Submitted:
05 July 2023
Posted:
07 July 2023
You are already at the latest version
Abstract
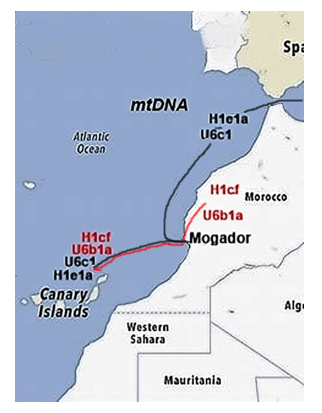
Keywords:
1. Introduction
2. Material and Methods
2.1. Samples
2.2. Sequence Classification
2.3. Population based statistical analyses
2.4. Coalescence Age Estimations
3. Results
3.1. Contamination and sequencing artefacts in the Canary Islands aboriginal maternal genetic pool
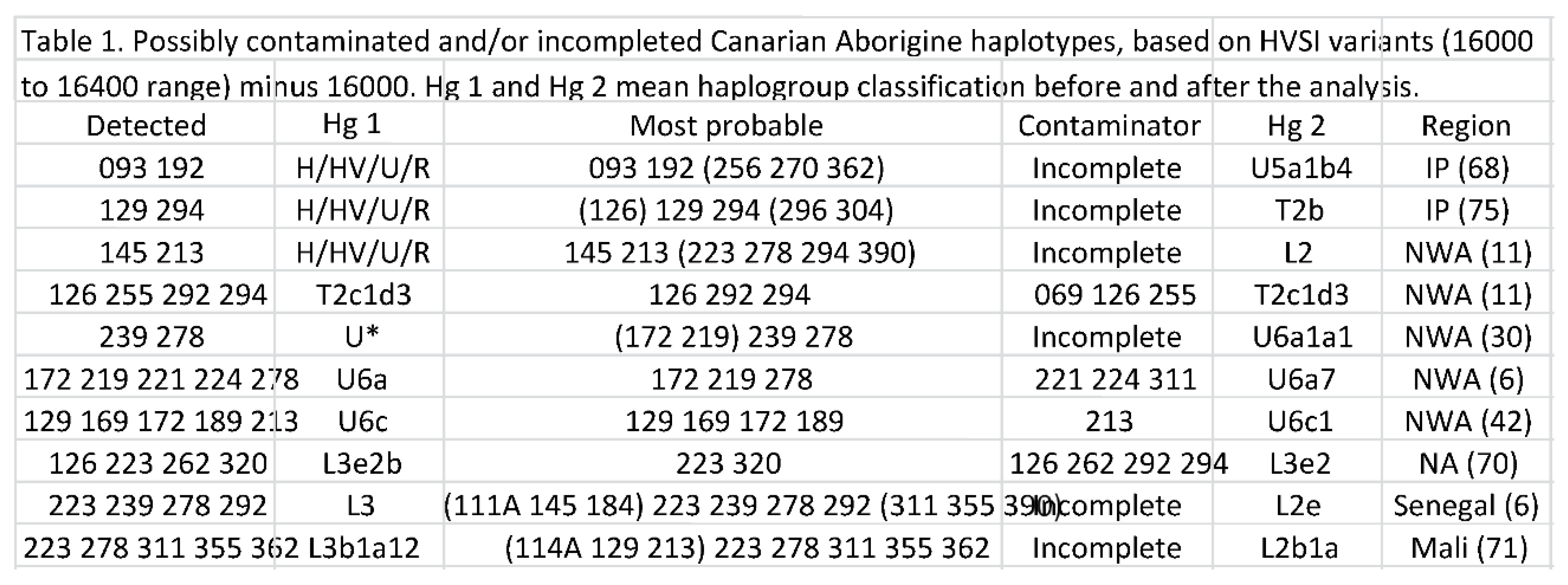
3.2. Persistency and phylogeographic origin of the Canary Islands aboriginal haplotypes
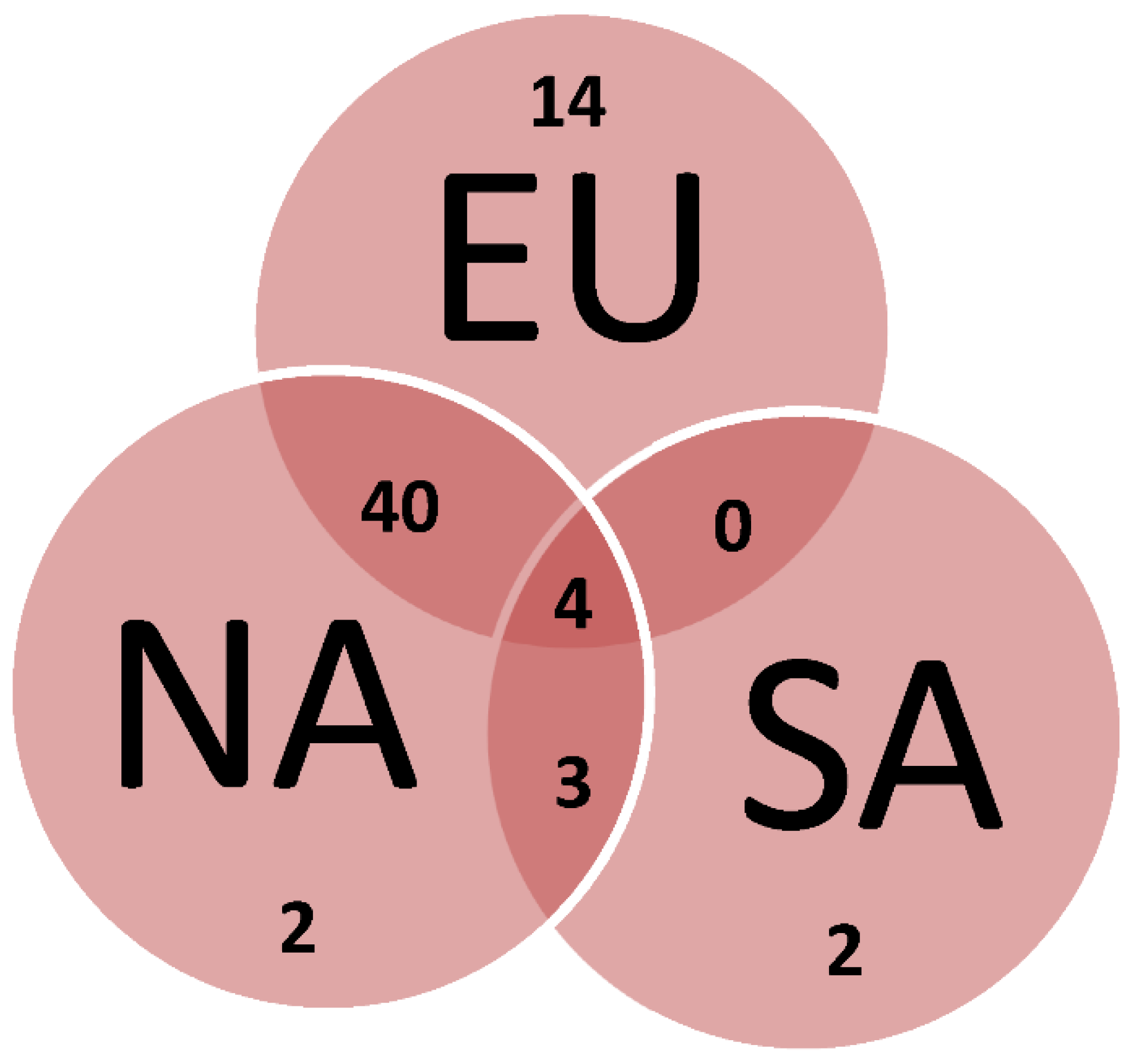
