Submitted:
11 November 2023
Posted:
14 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Cellular distribution of major inflammasomes in the nervous system
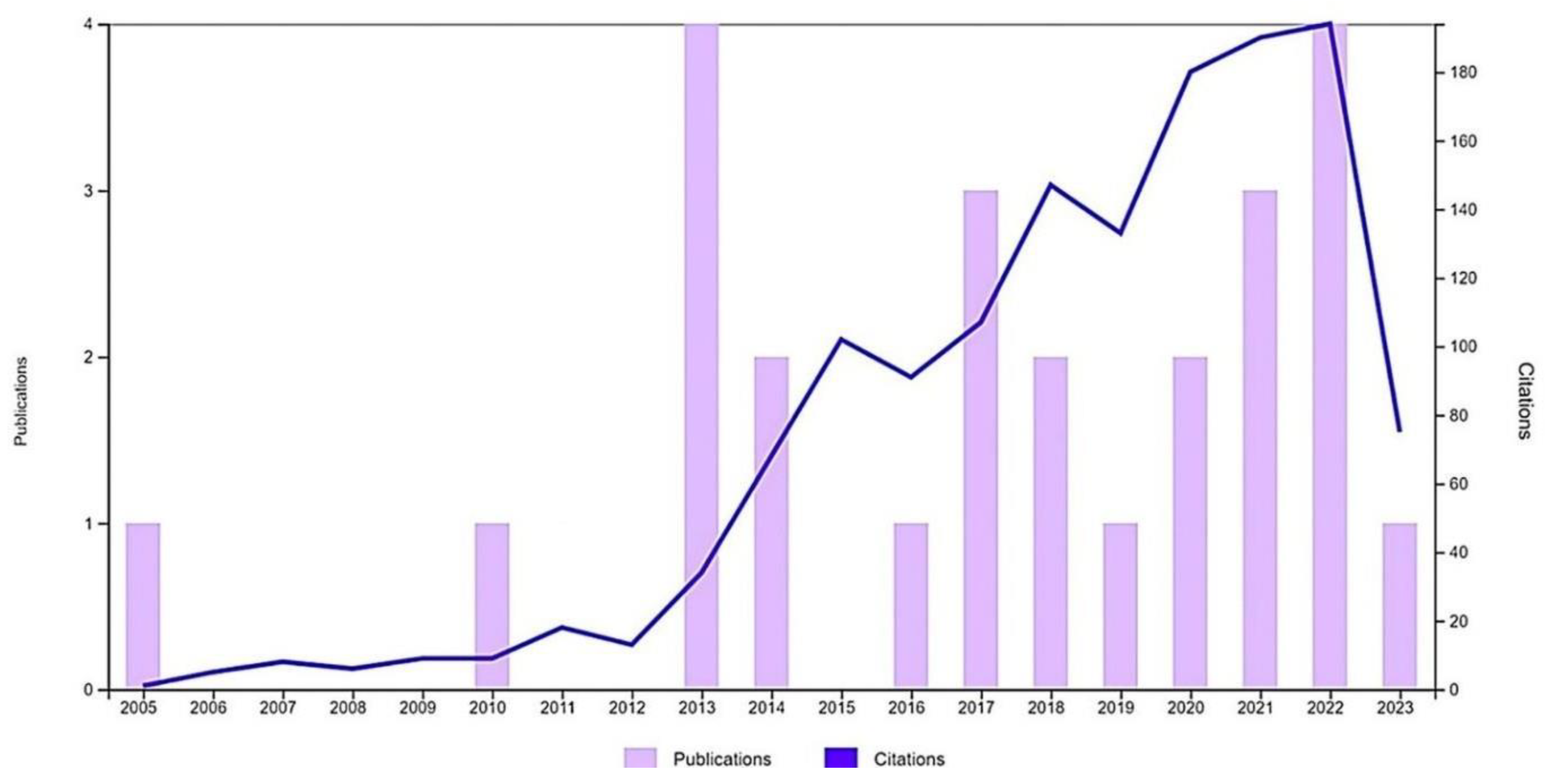
3. NLRP2 expression in the nervous system
3.1. Human NLRP2
3.2. Rodent NLRP2
3.3. Controversial findings of NLRP2 expression
4. Human NLRP2 connectome with STRING database
4.1. Src-like-adapter proteins (SLA proteins)
4.2. Beclin 1 (BECN1)
4.3. Protocadherin-11X (PCDH11X)
4.4. Glycogenin-2 (GYG-2)
4.5. Meiotic recombination 11A (MRE 11A)
4.6. Suppressor of G2 allele of Skp1 (SUGT1)
4.7. Epidermal growth factor receptor substrate 15 (EPS15)
4.8. Neurogenic differentiation factor 1 (NEUROD1)
4.9. Calpain-6 (CAPN6, CANPX, calpamodulin)
4.10. γ-taxilin (TXLNG, γ-TXLN)
4.11. Disks large-associated protein 2 (DLGAP2)
4.12. Apelin receptor (APLNR)
4.13. Solute carrier family 29A1 (SLC29A1, ENT1)
4.14. Arsenite methyltransferase (AS3MT)
4.15. Coiled-coil domain containing protein 50 (CCDC50)
5. Discussion and future prospects
- a)
- There are theories and evidence that inflammasomes can functionally take over each other’s roles, albeit these mostly unrevealed interplays among canonical and/or non-canonical inflammasomes need to be further investigated. Denes et al. [29] showed in their rodent model that NLRC4 (NLR family, CARD domain containing 4) and AIM2 contribute a lot to the pathogenesis of acute ischemic brain injury in the presence of pharmacologically blocked NLRP3.
- b) Above that, even myriad of inhibitors is mentioned with potential therapeutic benefits against inflammasomes like NLRP3 [46,227,228,229], these inhibitions mostly result from blocking related signaling pathways (such as dopamine receptor [243], adiponectin receptor [244], estrogen receptor [245], Angiotensin II receptor [246]) rather than direct inhibitory strategies. Thus, the same may be the case with NLRP2, if the major signaling pathways are precisely revealed. Data mining techniques like STRING can help us to target directly the potentially NLRP2-associated protein candidates and test them experimentally.
- c) Finally, despite accumulating experimental evidence, unfortunately, few experts in the field still take seriously the idea that the rodent and human NLRP2 inflammasome should be addressed more seriously.
6. Conclusions
Supplementary Materials
Author Contributions
Acknowledgements
Data Availability Statement
Competing interests
References
- Yi, Y.S. Functional crosstalk between non-canonical caspase-11 and canonical NLRP3 inflammasomes during infection-mediated inflammation. Immunology 2006, 159, 142–155. [Google Scholar] [CrossRef]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2002, 140, 821–832. [Google Scholar] [CrossRef]
- Ross, C.; Chan, A.H.; von Pein, J.B.; Maddugoda, M.P.; Boucher, D.; Schroder, K. Inflammatory Caspases: Toward a Unified Model for Caspase Activation by Inflammasomes. Annu. Rev. Immunol. 2022, 40, 249–269. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Akbal, A.; Dernst, A.; Lovotti, M.; Mangan, M.S.J.; McManus, R.M.; Latz, E. How location and cellular signaling combine to activate the NLRP3 inflammasome. Cell Mol. Immunol. 2022, 19, 1201–1214. [Google Scholar] [CrossRef]
- Minkiewicz, J.; de Rivero Vaccari, J.P.; Keane, R.W. Human astrocytes express a novel NLRP2 inflammasome. Glia 2013, 61, 1113–1121. [Google Scholar] [CrossRef]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host cell death and inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef]
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014, 14, 463–477. [Google Scholar] [CrossRef]
- Mamik, M.K.; Power, C. Inflammasomes in neurological diseases: Emerging pathogenic and therapeutic concepts. Brain 2017, 140, 2273–2285. [Google Scholar] [CrossRef] [PubMed]
- Voet, S.; Srinivasan, S.; Lamkanfi, M.; van Loo, G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 2019, 11, e10248. [Google Scholar] [CrossRef] [PubMed]
- de Rivero Vaccari, J.P.; Dietrich, W.D.; Keane, R.W. Activation and regulation of cellular inflammasomes: Gaps in our knowledge for central nervous system injury. J. Cereb. Blood Flow Metab. 2014, 34, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Enosi Tuipulotu, D.; Tan, W.H.; Kay, C.; Man, S.M. Emerging Activators and Regulators of Inflammasomes and Pyroptosis. Trends Immunol. 2019, 40, 1035–1052. [Google Scholar] [CrossRef] [PubMed]
- Sonnessa, M.; Cioffi, A.; Brunetti, O.; Silvestris, N.; Zito, F.A.; Saponaro, C.; et al. NLRP3 Inflammasome From Bench to Bedside: New Perspectives for Triple Negative Breast Cancer. Front. Oncol. 2020, 10, 1587. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Luc, N.; Dorfleutner, A.; Stehlik, C. Inflammasomes and their activation. Crit. Rev. Immunol. 2010, 30, 463–487. [Google Scholar] [CrossRef] [PubMed]
- Bruey, J.M.; Bruey-Sedano, N.; Newman, R.; Chandler, S.; Stehlik, C.; Reed, J.C. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J. Biol. Chem. 2004, 279, 51897–51907. [Google Scholar] [CrossRef] [PubMed]
- Fontalba, A.; Gutierrez, O.; Fernandez-Luna, J.L. NLRP2, an inhibitor of the NF-kappaB pathway, is transcriptionally activated by NF-kappaB and exhibits a nonfunctional allelic variant. J. Immunol. 2007, 179, 8519–8524. [Google Scholar] [CrossRef]
- Rossi, M.N.; Pascarella, A.; Licursi, V.; Caiello, I.; Taranta, A.; Rega, L.R.; et al. NLRP2 Regulates Proinflammatory and Antiapoptotic Responses in Proximal Tubular Epithelial Cells. Front. Cell Dev. Biol. 2019, 7, 252. [Google Scholar] [CrossRef]
- Peng, H.; Chang, B.; Lu, C.; Su, J.; Wu, Y.; Lv, P. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS ONE 2012, 7, e30344. [Google Scholar] [CrossRef]
- Kuchmiy, A.A.; D’Hont, J.; Hochepied, T.; Lamkanfi, M. NLRP2 controls age-associated maternal fertility. J. Exp. Med. 2016, 213, 2851–2860. [Google Scholar] [CrossRef]
- Huang, J.Y.; Su, M.; Lin, S.H.; Kuo, P.L. A genetic association study of NLRP2 and NLRP7 genes in idiopathic recurrent miscarriage. Hum. Reprod. 2012, 28, 1127–1134. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Das, N.; Chatterjee, D.; Banerjee, A.; Das, J.K.; Basu, S.; et al. Association of NALP2 polymorphism with arsenic induced skin lesions and other health effects. Mutat. Res. 2013, 755, 1–5. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Tremblay, M.È. A Diversity of Cell Types, Subtypes and Phenotypes in the Central Nervous System: The Importance of Studying Their Complex Relationships. Front. Cell Neurosci. 2020, 14, 628347. [Google Scholar] [CrossRef]
- Butt, A.; Verkhratsky, A. Neuroglia: Realising their true potential. Brain Neurosci. Adv. 2012, 2, 2398212818817495. [Google Scholar] [CrossRef]
- Dumas, A.A.; Prinz, M. The myeloid side of the CNS. Brain Pat. 2020, 30, 1158. [Google Scholar] [CrossRef]
- Cho, M.H.; Cho, K.; Kang, H.J.; Jeon, E.Y.; Kim, H.S.; Kwon, H.J.; et al. Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 2012, 10, 1761–1775. [Google Scholar] [CrossRef]
- Denes, A.; Coutts, G.; Lénárt, N.; Cruickshank, S.M.; Pelegrin, P.; Skinner, J.; et al. AIM2 and NLRC4 inflammasomes contribute with ASC to acute brain injury independently of NLRP3. Proc. Natl. Acad. Sci. USA 2015, 112, 4050–4055. [Google Scholar] [CrossRef]
