1. Introduction
Antibiotic resistance, the ability of bacteria to withstand antibiotics, is now recognized as one of the most serious global threats to human health today [
1,
2,
3]. The naturally occurring resistance that finally can lead to incurable bacterial infections could be accelerate by the improper use of antibiotics in human beings and animals [
4]. Except for the mistakes in the use of antibiotics in human medicine, antibiotics abuse in livestock is also a major contributor to the emergence of antibiotic resistance [
5]. To satisfy the growing global demand for animal protein, the massive and expending use of antibiotics in the farmed animal industries for different purposes, including overdose treatment for disease prevention and subtherapeutic doses for growth stimulation [
6]. In September 2017, a report from the World Health Organization (WHO) confirms that the world is running out of antibiotics [
7,
8]. Antibiotics abuse, if left unchecked, can drag human beings into a post-antibiotic era where minor injuries or common infections become fatal diseases again [
9,
10].
To protect the public from health risks, nations and related organizations have to establish more broader maximum-residue-limits (MRLs) for further surveillance of antibiotic residues in animal food[
11,
12]. Therefore, more efficient and robust detection methods were developed promptly to satisfy the more and more rigorous regulatory requirements during the past few years [
13,
14,
15,
16,
17,
18,
19]. The Triple Quadrupole Mass Spectrometer with multiple reaction monitoring (MRM) scan modes that go best with the requirement of ECD 2002/657/EC should be the preferred method of detection of antibiotic residues in animal food[
20,
21]. The Qtrap system (AB Sciex) with the scan mode of multiple reaction monitoring - information dependent acquisitim - enhenced product ion scan mode (MRM - IDA - EPI) was found to efficiently gather comprehensive information from samples in a single run. Consequently, the identification of antibiotic residues in locally sourced animal-derived food was reaffirmed through the successful comparison of antibiotic spectra from the samples with reference spectra. Quinolones and tetracyclines, being the most extensively employed veterinary antibiotics, have contributed to the development of antibiotic hyposensitivity, thereby adversely impacting the treatment of severe bacterial infections [
23,
24,
25,
26]. In this study, a diverse range of samples, fortified with fifteen quinolones and seven tetracyclines antibiotics at the Recommended Concentration (RC) of 1 µg L
−1,were subjected to analysis using liquid chromatography-tandem mass spectrometry on a QTRAP 5500 instrument.
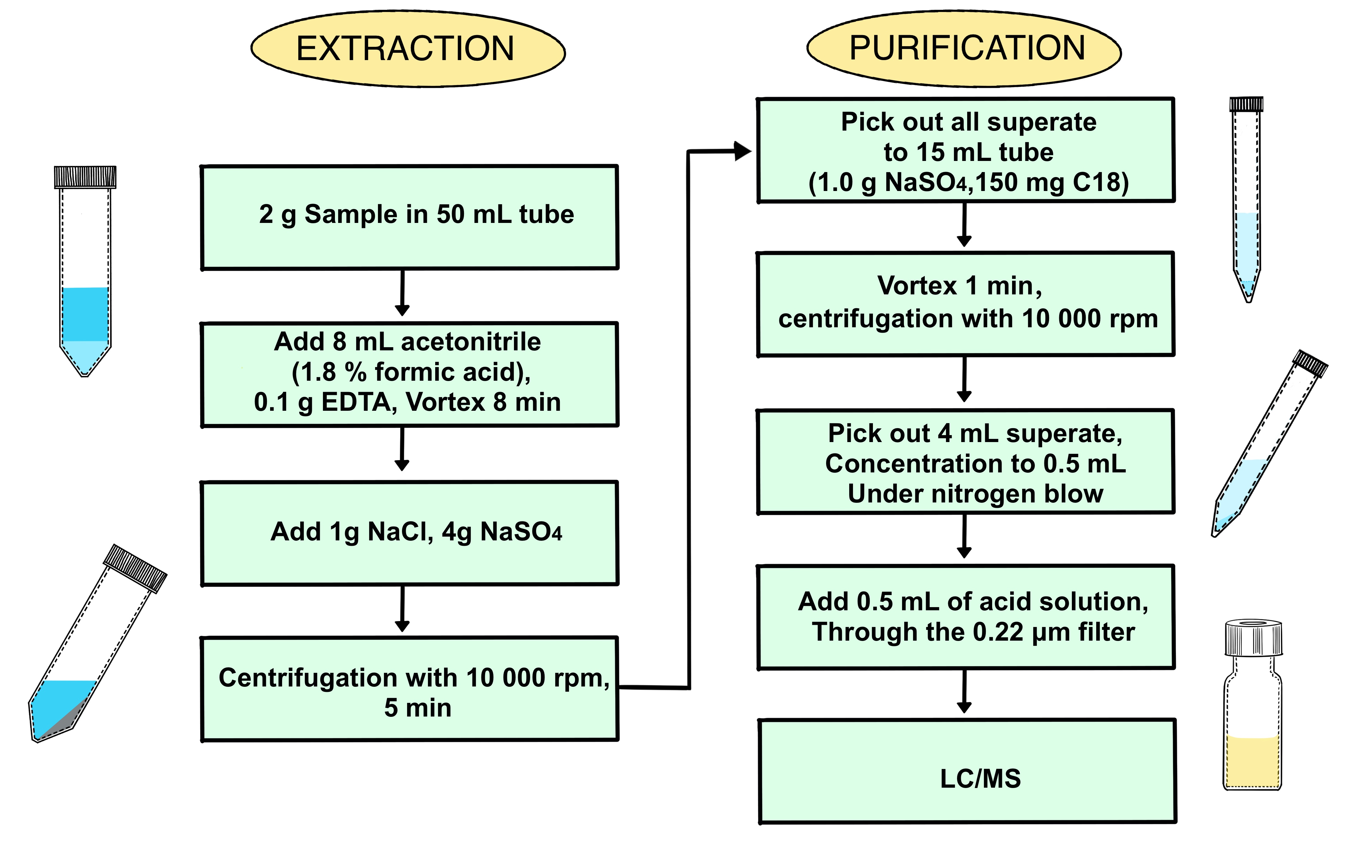
The sample pretreatment for the separation and concentration of antibiotic residues is also a critical step in the whole analysis process for quinolones and tetracyclines detection. There are alternatively two purification methods including QuEChERS and Solid-phase extraction. The solid-phase extraction using commercial cartridges have been widely used in daily work based on the former research (
Figure A1) [
27]. It could be performed in an automated SPE system (e.g., Reeko, Fotector plus, USA) on non-working hours and minimize human involvment (
Table A1), but be excluded by high-fat or high-protein samples that cause blockage in SPE cartridges frequently and prolong the pretreatment process heavily. Since its development in 2003, QuEChERS has gained widespread acceptance in various sample preparation techniques [
28,
29,
30,
31,
32]. It was initially introduced as cost-effective and time-efficient method for analyzing multi-residue samples containing relatively polar compounds. During the extraction process, the efficiency of the QuEChERS sample preparation method is known to be influenced by several factors. It is imperative to comprehensively optimized the QuEChERS method for the detection of quinolones and tetracyclines residues in this study. As a collection of statistical and mathematical techniques, Response surface methodology (RSM) has important applications in the design, development and improvement of new or existing product designs [
33,
34], especially in the multi-variable analysis[
35,
36]. In this study, response surface methodology with a multi-level-four-factor Box-Behnken design (BBD) was applied to simultaneously evaluate the rate of recovery for quinolones and tetracyclines residues in sample pretreatment.
