Architecture. 2023, 202–209. [Google Scholar] [CrossRef]
- Liao, J.-J.; Zhang, J.-W.; Liu, B.-E.; Lee, K.-C. Classification of Guide Rail Block by Xception Model. 2022, 2(4), 17–24. [Google Scholar] [CrossRef]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in Medical Image Analysis. Intelligent Medicine 2023, 3(1), 59–78. [Google Scholar] [CrossRef]
- Heidari, M. ; Amirhossein Kazerouni; Milad Soltany; Azad, R.; Ehsan Khodapanah Aghdam; Julien Cohen-Adad; Dorit Merhof. HiFormer: Hierarchical Multi-Scale Representations Using Transformers for Medical Image Segmentation. 2023. [Google Scholar] [CrossRef]
- Barulina, M.; Sanbaev, A.; Okunkov, S.; Ulitin, I.; Okoneshnikov, I. Deep Learning Approaches to Automatic Chronic Venous Disease Classification. Mathematics 2022, 10, 3571. [Google Scholar] [CrossRef]
- Gangsar, P.; Tiwari, R. Comparative Investigation of Vibration and Current Monitoring for Prediction of Mechanical and Electrical Faults in Induction Motor Based on Multiclass-Support Vector Machine Algorithms. Mechanical Systems and Signal Processing 2017, 94, 464–481. [Google Scholar] [CrossRef]
- Ceyhan, M.; Kartal, Y.; Özkan, K.; Seke, E. Classification of Wheat Varieties with Image-Based Deep Learning. 2023. [Google Scholar] [CrossRef]
- Jiang, Z.-P.; Liu, Y.-Y.; Shao, Z.-E.; Huang, K.-W. An Improved VGG16 Model for Pneumonia Image Classification. Appl. Sci. 2021, 11, 11185. [Google Scholar] [CrossRef]
- Jain, G.; Mittal, D.; Thakur, D.; Mittal, M. K. A Deep Learning Approach to Detect Covid-19 Coronavirus with X-Ray Images. Biocybernetics and Biomedical Engineering 2020, 40(4), 1391–1405. [Google Scholar] [CrossRef]
- Vianna, V. P. Study and development of a Computer-Aided Diagnosis system for classification of chest x-ray images using convolutional neural networks pre-trained for ImageNet and data augmentation. arXiv preprint arXiv 2018. submitted. [Google Scholar]
- Antonie, M. L.; Zaiane, O. R.; Coman, A. Application of data mining techniques for medical image classification. Proceedings of the Second International Conference on Multimedia Data Mining 2001, 94–101. [Google Scholar]
- Bellazzi, R.; Zupan, B. Predictive Data Mining in Clinical Medicine: Current Issues and Guidelines. International Journal of Medical Informatics 2008, 77(2), 81–97. [Google Scholar] [CrossRef]
- Tummala, S.; Kadry, S.; Bukhari, S. A. C.; Rauf, H. T. Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling. Current Oncology 2022, 29(10), 7498–7511. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.; Gan, S.; Zhang, L. Human-Computer Interaction Based Health Diagnostics Using ResNet34 for Tongue Image Classification. Computer Methods and Programs in Biomedicine 2022, 226, 107096. [Google Scholar] [CrossRef] [PubMed]
- None Galih Wasis Wicaksono; None Andreawan. ResNet101 Model Performance Enhancement in Classifying Rice Diseases with Leaf Images. 2023, 7(2), 345–352. [CrossRef]
- Faisal Dharma Adhinata; Nur Ghaniaviyanto Ramadhan; Akhmad Jayadi. DenseNet201 Model for Robust Detection on Incorrect Use of Mask. 2023, 251–263. [CrossRef]
- Chakkrit, Termritthikun; Umer, A.; Suwichaya, Suwanwimolkul; Xia, F.; Lee, I. Chakkrit Termritthikun; Umer, A.; Suwichaya Suwanwimolkul; Xia, F.; Lee, I. Explainable Knowledge Distillation for On-Device Chest X-Ray Classification. 2023, 1–12. [Google Scholar] [CrossRef]
- Murphy, Z. R.; Venkatesh, K.; Sulam, J.; Yi, P. H. Visual Transformers and Convolutional Neural Networks for Disease Classification on Radiographs: A Comparison of Performance, Sample Efficiency, and Hidden Stratification. Radiology: Artificial Intelligence. Radiology: Artificial Intelligence 2022, 4(6). [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).