- Lu, M.; Sun, X.L.; Qiao, C.; Liu, Y.; Ding, J.H.; Hu, G. Uncoupling protein 2 deficiency aggravates astrocytic endoplasmic reticulum stress and nod-like receptor protein 3 inflammasome activation. Neurobiol. Aging 2014, 35, 421–430. [Google Scholar] [CrossRef]
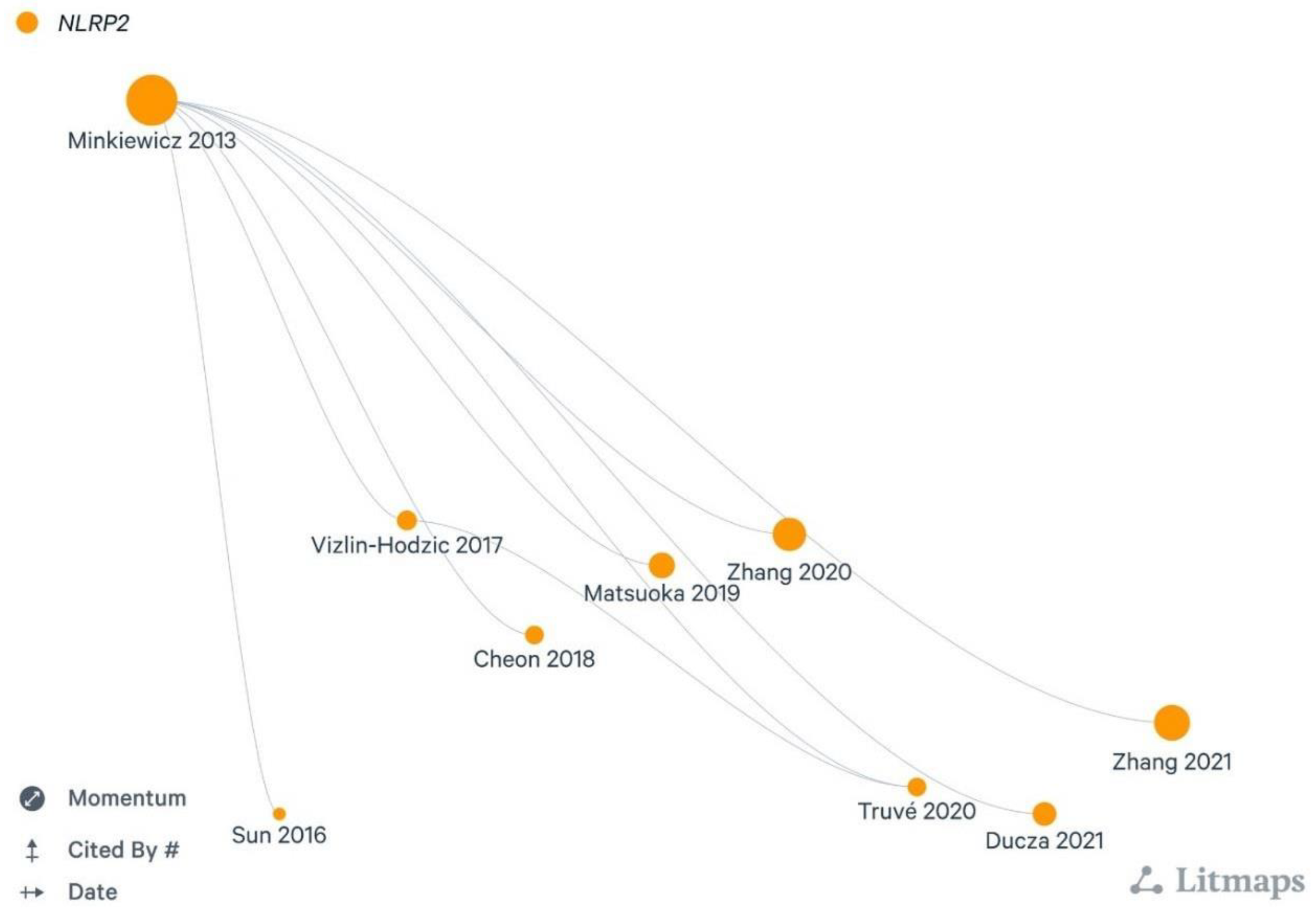
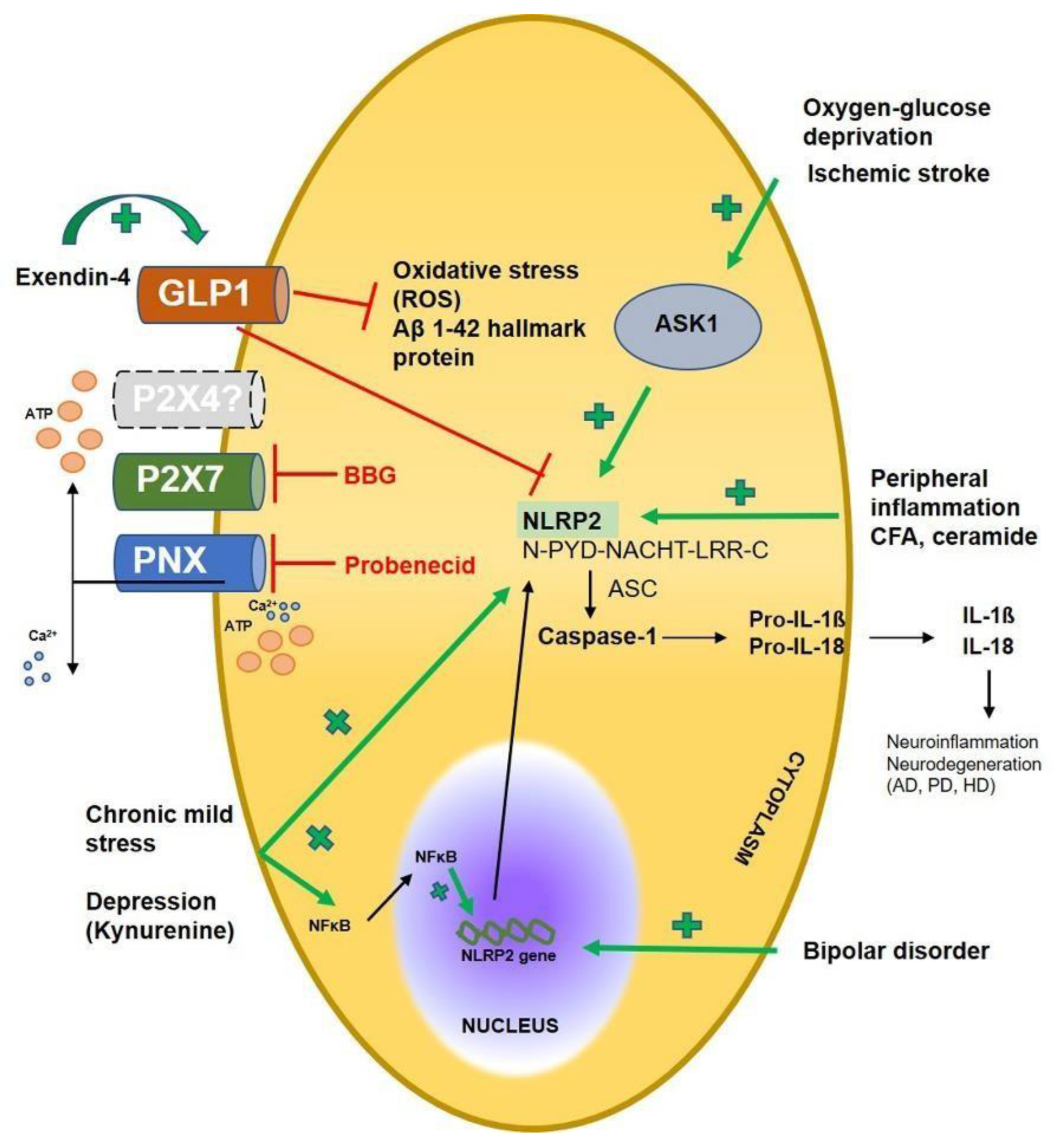
- Ducza, L.; Szücs, P.; Hegedűs, K.; Bakk, E.; Gajtkó, A.; Wéber, I.; et al. NLRP2 Is Overexpressed in Spinal Astrocytes at the Peak of Mechanical Pain Sensitivity during Complete Freund Adjuvant-Induced Persistent Pain. Int. J. Mol. Sci. 2021, 22, 11408. [Google Scholar] [CrossRef]
- Sun, X.; Song, X.; Zhang, L.; Sun, J.; Wei, X.; Meng, L.; et al. NLRP2 is highly expressed in a mouse model of ischemic stroke. Biochem. Biophys. Res. Commun. 2016, 479, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.Y.; Kim, E.J.; Kim, S.Y.; Kim, J.M.; Kam, E.H.; Park, J.K.; et al. Apoptosis Signal-regulating Kinase 1 Silencing on Astroglial Inflammasomes in an Experimental Model of Ischemic Stroke. Neuroscience 2018, 390, 218–230. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, Y.; He, Z.; Xu, Y.; Li, X.; Ding, J.; et al. Kynurenine regulates NLRP2 inflammasome in astrocytes and its implications in depression. Brain Behav. Immun. 2020, 88, 471–481. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, Y.; Gao, R.; Chen, X.; Chen, R.; Chen, Z. Glucagon-like peptide-1 analogs mitigate neuroinflammation in Alzheimer’s disease by suppressing NLRP2 activation in astrocytes. Mol. Cell Endocrinol. 2022, 542, 111529. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Yamashita, A.; Matsuda, M.; Kawai, K.; Sawa, T.; Amaya, F. NLRP2 inflammasome in dorsal root ganglion as a novel molecular platform that produces inflammatory pain hypersensitivity. Pain 2022, 160, 2149–2160. [Google Scholar] [CrossRef]
- Truvé, K.; Parris, T.Z.; Vizlin-Hodzic, D.; Salmela, S.; Berger, E.; Ågren, H.; et al. Identification of candidate genetic variants and altered protein expression in neural stem and mature neural cells support altered microtubule function to be an essential component in bipolar disorder. Transl. Psychiatry 2020, 10, 390. [Google Scholar] [CrossRef]
- Adamczak, S.E.; de Rivero Vaccari, J.P.; Dale, G.; Brand, F.J., 3rd; Nonner, D.; Bullock, M.R.; et al. Pyroptotic neuronal cell death mediated by the AIM2 inflammasome. J. Cereb. Blood Flow. Metab. 2014, 34, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.S.; Tan, L.; Jiang, T.; Zhu, X.C.; Wang, H.F.; Jia, C.D.; et al. Amyloid-β induces NLRP1-dependent neuronal pyroptosis in models of Alzheimer’s disease. Cell Death Dis. 2014, 5, e1382. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, R.; Hu, D.; Sun, X.; Fujioka, H.; Lundberg, K.; et al. Oligodendroglial glycolytic stress triggers inflammasome activation and neuropathology in Alzheimer’s disease. Sci. Adv. 2020, 6, eabb8680. [Google Scholar] [CrossRef]
- Yap, J.K.Y.; Pickard, B.S.; Chan, E.W.L.; Gan, S.Y. The Role of Neuronal NLRP1 Inflammasome in Alzheimer’s Disease: Bringing Neurons into the Neuroinflammation Game. Mol. Neurobiol. 2019, 56, 7741–7753. [Google Scholar] [CrossRef]
- Mi, L.; Min, X.; Chai, Y.; Zhang, J.; Chen, X. NLRP1 Inflammasomes: A Potential Target for the Treatment of Several Types of Brain Injury. Front. Immunol. 2022, 13, 863774. [Google Scholar] [CrossRef] [PubMed]
- Saresella, M.; La Rosa, F.; Piancone, F.; Zoppis, M.; Marventano, I.; Calabrese, E.; et al. The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol. Neurodegener. 2016, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Pei, L.; Yao, S.; Wu, Y.; Shang, Y. NLRP3 Inflammasome in Neurological Diseases, from Functions to Therapies. Front. Cell Neurosci. 2017, 11, 63. [Google Scholar] [CrossRef]
- Vizlin-Hodzic, D.; Zhai, Q.; Illes, S.; Södersten, K.; Truvé, K.; Parris, T.Z.; et al. Early onset of inflammation during ontogeny of bipolar disorder: The NLRP2 inflammasome gene distinctly differentiates between patients and healthy controls in the transition between iPS cell and neural stem cell stages. Transl. Psychiatry 2017, 7, e1010. [Google Scholar] [CrossRef]
- Chiarini, A.; Armato, U.; Gui, L.; Dal Prà, I. “Other Than NLRP3” Inflammasomes: Multiple Roles in Brain Disease. Neuroscientist 2022, 11, 10738584221106114. [Google Scholar] [CrossRef]
- Li, Q.; Tian, Y.; Wang, Z.F.; Liu, S.B.; Mi, W.L.; Ma, H.J.; et al. Involvement of the spinal NALP1 inflammasome in neuropathic pain and aspirin-triggered-15-epi-lipoxin A4 induced analgesia. Neuroscience 2013, 254, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Q.; Zhang, M.T.; Mao-Ying, Q.L.; Hu, L.Y.; Wu, G.C.; et al. Curcumin ameliorates neuropathic pain by down-regulating spinal IL-1β via suppressing astroglial NALP1 inflammasome and JAK2-STAT3 signalling. Sci. Rep. 2016, 6, 28956. [Google Scholar] [CrossRef]
- He, W.; Long, T.; Pan, Q.; Zhang, S.; Zhang, Y.; Zhang, D.; et al. Microglial NLRP3 inflammasome activation mediates IL-1β release and contributes to central sensitization in a recurrent nitroglycerin-induced migraine model. J. Neuroinflammation 2019, 16, 78. [Google Scholar] [CrossRef]
- Grace, P.M.; Strand, K.A.; Galer, E.L.; Urban, D.J.; Wang, X.; Baratta, M.V.; et al. Morphine paradoxically prolongs neuropathic pain in rats by amplifying spinal NLRP3 inflammasome activation. Proc. Natl. Acad. Sci. USA 2016, 113, E3441–E3450. [Google Scholar] [CrossRef]
- Chiarini, A.; Gui, L.; Viviani, C.; Armato, U.; Dal Prà, I. NLRP3 Inflammasome’s Activation in Acute and Chronic Brain Diseases-An Update on Pathogenetic Mechanisms and Therapeutic Perspectives with Respect to Other Inflammasomes. Biomedicines 2023, 11, 999. [Google Scholar] [CrossRef]
- Marton, N.; Baricza, E.; Érsek, B.; Buzás, E.I.; Nagy, G. The Emerging and Diverse Roles of Src-Like Adaptor Proteins in Health and Disease. Mediat. Inflamm. 2015, 2015, 952536. [Google Scholar] [CrossRef]
- Ge, M.M.; Zhou, Y.Q.; Tian, X.B.; Manyande, A.; Tian, Y.K.; Ye, D.W.; et al. Src-family protein tyrosine kinases: A promising target for treating chronic pain. Biomed. Pharmacother. 2020, 125, 110017. [Google Scholar] [CrossRef]
- Yang, H.; Wang, L.; Zang, C.; Wang, Y.; Shang, J.; Zhang, Z.; et al. Src Inhibition Attenuates Neuroinflammation and Protects Dopaminergic Neurons in Parkinson’s Disease Models. Front. Neurosci. 2020, 14, 45. [Google Scholar] [CrossRef]
- Giraud, F.; Pereira, E.; Anizon, F.; Moreau, P. Recent Advances in Pain Management: Relevant Protein Kinases and Their Inhibitors. Molecules 2021, 26, 2696. [Google Scholar] [CrossRef]
- Tran, S.; Fairlie, W.D.; Lee, E.F. BECLIN1: Protein Structure, Function and Regulation. Cells 2021, 10, 1522. [Google Scholar] [CrossRef]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, W.; Sun, X.; Xu, D.; Wang, C.; Zhang, Q.; et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy 2016, 12, 1447–1459. [Google Scholar] [CrossRef]
- Lucin, K.M.; O’Brien, C.E.; Bieri, G.; Czirr, E.; Mosher, K.I.; Abbey, R.J.; et al. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer’s disease. Neuron 2013, 79, 873–886. [Google Scholar] [CrossRef]
- Heppner, F.L.; Ransohoff, R.M.; Becher, B. Immune attack: The role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 2015, 16, 358–372. [Google Scholar] [CrossRef]