2. Materials and Methods
2.1. Standards and stock solutions
Standards of twenty-two antibiotics: Oxytetracycline (Oxytetracycline hydrochloride), Tetracycline (Tetracycline hydrochloride), Doxycycline (Doxycycline hyclate), Demeclocycline, Methacycline, Minocycline, Chlortetracycline (Chlortetracycline hydrochloride), Enrofloxacin, Norfloxacin, Pefloxacin (Pefloxacin methanesulfonate dehydrate), Ciprofloxacin (Ciprofloxacin hydrochloride), Ofloxacin, Sarafloxacin (Sarafloxacin hydrochloride), Enoxacin, Lomefloxacin (Lomefloxacin hydrochloride), Pipemidic acid, Nalidixic acid, Oxolinic acid, Flumequine, Cinoxacin, Danofloxacin (Danofloxacin mesylate) and Difloxacin (Difloxacin hydrochloride) were all purchased from Dr. Ehrenstorfer GmbH (Augsburg, Germany). Individual standards were weighed by an electronic balance (Metter Toledo, MS 205DU) and prepared in HPLC-grade methanol at a concentration of 1.0 mg mL−1 and stored at −28 °C temporarily, or ethanol solution of potassium hydroxide, for some antibiotics were practically insoluble in methanol.
2.2. Chemicals and Reagents
Acetonitrile and Methanol of HPLC grade were purchased from Merck (Darmstadt, Germany). Ethanol and formic acid (≥98%) of HPLC grade were purchased from Aladdin (Shanghai, China). Potassium hydroxide (G.R.) was purchased from Macklin (Shanghai, China). High purity water was obtained from a Milli-Q water purification system from Millipore (Bedford, USA). Cleanert® C18 for QuEChERS was purchased from Agela Technologies (Beijing, China). Oasis® HLB SPE cartridges (6cc, 200mg) was purchased from Waters (Milford, USA). Millipore filters of polytetrafluoroethylene (0.22 μm) were purchased from ANPEL Lab (Shanghai, China). Sodium chloride (A.R.), sodium sulphate (A.R.), citric acid (A.R.), disodium hydrogen phosphate (A.R.) and disodium ethylenediamine tetraacetic acid (EDTA, A.R.) for sample preparation were purchased from Sinopharm Chemical Reagent (Shanghai, China).
2.3. Instrumentation and software
For HPLC analysis, a Shimadzu LC-30AD system (Shimadzu, Japan) consisting of two interconnected pump units, a refrigerated autosampler, and a column oven compartment was used. A Waters BEH C18 column (1.7 µm 2.1 mm×100 mm, Waters, USA) was applied for analysis. Mass spectrometric detection was conducted on an AB SCIEX QTRAP® 5500 (AB SCIEX instruments, Foster City, Canada) in MRM mode and EPI mode. A Turbo V™ Ion Source (ESI) interface in positive ionization mode was used. Both the UPLC and mass spectrometer were controlled remotely using Analyst® software v. 1.6.2 (AB SCIEX instruments, Foster City, Canada). MS was optimized using a capillary voltage of 5.50 kV and desolvation temperature of 500 °C. The cone gas pressure and desolvation gas pressure were 50 psi. Nitrogen from generator (Claind Nitro35, Italy) was used as the cone and collision gasses, respectively. The raw data was analyzed using a MultiQuant® 3.0.1. The optimization of QuEChERS by Response Surface Method was performed on a Design Expert® 10.0.3.
2.4. Sample collection and processing
Samples were collected from local markets or supermarkets distributed randomly across neighborhoods of whole city, including swine, poultry, eggs, milk and eight cultured aquatic products (Parabramis pekinensis, Carassius auratus, Ctenopharyngodon idella, Ophiocephalus argus Cantor, Macrobranchium nipponense, Macrobrachium rosenbergii, Penaeus chinensis, Procambarus clarkia, Eriocheir sinensis, Larimichthys crocea). The edible part of each fresh sample was crushed and mixed into homogenate by a Mixer (BÜCHI, B400, Switzerland), and stored in polypropylene bottles at −28 °C until analysis.
3. Results
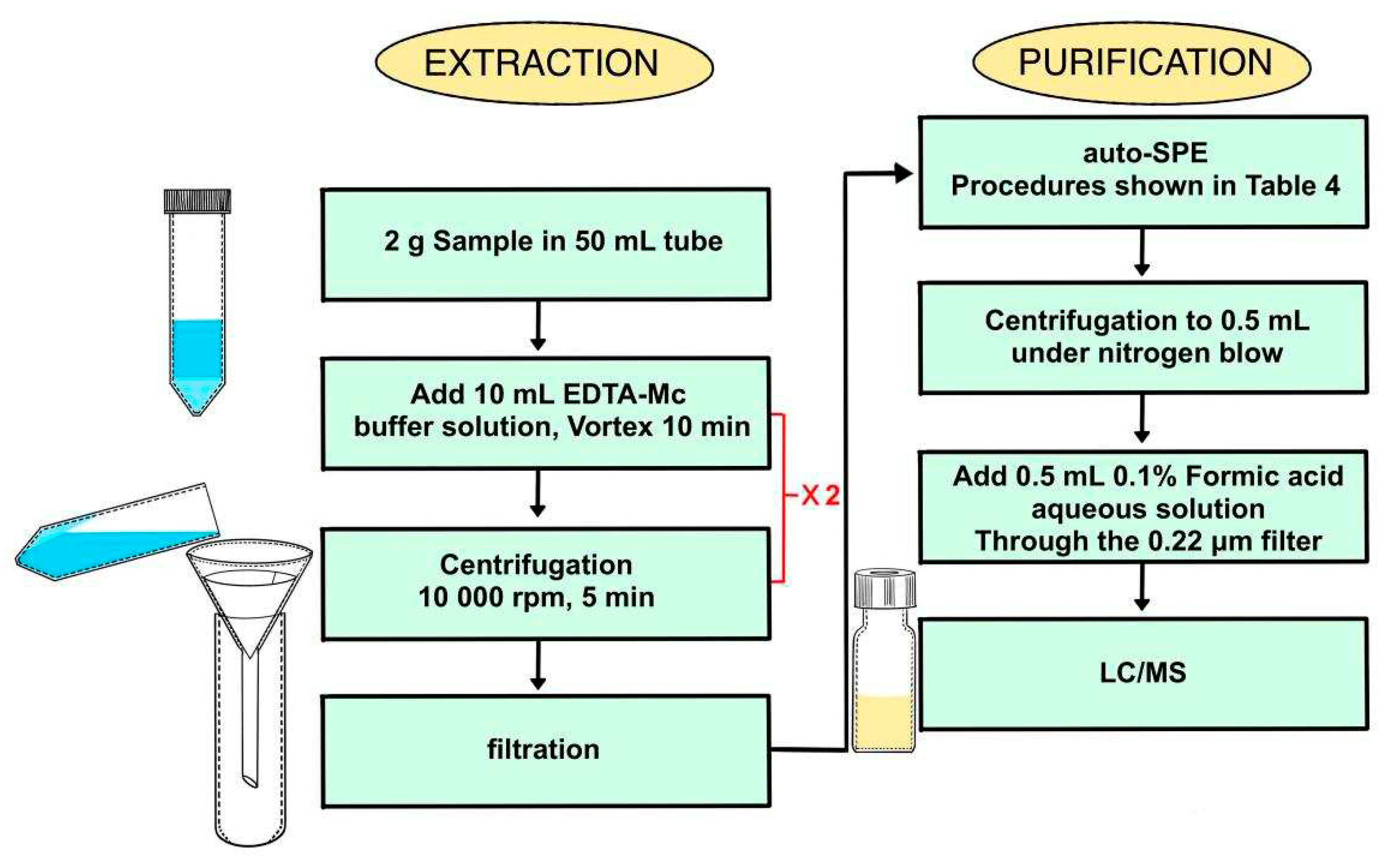
In the sample pretreatment process with SPE extraction method, 0.1 mol L
−1 EDTA-Mcllvaine buffer solution (pH=4.0) is applied as the extraction solution for quinolones and tetracyclines residue [
27,
37,
38,
39], which is non-volatile, non-poisonous, low-cost, eco-friendly. Furthermore, when handling a large amount of sample, the SPE method could enhance the method stability through increasing the level of automation, without adding man-hours (
Table A1). However, in the flowing sample purification procedure, the aqueous solution extracted from high-fat samples would cause blockage in SPE cartridges frequently and prolong the detection time heavily, especially when egg sample was handled.