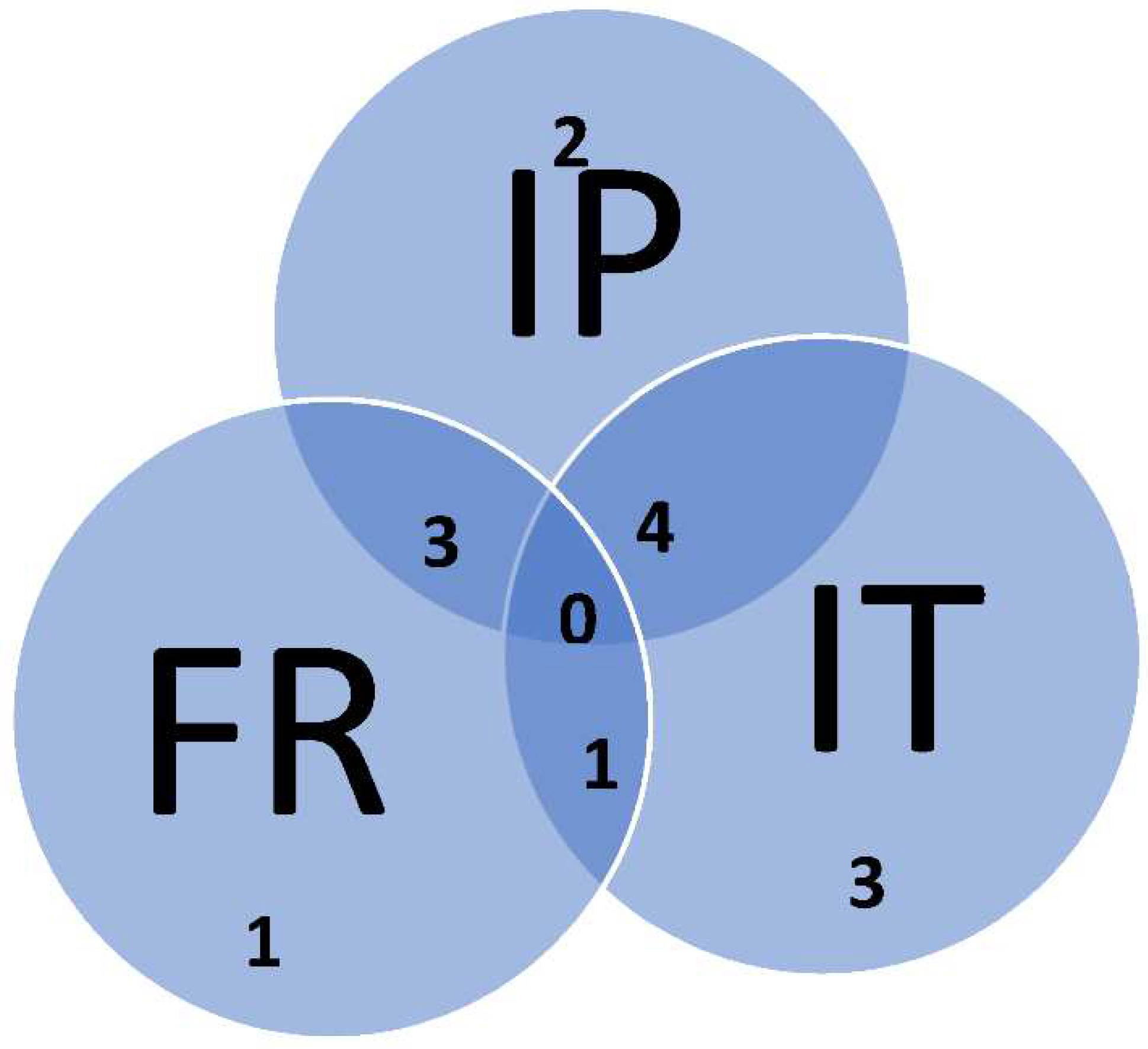
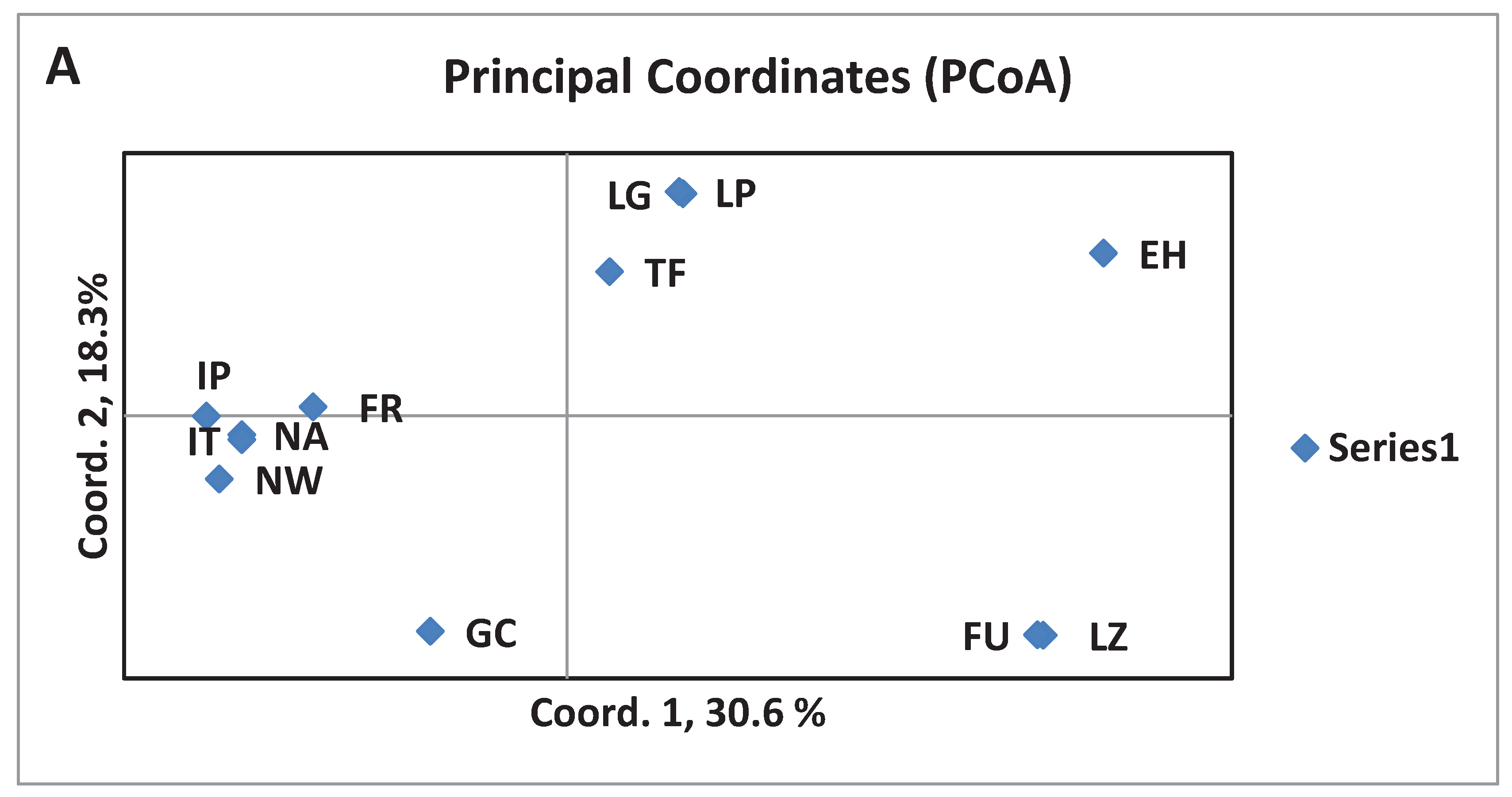
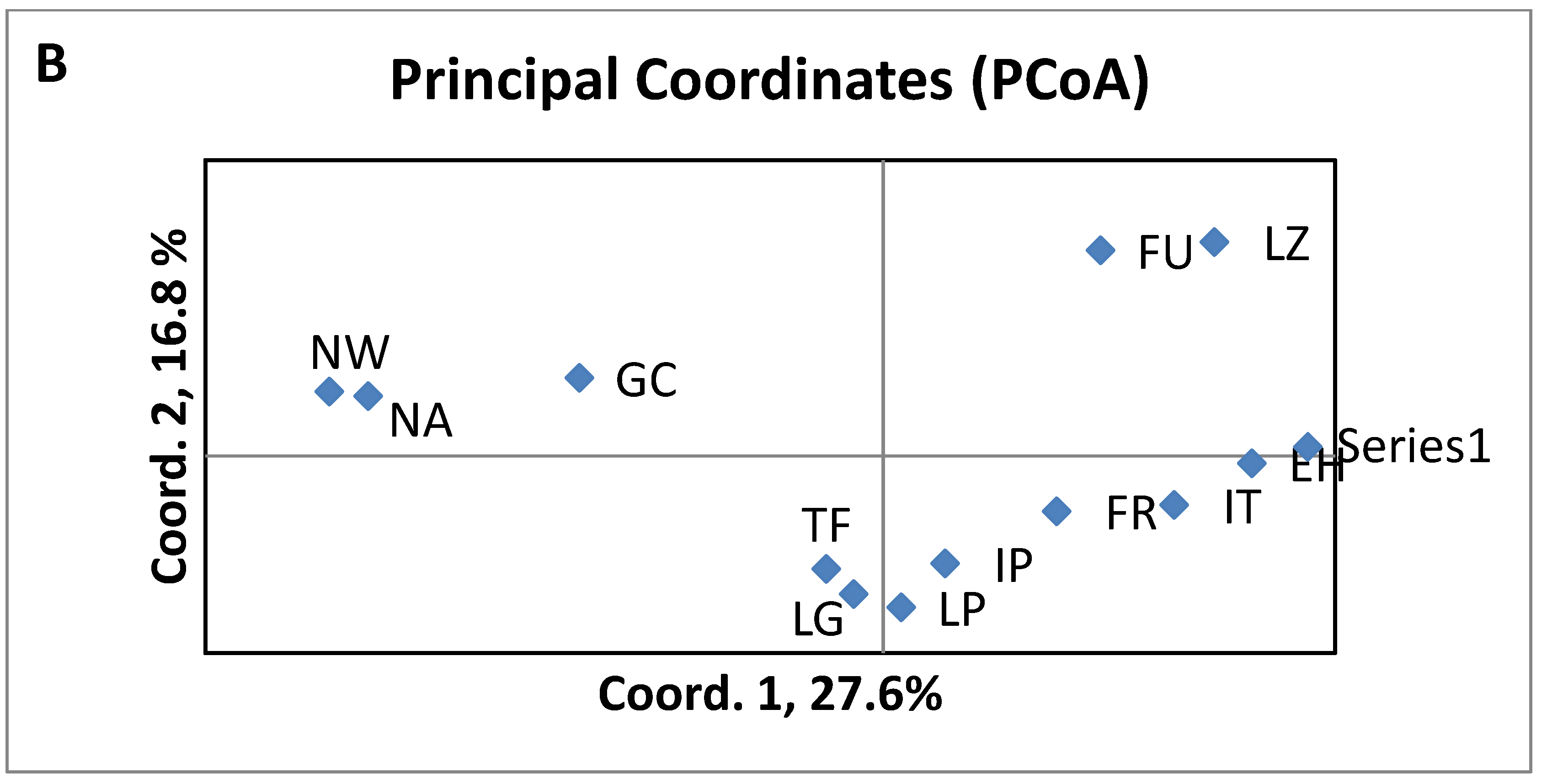
3.3. Divergence of aboriginal genetic pool among islands