- Houtman, J.; Freitag, K.; Gimber, N.; Schmoranzer, J.; Heppner, F.L.; Jendrach, M. Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J. 2019, 38, e99430. [Google Scholar] [CrossRef]
- Brattås, P.L.; Hersbach, B.A.; Madsen, S.; Petri, R.; Jakobsson, J.; Pircs, K. Impact of differential and time-dependent autophagy activation on therapeutic efficacy in a model of Huntington disease. Autophagy 2021, 17, 1316–1329. [Google Scholar] [CrossRef]
- Peek, S.L.; Mah, K.M.; Weiner, J.A. Regulation of neural circuit formation by protocadherins. Cell Mol. Life Sci. 2017, 74, 4133–4157. [Google Scholar] [CrossRef]
- Langfelder, P.; Cantle, J.P.; Chatzopoulou, D.; Wang, N.; Gao, F.; Al-Ramahi, I. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 2016, 19, 623–633. [Google Scholar] [CrossRef]
- Carrasquillo, M.M.; Zou, F.; Pankratz, V.S.; Wilcox, S.L.; Ma, L.; Walker, L.P. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat. Genet. 2019, 41, 192–198. [Google Scholar] [CrossRef]
- Beecham, G.W.; Naj, A.C.; Gilbert, J.R.; Haines, J.L.; Buxbaum, J.D.; Pericak-Vance, M.A. PCDH11X variation is not associated with late-onset Alzheimer disease susceptibility. Psychiatr. Genet. 2010, 20, 321–324. [Google Scholar] [CrossRef]
- Miar, A.; Alvarez, V.; Corao, A.I.; Alonso, B.; Díaz, M.; Menéndez, M. Lack of association between protocadherin 11-X/Y (PCDH11X and PCDH11Y) polymorphisms and late onset Alzheimer’s disease. Brain Res. 2011, 1383, 252–256. [Google Scholar] [CrossRef]
- Marr, L.; Biswas, D.; Daly, L.A.; Browning, C.; Vial, S.C.M.; Maskell, D.P. Mechanism of glycogen synthase inactivation and interaction with glycogenin. Nat. Commun. 2022, 13, 3372. [Google Scholar] [CrossRef]
- Fastman, N.M.; Liu, Y.; Ramanan, V.; Merritt, H.; Ambing, E.; DePaoli-Roach, A.A. The structural mechanism of human glycogen synthesis by the GYS1-GYG1 complex. Cell Rep. 2022, 40, 111041. [Google Scholar] [CrossRef]
- Imagawa, E.; Osaka, H.; Yamashita, A.; Shiina, M.; Takahashi, E.; Sugie, H. A hemizygous GYG2 mutation and Leigh syndrome: A possible link? Hum. Genet. 2014, 133, 225–234. [Google Scholar] [CrossRef]
- Duran, J.; Hervera, A.; Markussen, K.H.; Varea, O.; López-Soldado, I.; Sun, R.C.; et al. Astrocytic glycogen accumulation drives the pathophysiology of neurodegeneration in Lafora disease. Brain 2021, 144, 2349–2360. [Google Scholar] [CrossRef]
- Sayas, C.L.; Ávila, J. GSK-3 and Tau: A Key Duet in Alzheimer’s Disease. Cells 2021, 10, 721. [Google Scholar] [CrossRef]
- Leroy, K.; Yilmaz, Z.; Brion, J.P. Increased level of active GSK-3beta in Alzheimer’s disease and accumulation in argyrophilic grains and in neurones at different stages of neurofibrillary degeneration. Neuropathol. Appl. Neurobiol. 2007, 33, 43–55. [Google Scholar] [CrossRef]
- Ahmad, S.; Orellana, A.; Kohler, I.; Frölich, L.; de Rojas, I.; Gil, S. Association of lysophosphatidic acids with cerebrospinal fluid biomarkers and progression to Alzheimer’s disease. Alzheimers Res. Ther. 2020, 12, 124. [Google Scholar] [CrossRef]
- Madabhushi, R.; Pan, L.; Tsai, L.H. DNA damage and its links to neurodegeneration. Neuron 2014, 83, 266–282. [Google Scholar] [CrossRef]
- Kannan, A.; Bhatia, K.; Branzei, D.; Gangwani, L. Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res. 2018, 46, 8326–8346. [Google Scholar] [CrossRef]
- Suberbielle, E.; Sanchez, P.E.; Kravitz, A.V.; Wang, X.; Ho, K.; Eilertson, K. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013, 16, 613–621. [Google Scholar] [CrossRef]
- Thadathil, N.; Hori, R.; Xiao, J.; Khan, M.M. DNA double-strand breaks: A potential therapeutic target for neurodegenerative diseases. Chromosome Res. 2019, 27, 345–364. [Google Scholar] [CrossRef]
- Lu, R.; Zhang, H.; Jiang, Y.N.; Wang, Z.Q.; Sun, L.; Zhou, Z.W. Post-Translational Modification of MRE11: Its Implication in DDR and Diseases. Genes 2021, 12, 1158. [Google Scholar] [CrossRef]
- Miyamoto, R.; Morino, H.; Yoshizawa, A.; Miyazaki, Y.; Maruyama, H.; Murakami, N.; et al. Exome sequencing reveals a novel MRE11 mutation in a patient with progressive myoclonic ataxia. J. Neurol. Sci. 2014, 337, 219–223. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Miyamoto, T.; Sakamoto, H.; Izumi, H.; Nakazawa, Y.; Ogi, T. Two unrelated patients with MRE11A mutations and Nijmegen breakage syndrome-like severe microcephaly. DNA Repair. 2011, 10, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Sedghi, M.; Salari, M.; Moslemi, A.R.; Kariminejad, A.; Davis, M.; Goullée, H.; et al. Ataxia-telangiectasia-like disorder in a family deficient for MRE11A, caused by a MRE11 variant. Neurol. Genet. 2018, 4, e295. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Qing, X.; Zhang, G.; Baade-Büttner, C.; Gruber, R.; Lu, H. .The Essential DNA Damage Response Complex MRN Is Dispensable for the Survival and Function of Purkinje Neurons. Front. Aging Neurosci. 2022, 13, 786199. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, E.; Beach, T.; Shen, Y.; Li, R.; Chang, Y. Deficiency of the Mre11 DNA repair complex in Alzheimer’s disease brains. Brain Res. Mol. Brain Res. 2004, 128, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Skowyra, D.; Elledge, S.J.; Harper, J.W.; Hieter, P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 1999, 4, 21–33. [Google Scholar] [CrossRef]
- Willhoft, O.; Kerr, R.; Patel, D.; Zhang, W.; Al-Jassar, C.; Daviter, T. The crystal structure of the Sgt1-Skp1 complex: The link between Hsp90 and both SCF E3 ubiquitin ligases and kinetochores. Sci. Rep. 2017, 7, 41626. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Jacob, J.; Michowski, W.; Nowotny, M.; Kuznicki, J.; Chazin, W.J. Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 2004, 279, 16511–16517. [Google Scholar] [CrossRef] [PubMed]
- Eisele, F.; Eisele-Bürger, A.M.; Hao, X.; Berglund, L.L.; Höög, J.L.; Liu, B. An Hsp90 co-chaperone links protein folding and degradation and is part of a conserved protein quality control. Cell Rep. 2021, 35, 109328. [Google Scholar] [CrossRef]
- da Silva Correia, J.; Miranda, Y.; Leonard, N.; Ulevitch, R. SGT1 is essential for Nod1 activation. Proc. Natl. Acad. Sci. USA 2007, 104, 6764–6769. [Google Scholar] [CrossRef]
- Bohush, A.; Góral, A.; Sierant, M.; Nawrot, B.; Leśniak, W.; Filipek, A. Sgt1 Regulates α-Synuclein Subcellular Localization and Expression of Parkinson’s Disease Related Genes, PINK1 and PARK9. Biomolecules 2021, 11, 1675. [Google Scholar] [CrossRef]
- Bohush, A.; Niewiadomska, G.; Weis, S.; Filipek, A. HSP90 and Its Novel Co-Chaperones, SGT1 and CHP-1, in Brain of Patients with Parkinson’s Disease and Dementia with Lewy Bodies. J. Park. Dis. 2019, 9, 97–107. [Google Scholar] [CrossRef]
- Spiechowicz, M.; Bernstein, H.G.; Dobrowolny, H.; Leśniak, W.; Mawrin, C.; Bogerts, B.; et al. Density of Sgt1-immunopositive neurons is decreased in the cerebral cortex of Alzheimer’s disease brain. Neurochem. Int. 2006, 49, 487–493. [Google Scholar] [CrossRef]
- Fazioli, F.; Minichiello, L.; Matoskova, B.; Wong, W.T.; Di Fiore, P.P. eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol. Cell Biol. 1993, 13, 5814–5828. [Google Scholar] [PubMed]
- Benmerah, A.; Gagnon, J.; Bègue, B.; Mégarbané, B.; Dautry-Varsat, A.; Cerf-Bensussan, N. The tyrosine kinase substrate eps15 is constitutively associated with the plasma membrane adaptor AP-2. J. Cell Biol. 1995, 131, 1831–1838. [Google Scholar] [CrossRef] [PubMed]