3.1. Experimental design
The QuEChERS sample preparation method could be applied as an alternative to quinolones and tetracyclines residue quick analysis in high-fat samples. As former research, the efficiency of the QuEChERS sample preparation method for quinolones and tetracyclines residue could be relevant to multifarious factors including volume of acetonitrle, the pH value of the extract solvent, the duration of the extraction process and so on [
28,
29,
30,
31,
32,
33].
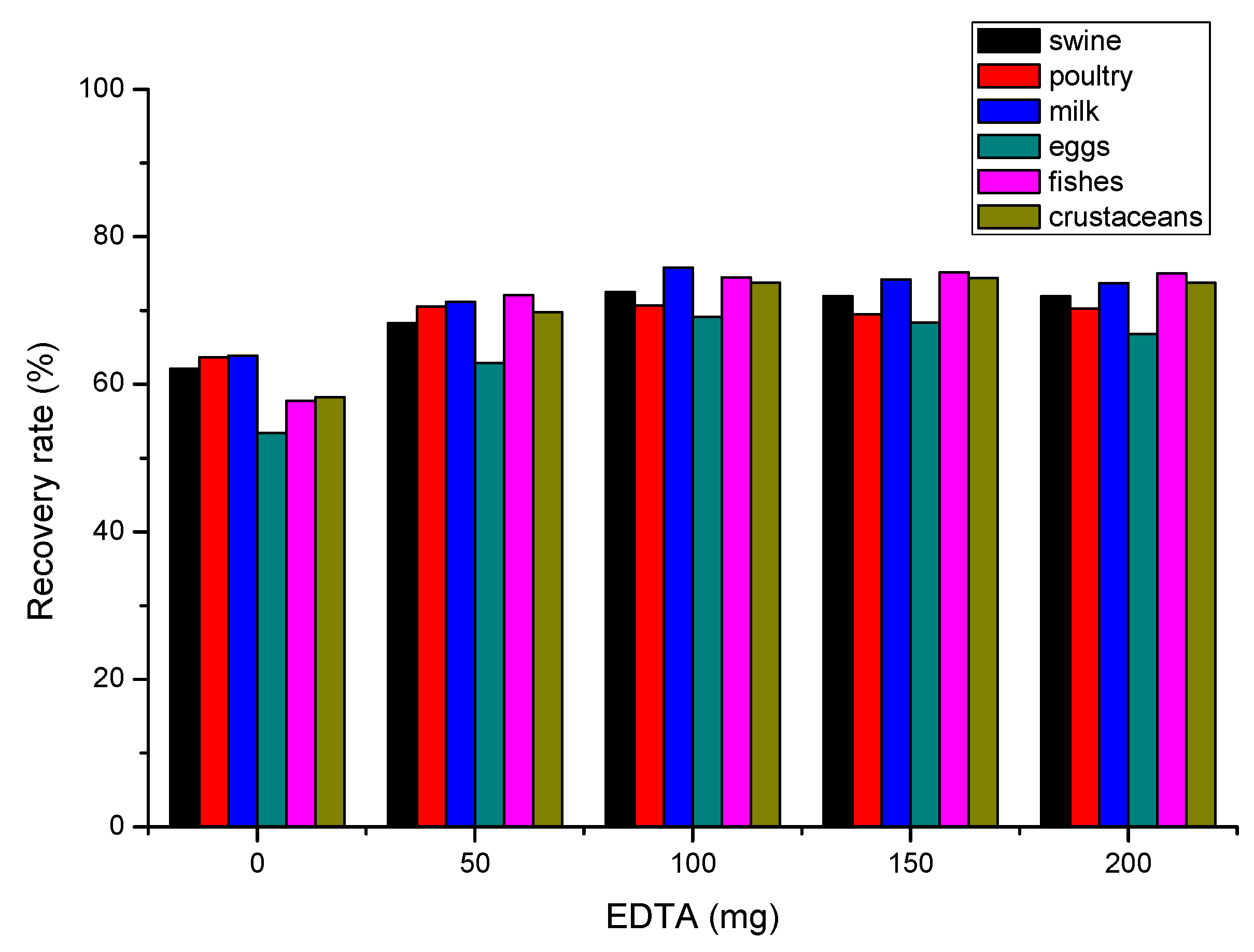
Because of the special chemical structure of quinolones and tetracyclines, the agent-agent interactions [
39,
40] between antibiotics and metal ion from experimental environment could have an effect on the recovery rate. As in EDTA-Mcllvaine buffer solution of SPE method, EDTA worked as screening agent to metal ions could also be added in the extraction procedure of QuEChERS method. As shown in
Figure 1.,when the addition of EDTA ranged from 0 to 0.2 g in 2 g of sample by QuEChERS preparation, the medium recovery rate of 22 antibiotics were slowly ascending to a plateau.
3.2. Optimization of QuEChERS method by Response Surface Method (RSM)
The QuEChERS sample preparation method is multivariable for optimization of the extraction process. The efficiency of the QuEChERS sample preparation method could be relevant to multifarious factors including volume of the acetonitrle (A), additive amount of formic acid (B), additive amount of EDTA (C) and extraction time (D). The univariate experiment has limited ability in evaluating the interactions of the extraction conditions in sample preparation. To avoid the interactions among all the extraction conditions while examining the most optimal extraction process, all the possible factors were comprehensively optimized by the response surface method (RSM) including volume of the acetonitrle (A), additive amount of formic acid (B), additive amount of EDTA (C) and extraction time (D). The median recovery rate of 22 antibiotics from standard samples (2.0 μg kg-1, n =3) was chosen as the response. The RSM analysis could model the relationship between the response (recovery rate) and the four factors. According to former research [
41], the respective low and high levels for factors were coded.
The Model
F-value of 15.79 implied the model was significant. This model can be used to navigate the design space. The final equation in terms of actual factors is:
Th
The
P-value was usually used to check the significance of variables and it also could reflect the interactions among all the independent variable [
42]. The smaller
P-value indicated the corresponding variable was more significant [
43]. As the ANOVA for response surface quadratic model shown in
Table 2, in this case, A, B, C, D, A
2, B
2, C
2, D
2 were all significant model terms, and the variable volume of the acetonitrle (A) and extraction time (D) were more significant for the recovery rate.