3.4. New coalescence ages for the Aboriginal lineages
4. Discussion
4.1. Contamination problems in ancient DNA studies

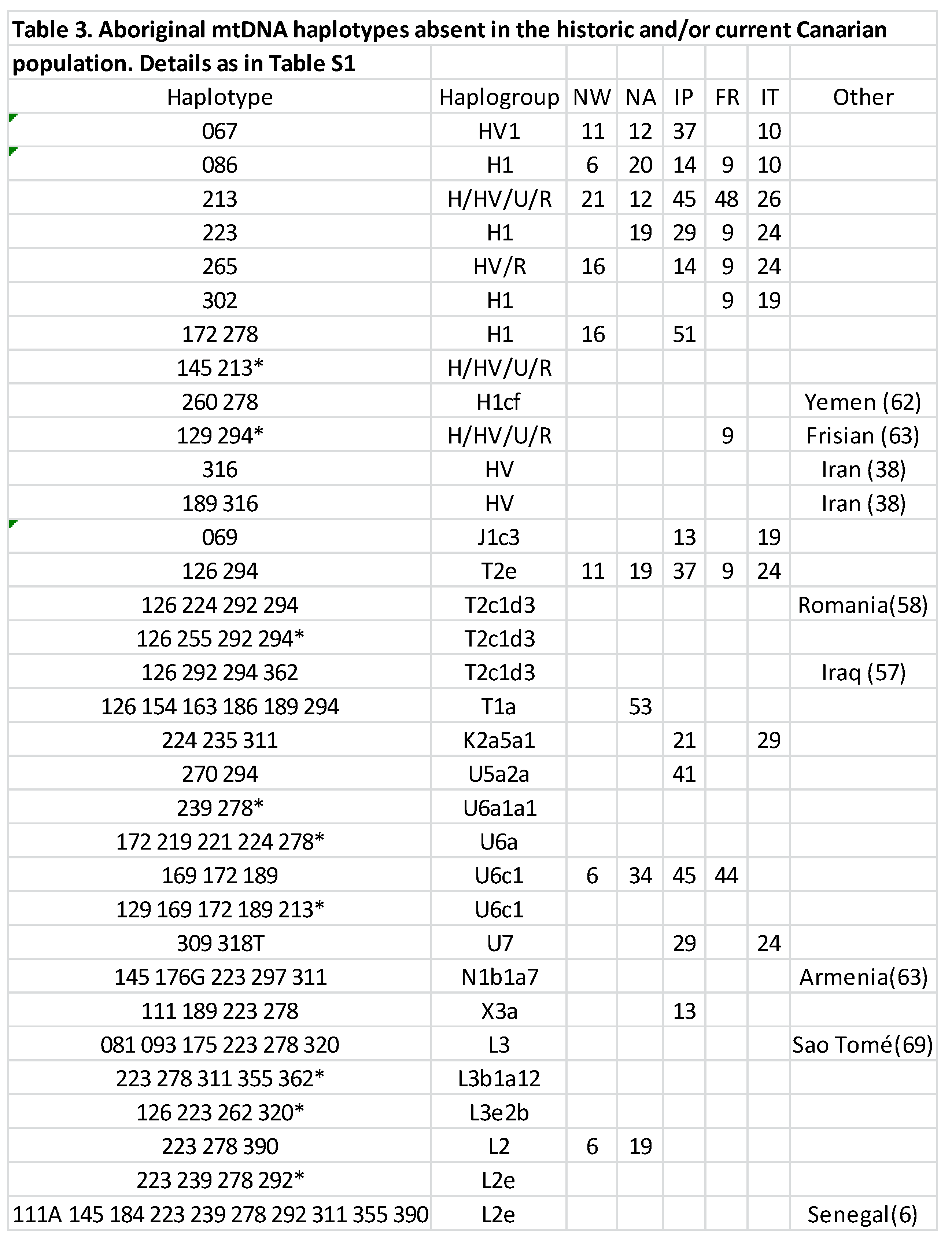
4.2. Lack of date and context of archaeological samples