- Carbone, R.; Fré, S.; Iannolo, G.; Belleudi, F.; Mancini, P.; Pelicci, P.G.; et al. eps15 and eps15R are essential components of the endocytic pathway. Cancer Res. 1997, 57, 5498–5504. [Google Scholar]
- Huang, F.; Khvorova, A.; Marshall, W.; Sorkin, A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 2004, 279, 16657–16661. [Google Scholar] [CrossRef]
- van Bergen En Henegouwen, P.M. Eps15: A multifunctional adaptor protein regulating intracellular trafficking. Cell Commun. Signal 2009, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Iwakura, Y.; Piao, Y.S.; Mizuno, M.; Takei, N.; Kakita, A.; Takahashi, H.; et al. Influences of dopaminergic lesion on epidermal growth factor-ErbB signals in Parkinson’s disease and its model: Neurotrophic implication in nigrostriatal neurons. J. Neurochem. 2005, 93, 974–983. [Google Scholar] [CrossRef]
- Atkin, G.; Paulson, H. Ubiquitin pathways in neurodegenerative disease. Front. Mol. Neurosci. 2014, 7, 63. [Google Scholar] [CrossRef]
- Conway, J.A.; Kinsman, G.; Kramer, E.R. The Role of NEDD4 E3 Ubiquitin-Protein Ligases in Parkinson’s Disease. Genes 2022, 13, 513. [Google Scholar] [CrossRef]
- Dokucu, M.E.; Zipursky, S.L.; Cagan, R.L. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 1996, 122, 4139–4147. [Google Scholar] [CrossRef] [PubMed]
- Sommer, L.; Ma, Q.; Anderson, D.J. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell Neurosci. 1996, 8, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, N.; Castro, D.S.; Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Tutukova, S.; Tarabykin, V.; Hernandez-Miranda, L.R. The Role of Neurod Genes in Brain Development, Function, and Disease. Front. Mol. Neurosci. 2002, 14, 662774. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.H.; Druffel-Augustin, S.; Gass, P.; Jung, M.; Klugmann, M.; Bartholomae, A.; et al. Neuronal basic helix-loop-helix proteins (NEX, neuroD, NDRF): Spatiotemporal expression and targeted disruption of the NEX gene in transgenic mice. J. Neurosci. 1998, 18, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Boulle, F.; Massart, R.; Stragier, E.; Païzanis, E.; Zaidan, L.; Marday, S.; et al. Hippocampal and behavioral dysfunctions in a mouse model of environmental stress: Normalization by agomelatine. Transl. Psychiatry 2014, 4, e485. [Google Scholar] [CrossRef]
- Rubio-Cabezas, O.; Minton, J.A.; Kantor, I.; Williams, D.; Ellard, S.; Hattersley, A.T. Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes 2010, 59, 2326–2331. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.H.; Li, W.; Zheng, J.J.; Xu, Y.G.; He, Q.; Chen, G. Differential neuronal reprogramming induced by NeuroD1 from astrocytes in grey matter versus white matter. Neural Regen. Res. 2020, 15, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Fedele, V.; Roybon, L.; Nordström, U.; Li, J.Y.; Brundin, P. Neurogenesis in the R6/2 mouse model of Huntington’s disease is impaired at the level of NeuroD1. Neuroscience 2011, 173, 76–81. [Google Scholar] [CrossRef]
- Satoh, J.; Yamamoto, Y.; Asahina, N.; Kitano, S.; Kino, Y. RNA-Seq data mining: Downregulation of NeuroD6 serves as a possible biomarker for alzheimer’s disease brains. Dis. Markers 2014, 2014, 123165. [Google Scholar] [CrossRef]
- Richetin, K.; Leclerc, C.; Toni, N.; Gallopin, T.; Pech, S.; Roybon, L.; et al. Genetic manipulation of adult-born hippocampal neurons rescues memory in a mouse model of Alzheimer’s disease. Brain 2015, 138, 440–455. [Google Scholar] [CrossRef]
- Lee, T.Y.; Cho, I.S.; Bashyal, N.; Naya, F.J.; Tsai, M.J.; Yoon, J.S.; et al. ERK Regulates NeuroD1-mediated Neurite Outgrowth via Proteasomal Degradation. Exp. Neurobiol. 2020, 29, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Pomeshchik, Y.; Klementieva, O.; Gil, J.; Martinsson, I.; Hansen, M.G.; de Vries, T.; et al. Human iPSC-Derived Hippocampal Spheroids: An Innovative Tool for Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and Developing Therapies. Stem Cell Rep. 2020, 15, 256. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhang, L.; Wu, Z.; Chen, Y.; Wang, F.; Chen, G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 2014, 14, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.J.; Yang, F.H.; Li, W.; Wang, T.; Lin, Y.; Feng, J.; et al. In vivo Neuroregeneration to Treat Ischemic Stroke Through NeuroD1 AAV-Based Gene Therapy in Adult Non-human Primates. Front. Cell Dev. Biol. 2020, 8, 590008. [Google Scholar] [CrossRef]
- Sorimachi, H.; Hata, S.; Ono, Y. Calpain chronicle—An enzyme family under multidisciplinary characterization. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 287–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiao, D.; Tang, F.; Gao, H.; Li, X. CAPN6 in disease: An emerging therapeutic target (Review). Int. J. Mol. Med. 2020, 46, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Dear, N.; Matena, K.; Vingron, M.; Boehm, T. A new subfamily of vertebrate calpains lacking a calmodulin-like domain: Implications for calpain regulation and evolution. Genomics 1997, 45, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Dear, T.N.; Boehm, T. Diverse mRNA expression patterns of the mouse calpain genes Capn5, Capn6 and Capn11 during development. Mech. Dev. 1999, 89, 201–209. [Google Scholar] [CrossRef]
- Moretti, D.; Del Bello, B.; Allavena, G.; Maellaro, E. Calpains and cancer: Friends or enemies? Arch. Biochem. Biophys. 2014, 564, 26–36. [Google Scholar] [CrossRef]
- Ono, Y.; Ojima, K.; Shinkai-Ouchi, F.; Hata, S.; Sorimachi, H. An eccentric calpain 2014, CAPN3/p94/calpain-3. Biochimie 2016, 122, 169–187. [Google Scholar] [CrossRef]
- Filali, H.; Vidal, E.; Bolea, R.; Márquez, M.; Marco, P.; Vargas, A.; et al. Gene and protein patterns of potential prion-related markers in the central nervous system of clinical and preclinical infected sheep. Vet. Res. 2013, 44, 14. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xiao, D.; Huang, L.; Li, S.; Ying, J.; Tong, Y.; et al. MicroRNA Alteration in Developing Rat Oligodendrocyte Precursor Cells Induced by Hypoxia-Ischemia. J. Neuropathol. Exp. Neurol. 2019, 78, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, R.; Kitazawa, M.; Chabrier, M.A.; Cheng, D.; Baglietto-Vargas, D.; Kling, A.; et al. Calpain inhibitor A-705253 mitigates Alzheimer’s disease-like pathology and cognitive decline in aged 3xTgAD mice. Am. J. Pathol. 2012, 181, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Mahaman, Y.A.R.; Huang, F.; Kessete Afewerky, H.; Maibouge, T.M.S.; Ghose, B.; Wang, X. Involvement of calpain in the neuropathogenesis of Alzheimer’s disease. Med. Res. Rev. 2019, 39, 608–630. [Google Scholar] [CrossRef] [PubMed]
- Nogami, S.; Satoh, S.; Nakano, M.; Shimizu, H.; Fukushima, H.; Maruyama, A.; et al. Taxilin; a novel syntaxin-binding protein that is involved in Ca2+-dependent exocytosis in neuroendocrine cells. Genes Cells 2003, 8, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Nogami, S.; Satoh, S.; Tanaka-Nakadate, S.; Yoshida, K.; Nakano, M.; Terano, A.; et al. Identification and characterization of taxilin isoforms. Biochem. Biophys. Res. Commun. 2004, 319, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Horii, Y.; Sakane, H.; Nogami, S.; Ohtomo, N.; Tomiya, T.; Shirataki, H. Expression of α-taxilin in the murine gastrointestinal tract: Potential implication in cell proliferation. Histochem. Cell Biol. 2014, 141, 165–180. [Google Scholar] [CrossRef]