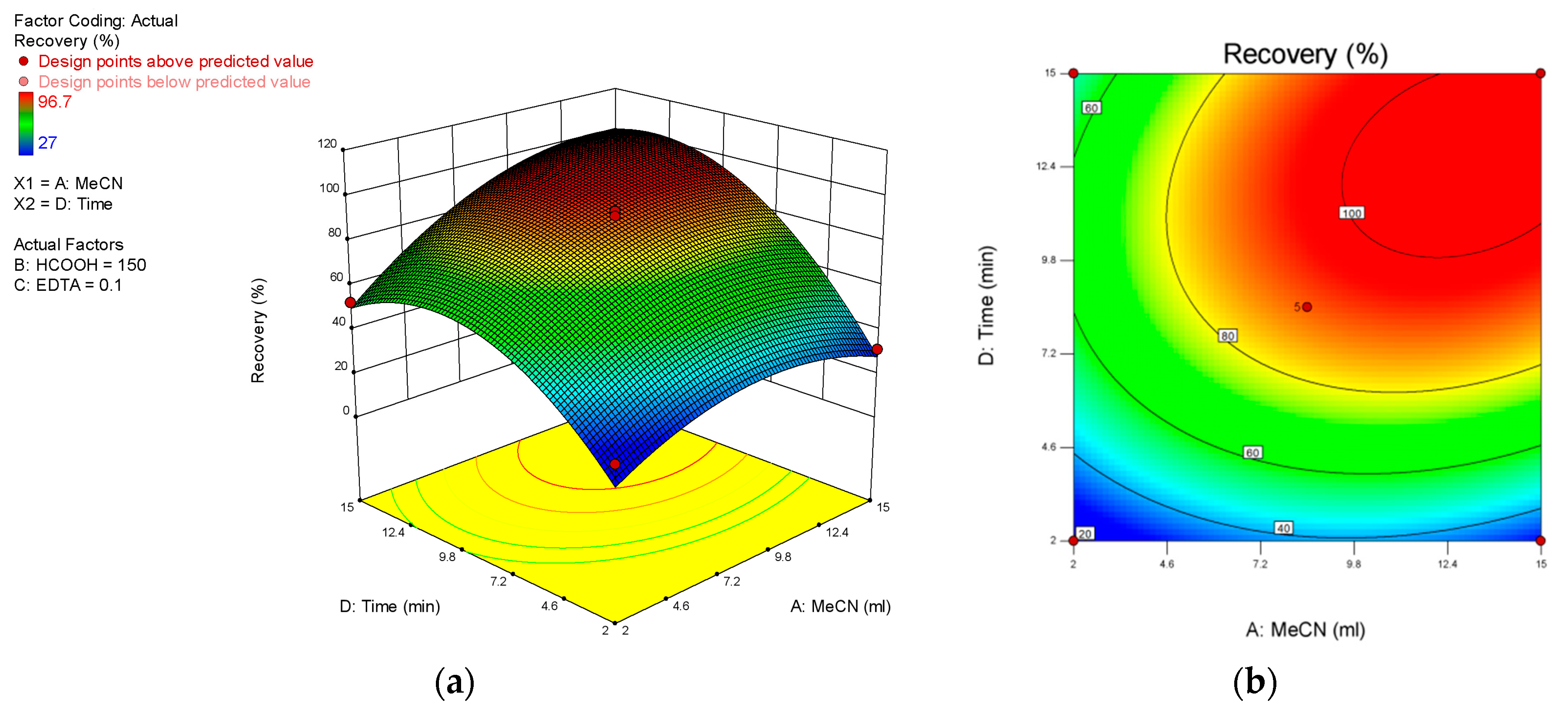
As shown in
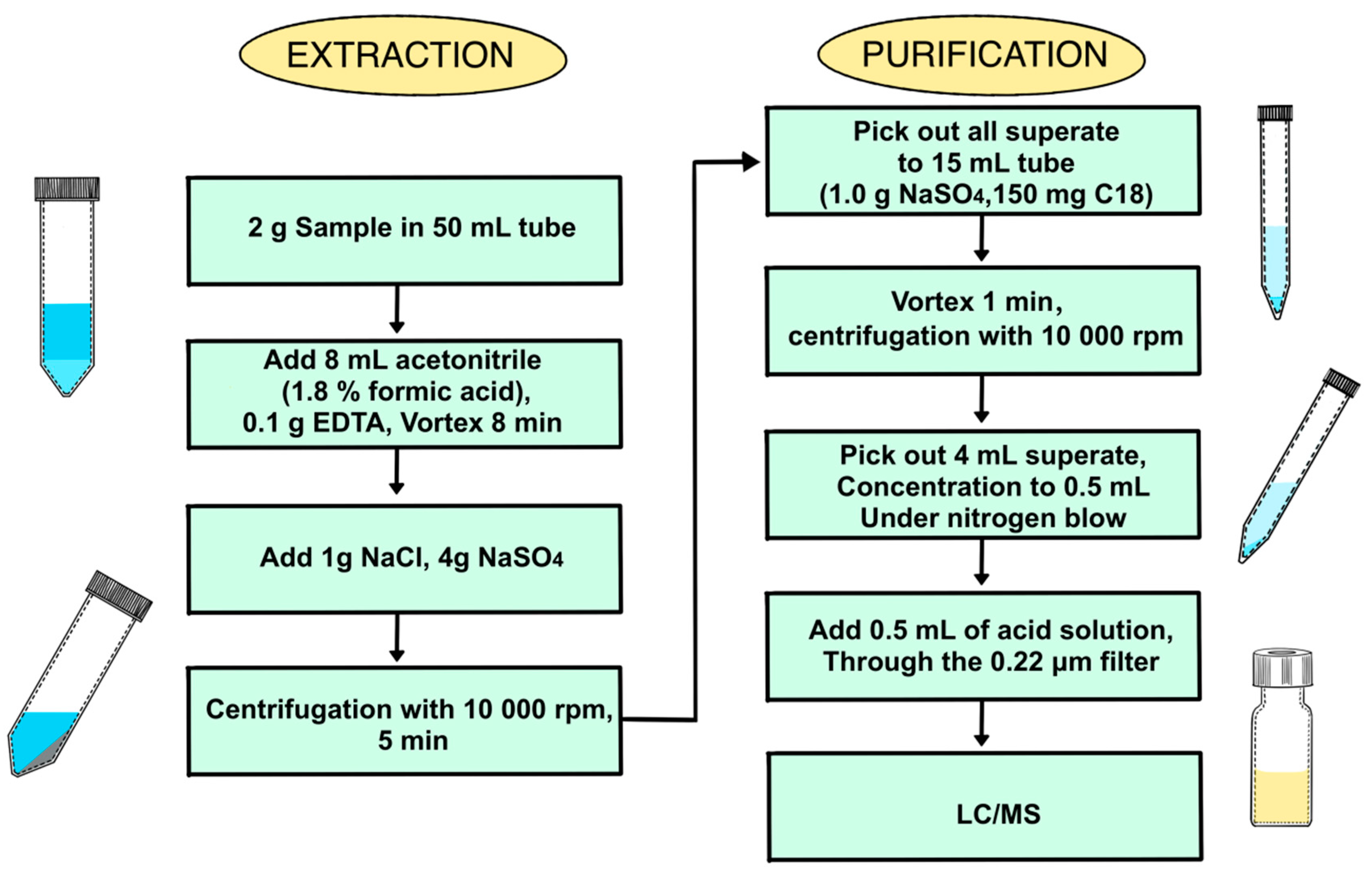
Figure 2, among all the solutions when A = 8 mL, B = 150 μL, C = 0.1 g, D = 8 min, the recovery rate of 22 antibiotics can reach the best. The verification tests were performed six times under the conditions optimized above. The median recovery rates of 22 antibiotics from six parallel tests were 75.4%, 81.6%, 85.9%, 73.6%, 77.9% and 82.5%, with errors range from 15.1% to 2.8%. The samples preparation of QuEChERS method was finally optimized as the best solution from RSM, and the method was presented as
Figure 3.