- Sakane, H.; Makiyama, T.; Nogami, S.; Horii, Y.; Akasaki, K.; Shirataki, H. β-Taxilin participates in differentiation of C2C12 myoblasts into myotubes. Exp. Cell Res. 2016, 345, 230–238. [Google Scholar] [CrossRef]
- Makiyama, T.; Higashi, S.; Sakane, H.; Nogami, S.; Shirataki, H. γ-Taxilin temporally regulates centrosome disjunction in a Nek2A-dependent manner. Exp. Cell Res. 2018, 362, 412–423. [Google Scholar] [CrossRef]
- Hotokezaka, Y.; Katayama, I.; van Leyen, K.; Nakamura, T. GSK-3β-dependent downregulation of γ-taxilin and αNAC merge to regulate ER stress responses. Cell Death Dis. 2015, 6, e1719. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Wang, F.; Wang, R.; Hu, Y.; Chen, Z.; Huang, N.; et al. α-/γ-Taxilin are required for centriolar subdistal appendage assembly and microtubule organization. Elife 2022, 11, e73252. [Google Scholar] [CrossRef] [PubMed]
- Higashi, S.; Makiyama, T.; Sakane, H.; Nogami, S.; Shirataki, H. Regulation of Hook1-mediated endosomal sorting of clathrin-independent cargo by γ-taxilin. J. Cell Sci. 2022, 135, jcs258849. [Google Scholar] [CrossRef] [PubMed]
- Kornau, H.C.; Schenker, L.T.; Kennedy, M.B.; Seeburg, P.H. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 1995, 269, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Naisbitt, S.; Kim, E.; Tu, J.C.; Xiao, B.; Sala, C.; Valtschanoff, J.; et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 1999, 23, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chetkovich, D.M.; Petralia, R.S.; Sweeney, N.T.; Kawasaki, Y.; Wenthold, R.J. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 2000, 408, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Steiner, P.; Higley, M.J.; Xu, W.; Czervionke, B.L.; Malenka, R.C.; Sabatini, B.L. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron 2008, 60, 788–802. [Google Scholar] [CrossRef]
- Naisbitt, S.; Kim, E.; Weinberg, R.J.; Rao, A.; Yang, F.C.; Craig, A.M. Characterization of guanylate kinase-associated protein, a postsynaptic density protein at excitatory synapses that interacts directly with postsynaptic density-95/synapse-associated protein 90. J. Neurosci. 1997, 17, 5687–5696. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.M.; Wang, D.; Feng, G. Differential mRNA expression and protein localization of the SAP90/PSD-95-associated proteins (SAPAPs) in the nervous system of the mouse. J. Comp. Neurol. 2004, 472, 24–39. [Google Scholar] [CrossRef]
- Yao, I.; Iida, J.; Nishimura, W.; Hata, Y. Synaptic localization of SAPAP1, a synaptic membrane-associated protein. Genes Cells 2003, 8, 121–129. [Google Scholar] [CrossRef]
- Rasmussen, A.H.; Rasmussen, H.B.; Silahtaroglu, A. The DLGAP family: Neuronal expression, function and role in brain disorders. Mol. Brain 2017, 10, 43. [Google Scholar] [CrossRef]
- Jiang-Xie, L.F.; Liao, H.M.; Chen, C.H.; Chen, Y.T.; Ho, S.Y.; Lu, D.H. Autism-associated gene Dlgap2 mutant mice demonstrate exacerbated aggressive behaviors and orbitofrontal cortex deficits. Mol. Autism 2014, 5, 32. [Google Scholar] [CrossRef]
- Li, J.M.; Lu, C.L.; Cheng, M.C.; Luu, S.U.; Hsu, S.H.; Hu, T.M. Role of the DLGAP2 gene encoding the SAP90/PSD-95-associated protein 2 in schizophrenia. PLoS ONE 2014, 9, e85373. [Google Scholar] [CrossRef]
- Gilbertson, M.W.; Shenton, M.E.; Ciszewski, A.; Kasai, K.; Lasko, N.B.; Orr, S.P.; et al. Smaller hippocampal volume predicts pathologic vulnerability to psychological trauma. Nat. Neurosci. 2002, 5, 1242–1247. [Google Scholar] [CrossRef]
- Ouellette, A.R.; Neuner, S.M.; Dumitrescu, L.; Anderson, L.C.; Gatti, D.M.; Mahoney, E.R.; et al. Cross-Species Analyses Identify Dlgap2 as a Regulator of Age-Related Cognitive Decline and Alzheimer’s Dementia. Cell Rep. 2020, 32, 108091. [Google Scholar] [CrossRef] [PubMed]
- O’Dowd, B.F.; Heiber, M.; Chan, A.; Heng, H.H.; Tsui, L.C.; Kennedy, J.L.; et al. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene 1993, 136, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, Z.; Zhang, R.; Sun, W.; Chen, X. The Role of Apelin/Apelin Receptor in Energy Metabolism and Water Homeostasis: A Comprehensive Narrative Review. Front. Physiol. 2021, 12, 632886. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Han, L.; Xu, J. Apelin/APJ system: A novel promising target for neurodegenerative diseases. J. Cell Physiol. 2021, 235, 638–657. [Google Scholar] [CrossRef]
- Pi, J.; Cheng, Y.; Sun, H.; Chen, X.; Zhuang, T.; Liu, J.; et al. Apln-CreERT:mT/mG reporter mice as a tool for sprouting angiogenesis study. BMC Ophthalmol. 2017, 17, 163. [Google Scholar] [CrossRef]
- Mughal, A.; O’Rourke, S.T. Vascular effects of apelin: Mechanisms and therapeutic potential. Pharmacol. Ther. 2018, 190, 139–147. [Google Scholar] [CrossRef]
- Tatemoto, K.; Takayama, K.; Zou, M.X.; Kumaki, I.; Zhang, W.; Kumano, K.; et al. The novel peptide apelin lowers blood pressure via a nitric oxide-dependent mechanism. Regul. Pept. 2001, 99, 87–92. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, A.M.; Lolait, S.J.; Harris, L.E.; Pope, G.R. The apelin receptor APJ: Journey from an orphan to a multifaceted regulator of homeostasis. J. Endocrinol. 2013, 219, R13–R35. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, F.; Zahediasl, S. Effects of exercise training on adipose tissue apelin expression in streptozotocin-nicotinamide induced diabetic rats. Gene 2018, 662, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.; Chun, H.J.; Quertermous, T. Opposing cardiovascular roles for the angiotensin and apelin signaling pathways. J. Mol. Cell Cardiol. 2016, 41, 778–781. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Murphy, K.; Cohen, M.; Sujkovic, E.; Kennedy, A.; Dhillo, W.; et al. The effects of centrally administered apelin-13 on food intake, water intake and pituitary hormone release in rats. Biochem. Biophys. Res. Commun. 2002, 291, 1208–1212. [Google Scholar] [CrossRef]
- Zeng, X.J.; Yu, S.P.; Zhang, L.; Wei, L. Neuroprotective effect of the endogenous neural peptide apelin in cultured mouse cortical neurons. Exp. Cell Res. 2010, 316, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.A.; Agrawal, A.; Sabnekar, P.; Dichter, M.A.; Lynch, D.R.; Kolson, D.L. Apelin, an endogenous neuronal peptide, protects hippocampal neurons against excitotoxic injury. J. Neurochem. 2007, 102, 1905–1917. [Google Scholar] [CrossRef] [PubMed]
- Aminyavari, S.; Zahmatkesh, M.; Farahmandfar, M.; Khodagholi, F.; Dargahi, L.; Zarrindast, M.R. Protective role of Apelin-13 on amyloid β25-35-induced memory deficit; Involvement of autophagy and apoptosis process. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 89, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Agostinho, P.; Cunha, R.A.; Oliveira, C. Neuroinflammation, oxidative stress and the pathogenesis of Alzheimer’s disease. Curr. Pharm. Des. 2010, 16, 2766–2778. [Google Scholar] [CrossRef]
- Xin, Q.; Cheng, B.; Pan, Y.; Liu, H.; Yang, C.; Chen, J. Neuroprotective effects of apelin-13 on experimental ischemic stroke through suppression of inflammation. Peptides 2015, 63, 55–62. [Google Scholar] [CrossRef]
- Yang, L.; Wang, H.; Liu, L.; Xie, A. The Role of Insulin/IGF-1/PI3K/Akt/GSK3β Signaling in Parkinson’s Disease Dementia. Front. Neurosci. 2018, 12, 73. [Google Scholar] [CrossRef] [PubMed]
- Haghparast, E.; Esmaeili-Mahani, S.; Abbasnejad, M.; Sheibani, V. Apelin-13 ameliorates cognitive impairments in 6-hydroxydopamine-induced substantia nigra lesion in rats. Neuropeptides 2018, 68, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Pouresmaeili-Babaki, E.; Esmaeili-Mahani, S.; Abbasnejad, M.; Ravan, H. Protective Effect of Neuropeptide Apelin-13 on 6-Hydroxydopamine-Induced Neurotoxicity in SH-SY5Y Dopaminergic Cells: Involvement of Its Antioxidant and Antiapoptotic Properties. Rejuvenation Res. 2018, 21, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Chai, J.; Xu, C.; Luo, H.; Zhang, Q. Apelin inhibits the activation of the nucleotide-binding domain and the leucine-rich, repeat-containing family, pyrin-containing 3 (NLRP3) inflammasome and ameliorates insulin resistance in severely burned rats. Surgery 2015, 157, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Nogales, M.; Santos-Galindo, M.; Hernández, I.H.; Cabrera, J.R.; Lucas, J.J. Faulty splicing and cytoskeleton abnormalities in Huntington’s disease. Brain Pathol. 2016, 26, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bai, Y.J.; Jiang, Y.R.; Yu, W.Z.; Shi, X.; Chen, L.; et al. Apelin-13 Is an Early Promoter of Cytoskeleton and Tight Junction in Diabetic Macular Edema via PI-3K/Akt and MAPK/Erk Signaling Pathways. Biomed. Res. Int. 2018, 2018, 3242574. [Google Scholar] [CrossRef] [PubMed]
- Bryan, M.R.; Bowman, A.B. Manganese and the Insulin-IGF Signaling Network in Huntington’s Disease and Other Neurodegenerative Disorders. Adv. Neurobiol. 2017, 18, 113–142. [Google Scholar] [PubMed]
- Zhang, Y.; Zhang, Y.; Sun, K.; Meng, Z.; Chen, L. The SLC transporter in nutrient and metabolic sensing, regulation, and drug development. J. Mol. Cell Biol. 2019, 11, 1–13. [Google Scholar] [CrossRef]
- César-Razquin, A.; Snijder, B.; Frappier-Brinton, T.; Isserlin, R.; Gyimesi, G.; Bai, X. A Call for Systematic Research on Solute Carriers. Cell 2015, 162, 478–487. [Google Scholar] [CrossRef]
- Brzica, H.; Abdullahi, W.; Ibbotson, K.; Ronaldson, P.T. Role of Transporters in Central Nervous System Drug Delivery and Blood-Brain Barrier Protection: Relevance to Treatment of Stroke. J. Cent. Nerv. Syst. Dis. 2017, 9, 1179573517693802. [Google Scholar] [CrossRef]
- Türková, A.; Zdrazil, B. Current Advances in Studying Clinically Relevant Transporters of the Solute Carrier (SLC) Family by Connecting Computational Modeling and Data Science. Comput. Struct. Biotechnol. J. 2019, 17, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Tao, L.; Cao, X.; Chen, L. The solute carrier transporters and the brain: Physiological and pharmacological implications. Asian J. Pharm. Sci. 2020, 15, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Young, J.D.; Yao, S.Y.; Baldwin, J.M.; Cass, C.E.; Baldwin, S.A. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Asp. Med. 2013, 34, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Anglada, M.; Urtasun, N.; Pérez-Torras, S. Intestinal Nucleoside Transporters: Function, Expression, and Regulation. Compr. Physiol. 2018, 8, 1003–1017. [Google Scholar] [PubMed]
- Kao, Y.H.; Lin, M.S.; Chen, C.M.; Wu, Y.R.; Chen, H.M.; Lai, H.L. Targeting ENT1 and adenosine tone for the treatment of Huntington’s disease. Hum. Mol. Genet. 2017, 26, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Chang, C.P.; Lin, C.J.; Lai, H.L.; Kao, Y.H.; Cheng, S.J.; et al. Adenosine Augmentation Evoked by an ENT1 Inhibitor Improves Memory Impairment and Neuronal Plasticity in the APP/PS1 Mouse Model of Alzheimer’s Disease. Mol. Neurobiol. 2018, 55, 8936–8952. [Google Scholar] [CrossRef]
- Chang, C.P.; Chang, Y.G.; Chuang, P.Y.; Nguyen, T.N.A.; Wu, K.C.; Chou, F.Y.; et al. Equilibrative nucleoside transporter 1 inhibition rescues energy dysfunction and pathology in a model of tauopathy. Acta Neuropathol. Commun. 2021, 9, 112. [Google Scholar] [CrossRef]
- Chung, J.Y.; Yu, S.D.; Hong, Y.S. Environmental source of arsenic exposure. J. Prev. Med. Public Health 2014, 47, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.A.; Khan, M.H.; Haque, M. Arsenic contamination in groundwater in Bangladesh: Implications and challenges for healthcare policy. Risk Manag. Healthc. Policy 2018, 11, 251–261. [Google Scholar] [CrossRef]
- Emadi, A.; Gore, S.D. Arsenic trioxide—An old drug rediscovered. Blood Rev. 2010, 24, 191–199. [Google Scholar] [CrossRef]
- Singh, A.P.; Goel, R.K.; Kaur, T. Mechanisms pertaining to arsenic toxicity. Toxicol. Int. 2011, 18, 87–93. [Google Scholar]
- Rahman, M.A.; Hannan, M.A.; Uddin, M.J.; Rahman, M.S.; Rashid, M.M.; Kim, B. Exposure to Environmental Arsenic and Emerging Risk of Alzheimer’s Disease: Perspective Mechanisms, Management Strategy, and Future Directions. Toxics 2021, 9, 188. [Google Scholar] [CrossRef]
- Chandravanshi, L.P.; Gupta, R.; Shukla, R.K. Developmental Neurotoxicity of Arsenic: Involvement of Oxidative Stress and Mitochondrial Functions. Biol. Trace Elem. Res. 2018, 186, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Hannan, M.A.; Dash, R.; Sohag, A.A.M.; Haque, M.N.; Moon, I.S. Neuroprotection Against Oxidative Stress: Phytochemicals Targeting TrkB Signaling and the Nrf2-ARE Antioxidant System. Front. Mol. Neurosci. 2020, 13, 116. [Google Scholar] [CrossRef]
- Dwivedi, N.; Flora, S.J. Concomitant exposure to arsenic and organophosphates on tissue oxidative stress in rats. Food Chem. Toxicol. 2011, 49, 1152–1159. [Google Scholar] [CrossRef]
- Dash, R.; Mitra, S.; Ali, M.C.; Oktaviani, D.F.; Hannan, M.A.; Choi, S.M.; et al. Phytosterols: Targeting Neuroinflammation in Neurodegeneration. Curr. Pharm. Des. 2021, 27, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Wisessaowapak, C.; Visitnonthachai, D.; Watcharasit, P.; Satayavivad, J. Prolonged arsenic exposure increases tau phosphorylation in differentiated SH-SY5Y cells: The contribution of GSK3 and ERK1/2. Environ. Toxicol. Pharmacol. 2021, 84, 103626. [Google Scholar] [CrossRef] [PubMed]
- Medda, N.; Patra, R.; Ghosh, T.K.; Maiti, S. Neurotoxic Mechanism of Arsenic: Synergistic Effect of Mitochondrial Instability, Oxidative Stress, and Hormonal-Neurotransmitter Impairment. Biol. Trace Elem. Res. 2020, 198, 8–15. [Google Scholar] [CrossRef]
- Prakash, C.; Soni, M.; Kumar, V. Mitochondrial oxidative stress and dysfunction in arsenic neurotoxicity: A review. J. Appl. Toxicol. 2016, 36, 179–188. [Google Scholar] [CrossRef]
- Prakash, C.; Soni, M.; Kumar, V. Biochemical and Molecular Alterations Following Arsenic-Induced Oxidative Stress and Mitochondrial Dysfunction in Rat Brain. Biol. Trace Elem. Res. 2015, 167, 121–129. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, M.S.; Uddin, M.J.; Mamum-Or-Rashid, A.N.M.; Pang, M.G.; Rhim, H. Emerging risk of environmental factors: Insight mechanisms of Alzheimer’s diseases. Environ. Sci. Pollut. Res. Int. 2020, 27, 44659–44672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Cui, X.; Gao, Y.; Sun, L.; Wang, J.; Yang, Y.; et al. Role of pigment epithelium-derived factor (PEDF) on arsenic-induced neuronal apoptosis. Chemosphere 2019, 215, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Jahan, I.; Ali, M.C.; Mitra, S.; Munni, Y.A.; Timalsina, B.; et al. Potential roles of natural products in the targeting of proteinopathic neurodegenerative diseases. Neurochem. Int. 2021, 145, 105011. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Ali, M.C.; Jahan, I.; Munni, Y.A.; Mitra, S.; Hannan, M.A.; et al. Emerging potential of cannabidiol in reversing proteinopathies. Ageing Res. Rev. 2021, 65, 101209. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Vazza, G.; Picelli, S.; Bozzato, A.; Mostacciuolo, M.L. Identification and characterization of C3orf6, a new conserved human gene mapping to chromosome 3q28. Gene 2003, 314, 113–120. [Google Scholar] [CrossRef]