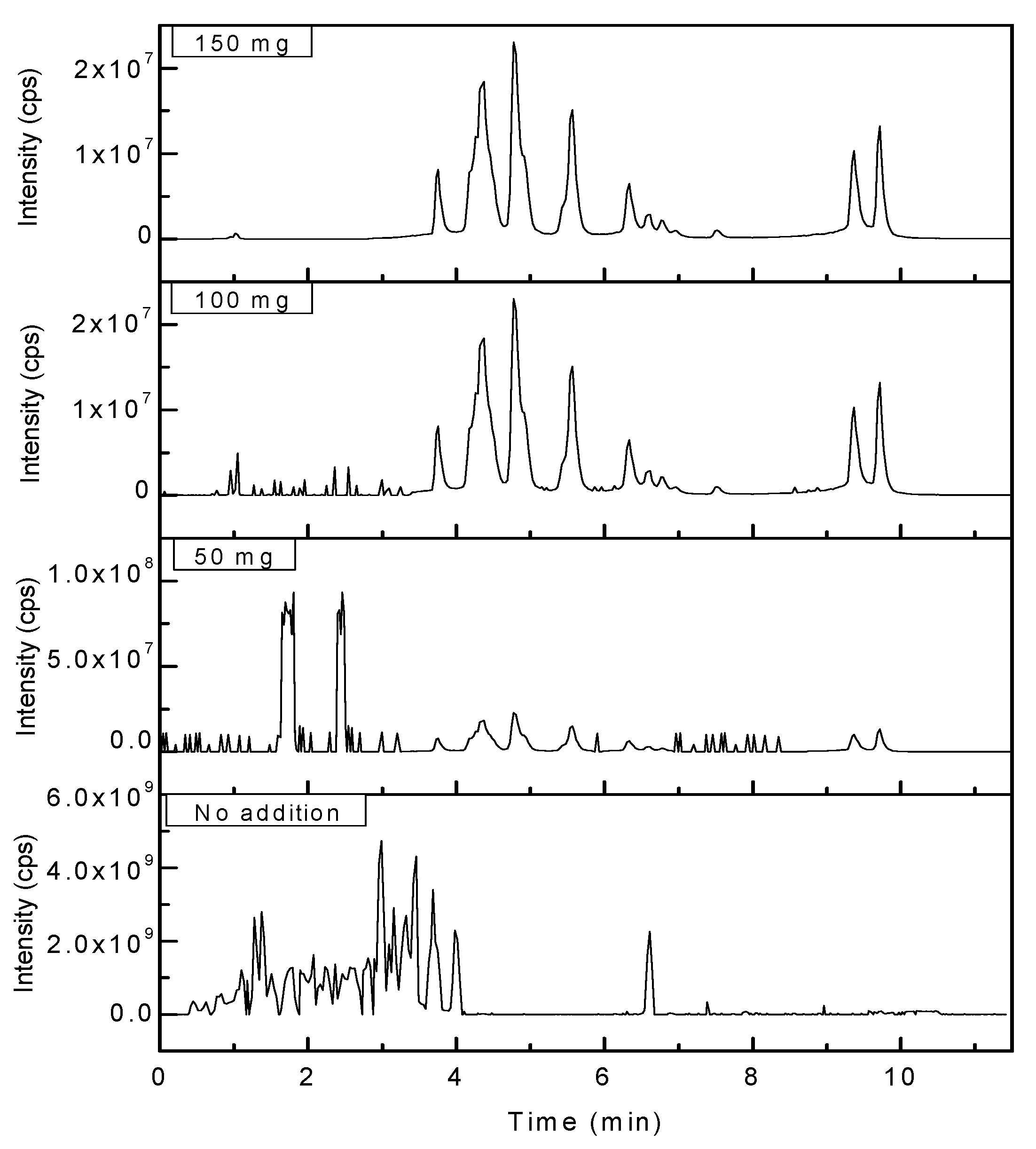
As for the process of purification, the addition of cleaning agents, namely, C18 powder was evaluated in fishes for the highest detection rate of antibiotic residue. With the right amount of cleaning agent the crude extraction of spiked sample was purified well, when 150 mg of C18 powder was added (see in
Figure 4).
3.3. Optimization of mass spectrometry and chromatographic conditions
The basic structures of quinolines and tetracyclines both contain several O and N atoms are easy to obtain protons and have high response in positive ion mode. To confirm the two ion pairs for quantification, the mixed standard solution of 22 antibiotics with a concentration of 100 µg L
-1 was infused into the QTRAP mass spectrometer at a flow rate of 7.0 µL min-1 to acquire automatic analytes optimization by the ESI positive mode. In the optimal mass spectrometry conditions including Declustering Potential and Collision Energy, every antibiotic was assigned two pairs of abundant ions for qualitative and quantitative with the high sentitivity. For the minimum retention time and the symmetric shape of the ionic peaks, this study optimized the elution type, flow rate, gradient on the basis of the most general chromatographic column C18. Therefore, several classical compositions of the mobile phase were performed including methanol, acetonitrile, water as well as water with ammonium acetates or formic acid. Finally, water and acetonitrile that both supplemented with 0.1% formic acid was chose as the optimal mobile phase. The final gradient elution with the total flow rate of 0.3 mL min-1 was as follows: 0-0.5 min,5-20% A; 0.5-2.0 min, 20-25% A; 2.0-7.0 min, 25-45% A; 7.0-10.0 min, 45-90% A 10.0-12.0 min, 90% A, and 12.1-13.0 min, 95-5%. The column oven was maintained at 40℃ and the injection volume was 10.0 µL. The representative LC-QTRAP-MS/MS chromatograms were merged in
Figure 4. The retention time (RT) and MS information for each antibiotic including precursor and product ions, DP and CE were shown in
Table 2.
The EPI scan mode could be activated in IDA experiment when the ionic intensity exceeded the threshold of 1000 cps. The scan time (including pauses) was totally 1.57 s for all MRM transitions. The EPI mass spectra were acquired over a mass range of m/z 50–500 at a scan rate of 10 000 Da s–1.