- Lin, Y.; Li, Z.; Wang, Y.; Tian, T.; Jia, P.; Ye, Y.; et al. CCDC50 suppresses NLRP3 inflammasome activity by mediating autophagic degradation of NLRP3. EMBO Rep. 2022, 23, e54453. [Google Scholar] [CrossRef] [PubMed]
- Farfsing, A.; Engel, F.; Seiffert, M.; Hartmann, E.; Ott, G.; Rosenwald, A.; et al. Gene knockdown studies revealed CCDC50 as a candidate gene in mantle cell lymphoma and chronic lymphocytic leukemia. Leukemia 2009, 23, 2018–2026. [Google Scholar] [CrossRef]
- Hou, P.; Yang, K.; Jia, P.; Liu, L.; Lin, Y.; Li, Z.; et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021, 31, 62–79. [Google Scholar] [CrossRef]
- Cruchaga, C.; Kauwe, J.S.; Harari, O.; Jin, S.C.; Cai, Y.; Karch, C.M.; et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron 2013, 78, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Skaria, A.P. The economic and societal burden of Alzheimer disease: Managed care considerations. Am. J. Manag. Care 2022, 28, S188–S196. [Google Scholar]
- Wong, W. Economic burden of Alzheimer disease and managed care considerations. Am. J. Manag. Care 2020, 26, S177–S183. [Google Scholar] [PubMed]
- Holbrook, J.A.; Jarosz-Griffiths, H.H.; Caseley, E.; Lara-Reyna, S.; Poulter, J.A.; Williams-Gray, C.H.; et al. Neurodegenerative Disease and the NLRP3 Inflammasome. Front. Pharmacol. 2021, 12, 643254. [Google Scholar] [CrossRef] [PubMed]
- Anderson, F.L.; Biggs, K.E.; Rankin, B.E.; Havrda, M.C. NLRP3 inflammasome in neurodegenerative disease. Transl. Res. 2023, 252, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Kaushik, D.K.; Gupta, M.; Basu, A. Inflammasome signaling at the heart of central nervous system pathology. J. Neurosci. Res. 2010, 88, 1615–1631. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.C.; Choi, C.H.; Said-Sadier, N.; Johnson, L.; Atanasova, K.R.; Sellami, H.; et al. P2X4 assembles with P2X7 and pannexin-1 in gingival epithelial cells and modulates ATP-induced reactive oxygen species production and inflammasome activation. PLoS ONE 2013, 8, e70210. [Google Scholar] [CrossRef] [PubMed]
- Kanellopoulos, J.M.; Almeida-da-Silva, C.L.C.; Rüütel Boudinot, S.; Ojcius, D.M. Structural and Functional Features of the P2X4 Receptor: An Immunological Perspective. Front. Immunol. 2021, 12, 645834. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Fu, W.; Yao, J.; Ji, Z.; Wang, Y.; Zhou, Z.; et al. Heme induces IL-1β secretion through activating NLRP3 in kidney inflammation. Cell Biochem. Biophys. 2014, 69, 495–502. [Google Scholar] [CrossRef]
- Ducza, L.; Gajtkó, A.; Hegedűs, K.; Bakk, E.; Kis, G.; Gaál, B.; et al. Neuronal P2X4 receptor may contribute to peripheral inflammatory pain in rat spinal dorsal horn. Front. Mol. Neurosci. 2023, 16, 1115685. [Google Scholar] [CrossRef]
- Du, R.H.; Wu, F.F.; Lu, M.; Shu, X.D.; Ding, J.H.; Wu, G.; et al. Uncoupling protein 2 modulation of the NLRP3 inflammasome in astrocytes and its implications in depression. Redox Biol. 2016, 9, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xiao, Q.; Xie, L.; Yang, F.; Wang, L.; Tu, J. Astrocyte, a Promising Target for Mood Disorder Interventions. Front. Mol. Neurosci. 2019, 12, 136. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Zheng, X.; Hou, Y.; Hu, M.; Wang, H.; Bao, X.; et al. Regulation of proinflammatory monocyte activation by the kynurenine-AhR axis underlies immunometabolic control of depressive behavior in mice. FASEB J. 2018, 32, 1944–1956. [Google Scholar] [CrossRef] [PubMed]
- Rolstad, S.; Jakobsson, J.; Sellgren, C.; Isgren, A.; Ekman, C.J.; Bjerke, M.; et al. CSF neuroinflammatory biomarkers in bipolar disorder are associated with cognitive impairment. Eur. Neuropsychopharmacol. 2015, 25, 1091–1098. [Google Scholar] [CrossRef]
- Tsai, P.J.; Lai, Y.H.; Manne, R.K.; Tsai, Y.S.; Sarbassov, D.; Lin, H.K. Akt: A key transducer in cancer. J. Biomed. Sci. 2023, 29, 76. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Gu, J.H.; Dai, C.L.; Iqbal, K.; Liu, F.; Gong, C.X. AKT/GSK-3β signaling is altered through downregulation of mTOR during cerebral Ischemia/Reperfusion injury. Mol. Biol. Rep. 2022, 49, 3955–3964. [Google Scholar] [CrossRef]
- Stefaniak, J.; O’Brien, J. Imaging of neuroinflammation in dementia: A review. J. Neurol. Neurosurg. Psychiatry 2016, 87, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, L.; Tsvetanov, K.A.; Jones, P.S.; Bevan-Jones, W.R.; Arnold, R.; Borchert, R.J. Neuroinflammation and Functional Connectivity in Alzheimer’s Disease: Interactive Influences on Cognitive Performance. J. Neurosci. 2019, 39, 7218–7226. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.; Tam, W.W.; Zhang, M.W.; Ho, C.S.; Husain, S.F.; McIntyre, R.S.; et al. IL-1β, IL-6, TNF-α and CRP in Elderly Patients with Depression or Alzheimer’s disease: Systematic Review and Meta-Analysis. Sci Rep 2018, 8, 12050. [Google Scholar] [CrossRef]
- Cuello, A.C. Early and Late CNS Inflammation in Alzheimer’s Disease: Two Extremes of a Continuum? Trends Pharmacol Sci 2017, 38, 956–966. [Google Scholar] [CrossRef]
- Aziz, N. Measurement of Circulating Cytokines and Immune-Activation Markers by Multiplex Technology in the Clinical Setting: What Are We Really Measuring? For. Immunopathol. Dis. Ther. 2015, 6, 19–22. [Google Scholar] [CrossRef]
- Koelman, L.; Pivovarova-Ramich, O.; Pfeiffer, A.F.H.; Grune, T.; Aleksandrova, K. Cytokines for evaluation of chronic inflammatory status in ageing research: Reliability and phenotypic characterisation. Immun. Ageing 2019, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Zuliani, G.; Ranzini, M.; Guerra, G.; Rossi, L.; Munari, M.R.; Zurlo, A.; et al. Plasma cytokines profile in older subjects with late onset Alzheimer’s disease or vascular dementia. J. Psychiatr. Res. 2007, 41, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Belkhelfa, M.; Beder, N.; Mouhoub, D.; Amri, M.; Hayet, R.; Tighilt, N.; et al. The involvement of neuroinflammation and necroptosis in the hippocampus during vascular dementia. J. Neuroimmunol. 2018, 320, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, M.J.; Geerlings, M.I.; Meijer, J.; Kiliaan, A.; Ruitenberg, A.; van Swieten, J.C.; et al. Inflammatory proteins in plasma and the risk of dementia: The rotterdam study. Arch. Neurol. 2004, 61, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Kummer, M.P.; Stutz, A.; Delekate, A.; Schwartz, S.; Vieira-Saecker, A.; et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 2013, 493, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.G.; Brough, D.; Freeman, S. Inhibiting the Inflammasome: A Chemical Perspective. J. Med. Chem. 2016, 59, 1691–1710. [Google Scholar] [CrossRef]