3.4. Fragmentation manner of Quinolones and Tetracyclines
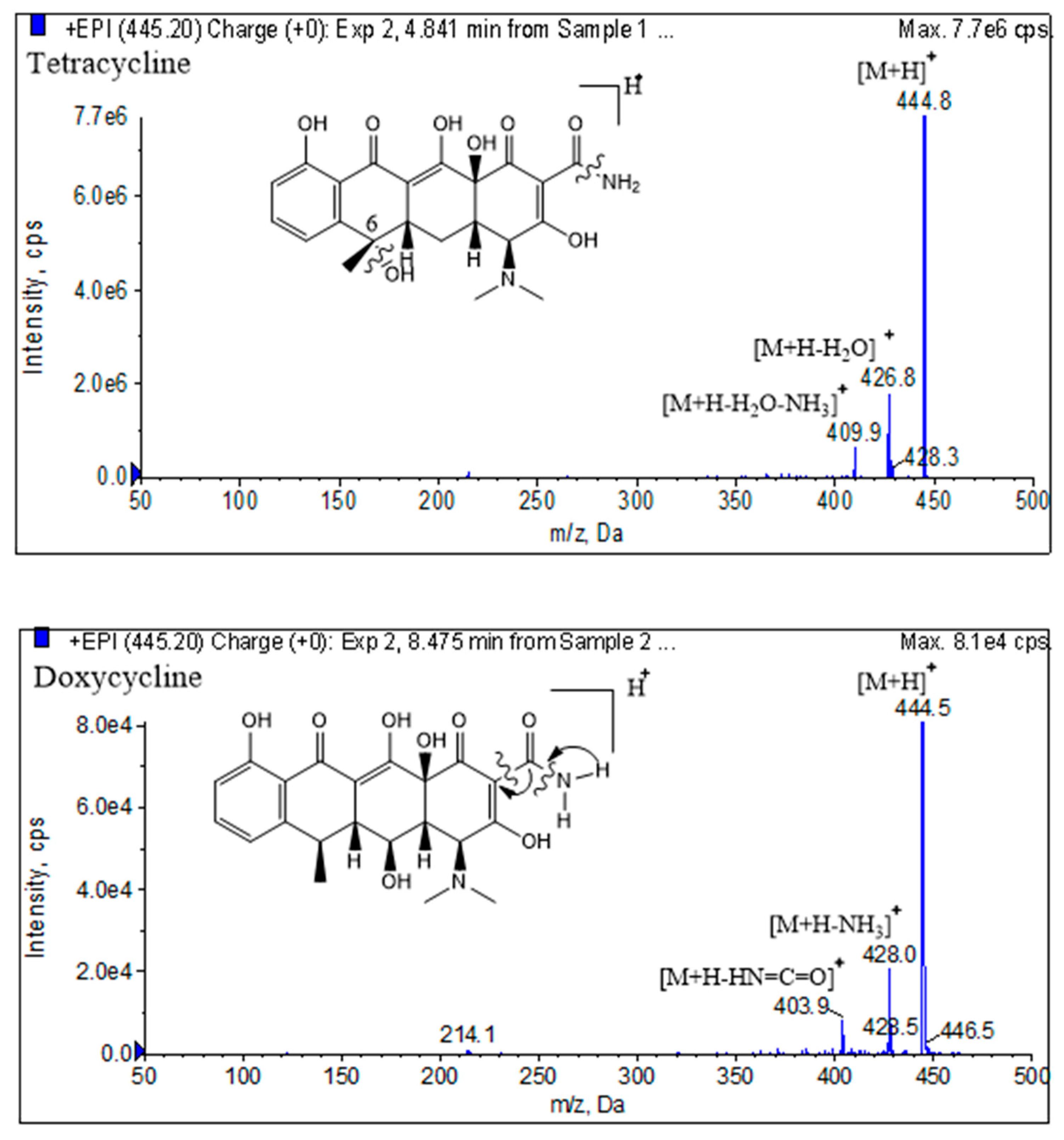
In the positive mode of electrospray ionization (ESI) mass spectrometry, the proton first attaches to the protonation site, usually at N atom or O atom, and then triggers the cleavages by migrating to the reactive center. Although the most basic site in the tetracyclines is the dimethylamino group, the protonated tetracyclines dissociate initially by loss of H
2O or NH
3 from acylamino group. For the Tetracycline, Demeclocycline and Chlortetracycline, there are no OH at C–6 site, are all present losses of H
2O and NH
3 successively. As shown in
Figure 5., tetracyclines without the tertiary OH at C–6 lose only NH
3 initially [
43].
According to the spectra from EPI mode, the reactive center of quinolines is located in carboxylic acid group. The abundant fragment ion [M+H–H
2O]
+ is formed by the dehydration of –COOH, while another abundant fragment ion characterized the decarboxylation of this group. The neutral loss of
m/z 20 Da and
m/z 30 Da are most probably formed from the dissociation of –HF or –CH
2CH
3. And another characteristic neutral loss of –CH=CH–NH
2 (
m/z 20 Da) is produced from the cracking of the azine ring [
44].
3.5. Method validation
3.5.1. Matrix Effect
To evaluate the matrix effects in LC-MS detection, six distinct types of antibiotic-free samples were used as matrix-matched blank on the ionization of 22 antibiotics residues. It was calculated using the following equation [
45]:
where AMatrix is the peak area of the standard solution with the matrix-matched blank and the AS is the peak area of the standard solution with initial mobile phase. The percentages of the matrix effects of 22 antibiotics at three different concentrations (2, 20, 200 ng mL-1) were ranged from 84.7 % to 119.3%. When ion suppression and ion enhancement at chosen levels were considered, the blank matrix matching standard curve should be adopted to eliminate the effect of matrix.
3.5.2. Linearity and sensitivity
Satisfactory linearities (R>0.99) were obtained for 22 antibiotics in blank matrix-matched curves over the concentration range of 0.5–200.0 ng mL-1. The sensitivity of the proposed method was measure by the limit of detection (LOD) and the limit of quantification (LOQ) values. The LOD and the LOQ were calculated using the following equations [
45]:
where S/N is the average signal to noise ratio; and C
S is the concentration of the specific antibiotic. The estimated values were testified by suitable spiked samples containing the 22 antibiotics at the corresponding concentrations. When the method concentration range over 0.5–200.0 ng mL
-1, the LOD and LOQ values were 0.3 μg kg
-1 and 1.0 μg kg
-1, respectively, it demonstrated the sensitivity of the method for antibiotic residues.
3.5.3. Accuracy and precision
The accuracy and precision of the method were measured by the recoveries and coefficients of variation (CVs) for intra- and inter-day. Therefore, the standard mixtures solution of 22 antibiotics was spiked into distinct types of samples including swine, poultry, eggs, milk, fishes and crustacea, and 18 spiked samples (six types at three concentrations of 2, 20, 200 μg kg-1) were obtained. All these spiked samples were detected three times intra- day and on five inter- days. The recovery of 22 antibiotic residues (73.8-98.5%) fell within the recommended guidelines of 60%–120% (GB/T 27404-2008,China). The analysis precision, measured as the coefficient of variation percentage (%CV) of the recovery (5.8%-12.4%), was well under the criteria of 30% (GB/T 27404-2008, China).
3.6. Sample Analyses
After validation of the analytical methodology through above experimentation, it was applied to detect numerous types of food samples including swine, poultry, eggs, milk and nine cultured aquatic products (Parabramis pekinensis, Carassius auratus, Ctenopharyngodon idella, Ophiocephalus argus Cantor, Macrobranchium nipponense, Macrobrachium rosenbergii, Penaeus chinensis, Procambarus clarkia, Eriocheir sinensis, Larimichthys crocea). In the recent six years of detection (2017-2022, total 781 samples), the quinolines or tetracyclines residues from swine, eggs, milk and Eriocheir sinensis were basically not detected. The tetracylines residues were occasionally detected but never exceed the MRL (GB31650-2019,China) from poultry, Macrobranchium nipponense, Macrobrachium rosenbergii, Penaeus chinensis and Procambarus clarkia. Quinoline residues were generally detected from the cultured aquatic products except for the Eriocheir sinensis. The detection rate of quinolines residues was highest in fishes (Parabramis pekinensis, Carassius auratus, Ctenopharyngodon idella, Ophiocephalus argus Cantor) ranged from 11.36% to 37.51%, and the over limit rate (MRL) ranged from 1.85% to 9.07%. Furthermore, Enrofloxacin and Ciprofloxacin were the dominant detected residues of all the 22 antibiotics. Meanwhile, among the total 12 kinds of samples, Parabramis pekinensis, shown medium detected concentration of Enrofloxacin and Ciprofloxacin respectively at 179.9 μg kg-1 and 21.4 μg kg-1,contributed the maximum detected frequency and value.
4. Conclusion
The analysis method for antibiotic residue by LC-QTRAP-MS validated in this manuscript is reliable and appropriate in the daily risk monitoring and assessment for food safety. The mass spectrum of each antibiotic obtained from EPI mode could be used as a corroboration of positive sample. The optimization of the sample pretreatment by Response Surface Method (RSM) enhanced the work efficiency. The analysis of real food origin samples proved the robustness and applicability of the modified QuEChERS method.
Author Contributions
Conceptualization, Ying Zhou; methodology, Ying Zhou; software,Ying Zhou; validation, Xiaoqiong Wu; formal analysis, Ying Zhou, Xiaoqiong Wu, Xiang Zhang; investigation, Yun Lin; resources, Yun Lin, Xiaoqiong Wu; data curation, Ying Zhou; writing, Ying Zhou. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Jiaxing Science and Technology Bureau,grant number 2019AD32050.
Data Availability Statement
Not applicable.
Acknowledgments
The authers would like to thank each other for the support in daily work and thank the help from Zhejiang Center for Disease Control and Prevention.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Figure A1.
Scheme of Solid-Phase Extraction.
Figure A1.
Scheme of Solid-Phase Extraction.
Table A1.
Procedures for automated Solid-Phase Extraction.
Table A1.
Procedures for automated Solid-Phase Extraction.
| NO. |
Step |
Source |
Output |
Flow Rate (mL/min) |
Volume(mL) |
Time(min) |
| 1 |
Rinse sample path |
CH3OH |
|
|
|
2.8 |
| 2 |
Rinse sample path |
H2O |
|
|
|
2.8 |
| 3 |
Rinse plunger |
CH3OH |
Solvent |
10 |
6 |
1.1 |
| 4 |
Rinse plunger |
H2O |
Solvent |
10 |
6 |
1.1 |
| 5 |
Load sample |
|
Waste |
2 |
20 |
22.3 |
| 6 |
Rinse |
5% CH3OH |
Solvent |
3 |
3 |
4.3 |
| 7 |
Rinse syringe |
CH3OH |
|
10 |
10 |
1.6 |
| 8 |
Elute |
CH3OH |
Collect |
10 |
5 |
0.9 |
| 9 |
Air push |
|
Collect |
10 |
5 |
1.1 |
| 10 |
End |
|
|
|
|
|
References
- Qiao, M.; Ying, G. G.; Singer, A. C.; Zhu, Y. G. Review of antibiotic resistance in China and its environment. Environ Int 2018, 110, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T. R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; Yu, L.-F.; Gu, D.; Ren, H.; Chen, X.; Lv, L.; He, D.; Zhou, H.; Liang, Z.; Liu, J.-H.; Shen, J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. The Lancet Infectious Diseases 2016, 16, (2), 161–168. [Google Scholar] [CrossRef]
- Zhang, Q. Q.; Ying, G. G.; Pan, C. G.; Liu, Y. S.; Zhao, J. L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environmental Science & Technology 2015, 49, (11), 6772-6782.
- Da, V. Origins and Evolution of Antibiotic Resistance. Microbiologia, 1996, 12, 9. [Google Scholar]
- Hao, R.-Z.; Zhao, R.-T.; Qiu, S.-F.; Wang, L.-G.; Song, H.-B. Antibiotics crisis in China. Science 2015, 348, 1100–1101. [Google Scholar] [CrossRef]
- Nelson, F.E.; Jensen, L.S.; Mcginns, J. Studies on the Stimulation of Growth by Dietary Antibiotics: 1. Changes in Growth Response of Chicks to Antibiotics over a three year Period. Poultry Science, 1963, 42, 906–909. [Google Scholar] [CrossRef]
- Carlet, J. Rambaud, C. &Pulcini, C., Save Antibiotics: a call for action of the World Alliance Against Antibiotic Resistance (WAAAR). BMC Infectious Diseases 2014, 14, 1–4. [Google Scholar]
- The world is running out of antibiotics, WHO report confirms. https://www.who.int/news/item/20-09-
2017-the-world-is-running-out-of-antibiotics-who-report-confirms.
- Merkatoris, P.; Schleining, J.; Kruil, A.; Borts, D.; Fajt, V. In vitro Elution of Penicillin, Ampicillin, Tetracycline, Tulathromycin, and Florfenicol From Plaster of Paris Beads. Front Vet Sci, 2020; 7, 585423. [Google Scholar]
- Woman Killed by a Superbug Resistant to Every Available Antibiotic - Scientific American
https://www.scientificamerican.com/article/woman-killed-by-a-superbug-resistant-to-every-availableantibiotic/.
- Commission, E. COMMISSION DECISION (EU) No 37/2010 of December 2009 on pharmacologically
active substances and their classification regarding maximum residue limits in foodstuffs of animal origin.
2010.
- Government of Canada, H.C. Health products &food branch, V. D. D. List of Maximum Residue Limits (MRLs) for Veterinary Drugs in Foods - Veterinary Drugs - Health Canada. 2015.
- Chung, H.-H.; Lee, J.-B.; Chung, Y.-H.; Lee, K.-G. Analysis of sulfonamide and quinolone antibiotic residues in Korean milk using microbial assays and high performance liquid chromatography. Food Chemistry 2009, 113, (1), 297–301. [Google Scholar] [CrossRef]
- Hernandez-Arteseros, J. A.; Barbosa, J.; Compaó, R.; Prat, M. Analysis of quinolone residues in edible
animal products. Journal of Chromatography A 2002, 945, (1-2), 1-24. [CrossRef]
- Muscarella, M.; Magro, S. L.; Palermo, C.; Centonze, D. Validation according to European Commission Decision 2002/657/EC of a confirmatory method for aflatoxin M 1 in milk based on immunoaffinity columns and high performance liquid chromatography with fluorescence detection. Analytica Chimica Acta 2007, 594, (2), 257–264. [Google Scholar] [CrossRef]
- Önal, A. , Overview on liquid chromatographic analysis of tetracycline residues in food matrice. Food Chemistry 2011. [Google Scholar] [CrossRef]
- Sinem,R.; Sezer, A.; Zge, E.; Emir,A.S.,Validation of two UHPLC-MS/MS methods for fast and reliable determination of quinolone residues in honey, Food Additives & Contaminants: Part A. 2021.