- Bertinaria, M. Inflammasome Inhibitors. Molecules 2021, 26, 6912. [Google Scholar] [CrossRef] [PubMed]
- Coll, R.C.; Robertson, A.A.; Chae, J.J.; Higgins, S.C.; Muñoz-Planillo, R.; Inserra, M.C.; et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015, 21, 248–255. [Google Scholar] [CrossRef]
- Dempsey, C.; Rubio Araiz, A.; Bryson, K.J.; Finucane, O.; Larkin, C.; Mills, E.L.; et al. Inhibiting the NLRP3 inflammasome with MCC950 promotes non-phlogistic clearance of amyloid-β and cognitive function in APP/PS1 mice. Brain Behav. Immun. 2017, 61, 306–316. [Google Scholar] [CrossRef]
- Gordon, R.; Albornoz, E.A.; Christie, D.C.; Langley, M.R.; Kumar, V.; Mantovani, S.; et al. Inflammasome inhibition prevents α-synuclein pathology and dopaminergic neurodegeneration in mice. Sci. Transl. Med. 2018, 10, eaah4066. [Google Scholar] [CrossRef]
- Piancone, F.; La Rosa, F.; Marventano, I.; Saresella, M.; Clerici, M. The Role of the Inflammasome in Neurodegenerative Diseases. Molecules 2021, 26, 953. [Google Scholar] [CrossRef]
- La Rosa, F.; Saresella, M.; Marventano, I.; Piancone, F.; Ripamonti, E.; Al-Daghri, N.; et al. Stavudine Reduces NLRP3 Inflammasome Activation and Modulates Amyloid-β Autophagy. J. Alzheimers Dis. 2019, 72, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Gastaldi, S.; Boscaro, V.; Gianquinto, E.; Sandall, C.F.; Giorgis, M.; Marini, E.; et al. Chemical Modulation of the 1-(Piperidin-4-yl)-1,3-dihydro-2H-benzo[d]imidazole-2-one Scaffold as a Novel NLRP3 Inhibitor. Molecules 2021, 26, 3975. [Google Scholar] [CrossRef]
- Poli, G.; Fabi, C.; Bellet, M.M.; Costantini, C.; Nunziangeli, L.; Romani, L.; et al. Epigenetic Mechanisms of Inflammasome Regulation. Int. J. Mol. Sci. 2020, 21, 5758. [Google Scholar] [CrossRef]
- Li, D.; Yang, H.; Ma, J.; Luo, S.; Chen, S.; Gu, Q. MicroRNA-30e regulates neuroinflammation in MPTP model of Parkinson’s disease by targeting Nlrp3. Hum Cell 2018, 31, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lu, M.; Du, R.H.; Qiao, C.; Jiang, C.Y.; Zhang, K.Z.; et al. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 2016, 11, 28. [Google Scholar] [CrossRef]
- Kaushal, V.; Dye, R.; Pakavathkumar, P.; Foveau, B.; Flores, J.; Hyman, B.; et al. Neuronal NLRP1 inflammasome activation of Caspase-1 coordinately regulates inflammatory interleukin-1-beta production and axonal degeneration-associated Caspase-6 activation. Cell Death Differ. 2015, 22, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- de Souza, N. Model organisms: Mouse models challenged. Nat. Methods 2013, 10, 288. [Google Scholar] [CrossRef]
- Pound, P.; Ritskes-Hoitinga, M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J. Transl. Med. 2018, 16, 304. [Google Scholar] [CrossRef]
- Chiarini, A.; Armato, U.; Hu, P.; Dal Prà, I. Danger-Sensing/Patten Recognition Receptors and Neuroinflammation in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 9036. [Google Scholar] [CrossRef] [PubMed]
- van Dyck, C.H.; Swanson, C.J.; Aisen, P.; Bateman, R.J.; Chen, C.; Gee, M.; et al. Lecanemab in Early Alzheimer’s Disease. N. Engl. J. Med. 2023, 388, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Li, M.; He, F.; Bian, Z.; Liu, J.; He, Q.; et al. Dopamine D1 receptor agonist A-68930 inhibits NLRP3 inflammasome activation and protects rats from spinal cord injury-induced acute lung injury. Spinal Cord. 2016, 54, 951–956. [Google Scholar] [CrossRef]
- Wang, S.; Yao, Q.; Wan, Y.; Wang, J.; Huang, C.; Li, D.; et al. Adiponectin reduces brain injury after intracerebral hemorrhage by reducing NLRP3 inflammasome expression. Int. J. Neurosci. 2020, 130, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Aryanpour, R.; Zibara, K.; Pasbakhsh, P.; Jame’ei, S.B.; Namjoo, Z.; Ghanbari, A.; et al. 17β-Estradiol Reduces Demyelination in Cuprizone-fed Mice by Promoting M2 Microglia Polarity and Regulating NLRP3 Inflammasome. Neuroscience 2021, 463, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sun, J.; Kim, T.J.; Kim, Y.J.; Ko, S.B.; Kim, C.K.; et al. Pretreatment with low-dose fimasartan ameliorates NLRP3 inflammasome-mediated neuroinflammation and brain injury after intracerebral hemorrhage. Exp. Neurol. 2018, 310, 22–32. [Google Scholar] [CrossRef]
- Soriano-Teruel, P.M.; García-Laínez, G.; Marco-Salvador, M.; Pardo, J.; Arias, M.; DeFord, C.; et al. Identification of an ASC oligomerization inhibitor for the treatment of inflammatory diseases. Cell Death Dis. 2021, 12, 1155. [Google Scholar] [CrossRef]
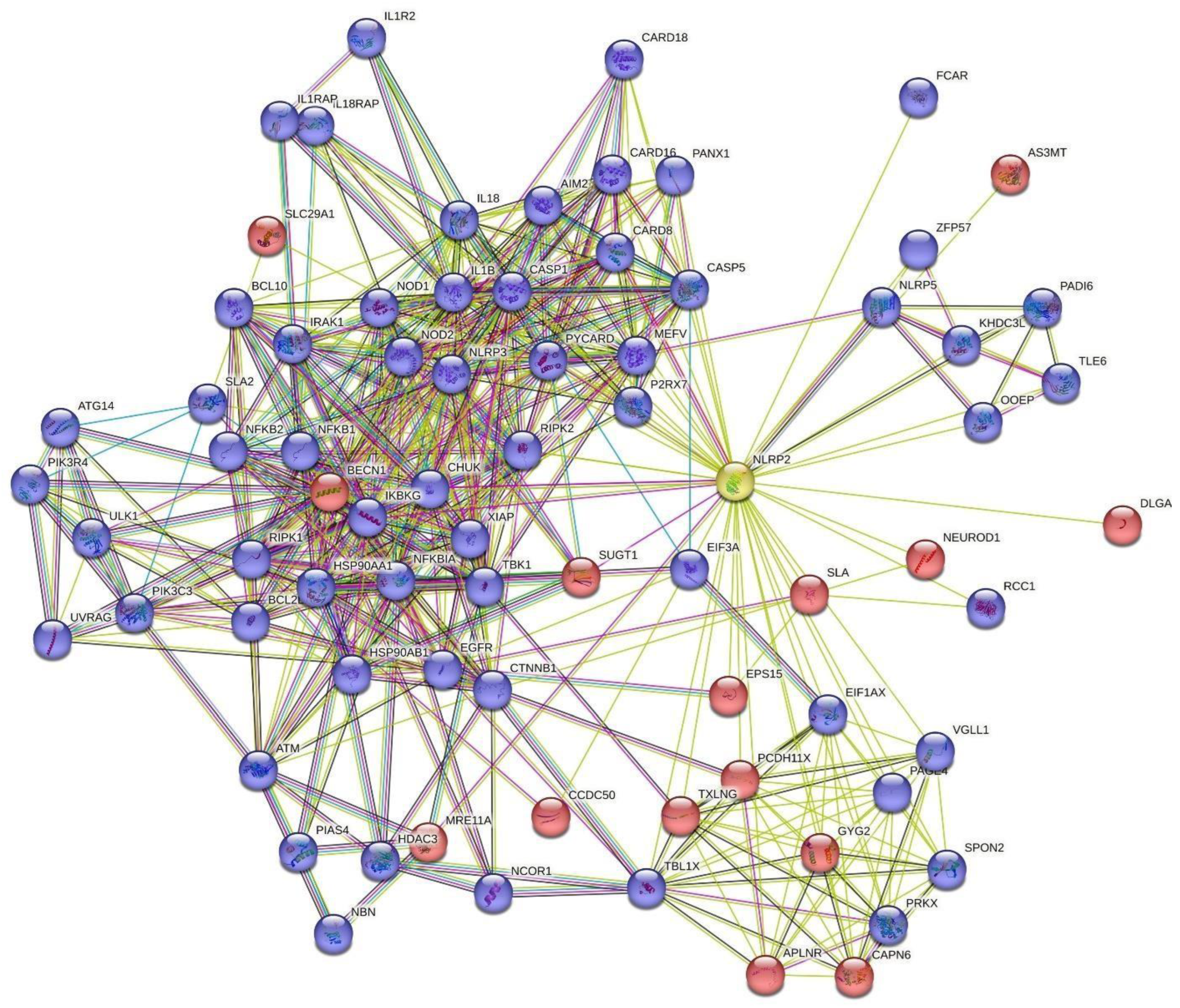
| Node protein hits | Full name | Interaction score | |
|---|---|---|---|
| 1 | CASP1 | Caspase-1 | 0.862 |
| 2 | PYCARD | Apoptosis-associated speck-like protein containing a CARD | 0.825 |
| 3 | CASP5 | Caspase-5 | 0.811 |
| 4 | MEFV | Pyrin | 0.798 |
| 5 | CARD8 | Caspase recruitment domain-containing protein 8 | 0.771 |
| 6 | SLA | Src-like-adapter; Adapter protein | 0.752 |
| 7 | KHDC3L | KHDC3-like protein; KH domain containing 3 like, subcortical maternal complex member | 0.739 |
| 8 | BECN1 | Beclin-1 | 0.664 |
| 9 | AIM2 | Interferon-inducible protein AIM2 | 0.645 |
| 10 | P2RX7 | P2X purinoceptor 7; Receptor | 0.638 |
| 11 | CARD16 | Caspase recruitment domain-containing protein 16 | 0.629 |
| 12 | PANX1 | Pannexin-1 | 0.600 |
| 13 | IL18 | Interleukin-18 | 0.597 |
| 14 | SLA2 | Src-like-adapter 2; Adapter protein | 0.590 |
| 15 | PAGE4 | P antigen family member 4 | 0.575 |
| 16 | EIF1AX | Eukaryotic translation initiation factor 1A | 0.567 |
| 17 | IL1B | Interleukin-1 beta | 0.563 |
| 18 | ZFP57 | Krab domain-containing zinc finger protein | 0.563 |
| 19 | VGLL1 | Transcription cofactor vestigial-like protein 1 | 0.562 |
| 20 | CHUK | Inhibitor of nuclear factor kappa-B kinase subunit alpha | 0.560 |
| 21 | NLRP5 | NACHT, LRR and PYD domains-containing protein 5 | 0.556 |
| 22 | PCDH11X | Protocadherin-11 X-linked; Potential calcium-dependent cell-adhesion protein | 0.547 |
| 23 | GYG2 | Glycogenin-2 | 0.515 |
| 24 | OOEP | Oocyte-expressed protein homolog | 0.514 |
| 25 | MRE11A | Double-strand break repair protein MRE11 | 0.490 |
| 26 | TLE6 | Transducin-like enhancer protein 6 | 0.487 |
| 27 | SUGT1 | SGT1 homolog, MIS12 kinetochore complex assembly cochaperone | 0.485 |
| 28 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | 0.473 |
| 29 | SPON2 | Spondin-2 | 0.471 |
| 30 | IKBKG | NF-kappa-B essential modulator | 0.470 |
| 31 | EPS15 | Epidermal growth factor receptor substrate 15 | 0.468 |
| 32 | NEUROD1 | Neurogenic differentiation factor 1 | 0.466 |
| 33 | CAPN6 | Calpain-6 | 0.462 |
| 34 | PRKX | cAMP-dependent protein kinase catalytic subunit PRKX | 0.456 |
| 35 | TXLNG | Gamma-taxilin | 0.455 |
| 36 | DLGAP2 | Disks large-associated protein 2 | 0.449 |
| 37 | APLNR | Apelin receptor | 0.445 |
| 38 | SLC29A1 | Solute carrier family 2 | 0.444 |
| 39 | CARD18 | Caspase recruitment domain-containing protein 18 | 0.437 |
| 40 | AS3MT | Arsenite methyltransferase | 0.423 |
| 41 | CCDC50 | Coiled-coil domain-containing protein 50 | 0.421 |
| 42 | TBL1X | F-box-like/WD repeat-containing protein TBL1X | 0.418 |
| 43 | PADI6 | Protein-arginine deiminase type-6 | 0.418 |
| 44 | FCAR | Immunoglobulin alpha Fc receptor | 0.408 |
| 45 | RCC1 | Regulator of chromosome condensation | 0.400 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