- Yamaguchi, T.; Okihashi, M.; Harada, K.; Uchida, K.; Konishi, Y.; Kajimura, K.; Hirata, K.; Yamamoto, Y. Rapid and easy multiresidue method for the analysis of antibiotics in meats by ultrahigh-performance
liquid chromatography-tandem mass spectrometry. Journal of Agricultural & Food Chemistry 2015, 63, (21),
5133. [CrossRef]
- Zhang, L.; Shi, L.; He, Q.; Li, Y. A rapid multiclass method for antibiotic residues in goat dairy products
by UPLC-quadrupole/electrostatic field orbitrap high-resolution mass spectrometry. Journal of Analytical
Science & Technology 2021, 12, (1). [CrossRef]
- Zhou, Y.; Guan, J.; Gao, W.; Lv, S.; Ge, M. Quantification and Confirmation of Fifteen Carbamate Pesticide
Residues by Multiple Reaction Monitoring and Enhanced Product Ion Scan Modes via LC-MS/MS QTRAP
System. Molecules 2018, 23, (10). [CrossRef]
- Yuan-Yuan; Li; Haixing; Wang; Chunyan; Zhao; Ye-Qing; Huang; Xiaowei; Tang, Identification and Characterization of Kukoamine Metabolites by Multiple Ion Monitoring Triggered Enhanced Product Ion Scan Method with a Triple-Quadruple Linear Ion Trap Mass Spectrometer. Journal of Agricultural and Food Chemistry 2015, 63, (50), 10785–10790.
- Ferech, M. European Surveillance of Antimicrobial Consumption (ESAC): outpatient antibiotic use in Europe. Journal of Antimicrobial Chemotherapy 2006, 58, 401. [Google Scholar] [CrossRef]
- Lode, H. Development of clinically important resistance to quinolones and other antibiotic groups. Journal of International Medical Research 1990, 18, 27–36. [Google Scholar]
- Roberts, M.C. Tetracyclines: Mode of Action and their Bacterial Mechanisms of Resistance, (Bacterial Resistance to Antibiotics – From Molecules to Man).2019.
- Yan, M.; Chen, X.; Huang, Y.; Nie, H.; Wang, J. Tetracyclines, sulfonamides and quinolones and their
corresponding resistance genes in the Three Gorges Reservoir, China. Science of The Total Environment, 2018
631–632, 840-848. [CrossRef]
- Bing, S.; Xiaofei, J.; Yongning, W. .; Jianying H.; Xiaoming T.; Jing Z., Multi-class confirmatory method for analyzing trace levels of tetracyline and quinolone antibiotics in pig tissues by ultra-performance liquid chromatography coupled with tandem mass spectrometry Rapid Commun. Mass Spectrom. 2007; 21: 3487–3496.
- Ajibola, A. S.; Tisler, S.; Zwiener, C. Simultaneous determination of multiclass antibiotics in sewage sludge
based on QuEChERS extraction and liquid chromatography-tandem mass spectrometry. Analytical Methods
2020, 12, (4).
- Chen, L.; Qi, W. Y.; Zhao, Z. Y.; Zhao, X. Y.; Zhou, C. Y. , Rapid Determination of 19 Fluoroquinolones Antibiotic Residues in Soil by QuEChERS/Ultra Performance Liquid Chromatography-Tandem Mass Spectrometry. Journal of Instrumental Analysis 2019.
- Frenich, A. G.; Romero-González, R.; Gómez-Pérez, M.; Vidal, J. , Multi-mycotoxin analysis in eggs using a QuEChERS-based extraction procedure and ultra-high-pressure liquid chromatography coupled to triple quadrupole mass spectrometry. Journal of Chromatography A 2011, 1218, (28), 4349–4356. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, T.; Huang, S.; Lin, X.; Chen, M. , Screening of 49 antibiotic residues in aquatic products using modified QuEChERS sample preparation and UPLC-QToFMS analysis. 2021.
- Wilkowska, A.; Biziuk, M. , Determination of pesticide residues in food matrices using the QuEChERS methodology. Food Chemistry 2011, 125, (3), 803–812. [Google Scholar] [CrossRef]
- Zhang, C.; Deng, Y.; Zheng, J.; Zhang, Y.; Luo, A. , The Application of the QuEChERS Methodology in the Determination of Antibiotics in Food: a review. TrAC Trends in Analytical Chemistry 2019, 118. [Google Scholar] [CrossRef]
- Andre, I. K.; Siuli, M. ,Response Surface Methodology. Computational Statistics 2010, 2. [Google Scholar]
- Raymond, H.; Myers, D. C.; Montgomery,; Christine M. A.-C.,Response Surface Methodology: Process and
Product Optimization Using Designed Experiments, Fourth Edition. 2016.
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef]
- Ferreira, S.L.C.; Bruns, R.E.; Ferreira, H.S.; Matos, G.D.; David, J.M.; Brandao, G.C.; da Silva, E.G.P.; Portugal, L.A.; Reis, P.S.; Souza, A.S.; et al. Box-Behnken design An alternative for the optimization of analytical methods. Anal. Chim. Acta 2007, 597, 179–186. [Google Scholar] [CrossRef]
- Li, Y. W.; Mo, C. H.; Zhao, N.; Zhang, R. J.; Yi, R. H. , Determination of Sulfonamides Antibiotics in Water and Soil Using High Performance Liquid Chromatography. Chinese Journal of Analytical Chemistry 2008. [Google Scholar]
- Luo, H. T.; Huang, X. L.; Wu, H. Q.; Zhu, Z. X.; Lin, X. S. , Simultaneous Determination of 33 Medicine Residues in Aquatic Products by Rapid Resolution Liquid Chromatography-Tandem Mass Spectrometry. Chinese Journal of Analytical Chemistry 2012, 40, 273–279. [Google Scholar] [CrossRef]
- Yue, Z.; Qiu, Y.; Liu, X.; Ji, C. , Determination of Multi-Residues of Tetracyclines and Their Metabolites in Milk by High Performance Liquid Chromatography-Tandem Positive-ion Electrospray Ionization Mass Spectrometry. Chinese Journal of Analytical Chemistry 2006, 34, 1255–1259. [Google Scholar] [CrossRef]
- Eric Scholar. Tetracyclines. Elsevier Inc, U.S.A., 2007.
- Gao, J.; Xu, H.; Li, Q. J.; Feng, X. H.; Li, S. , Optimization of medium for one-step fermentation of inulin extract from Jerusalem artichoke tubers using Paenibacillus polymyxa ZJ-9 to produce R,R-2,3-butanediol. Bioresour Technol 2010, 101, (18), 7076–7082. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Weng, L.P.; Zhang, Q.L.; Xu, H.; Ji, L.N. , Optimization of glucose oxidase production by Aspergillus niger in a benchtop bioreactor using response surface methodology. World Journal of Microbiology & Biotechnology 2003, 19, 317–323. [Google Scholar]
- Amin, M. K.; Hassan, G. F.; Phyllis, R. B.; Burnaby, M. Mass spectral characterization of tetracyclines by electrospray ionization, H/D exchange, and multiple stage mass spectrometry. J Am Soc Mass Spectrom 2002, 13, 543–557. [Google Scholar]
- Maksimovi, M.; Ober, M.; Leki, M.; Nikolin, A. , Mass Spectral Characteristics of some 4-Quinolinecarboxylic Acid Derivatives. 2006.
- Matraszek-Zuchowska; I.; Wozniak; B.; Posyniak; A., Comparison of the Multiple Reaction Monitoring and Enhanced Product Ion Scan Modes for Confirmation of Stilbenes in Bovine Urine Samples Using LC–MS/MS QTRAP®System. Chromatographia Wiesbaden 2016.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).