4.3. Molecular age for a permanent aboriginal settlement
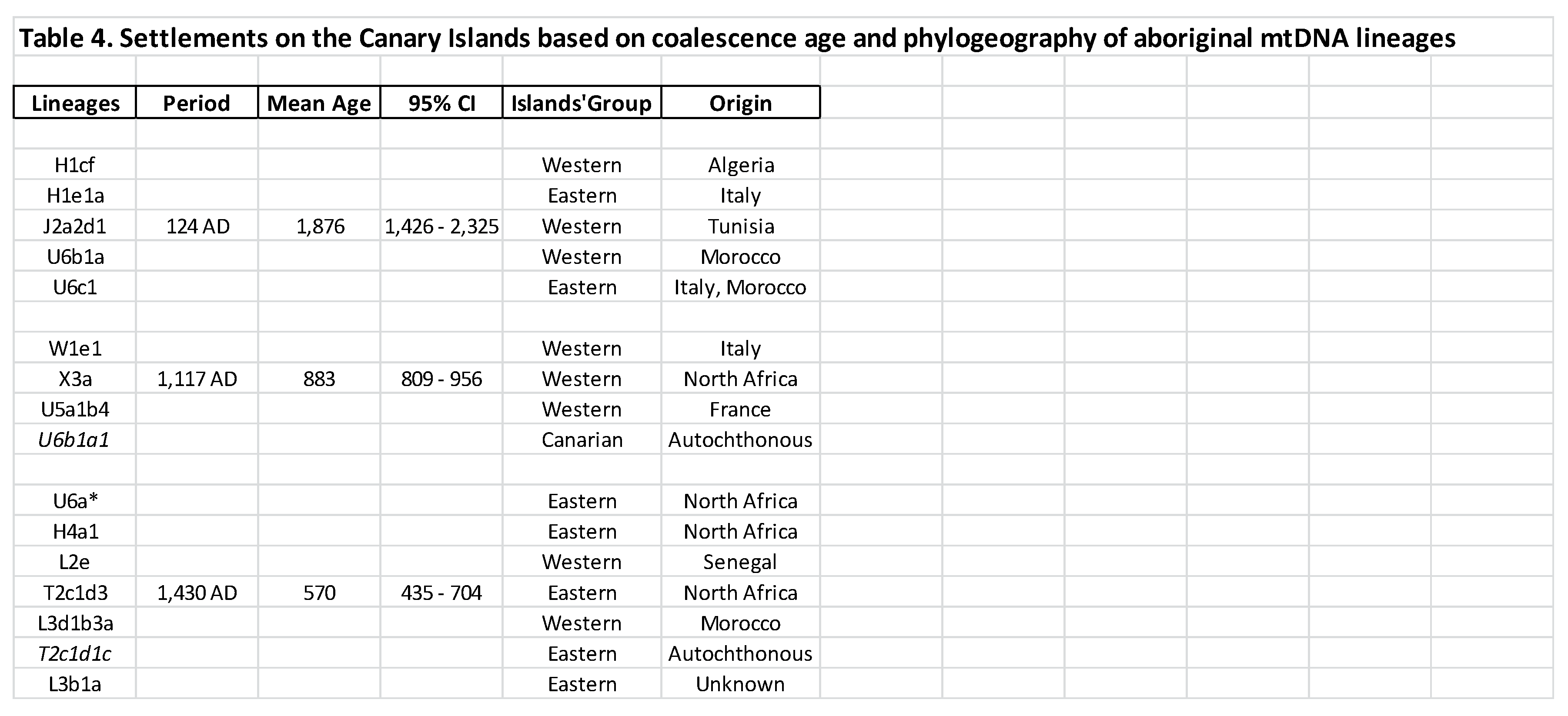
4.4. In support of a Roman-mediated aboriginal settlement of the Canary Islands
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blench R. The peopling of the Canaries by the Berbers: new data and new hypotheses. Études et documents berbères. 2022;45(2):149-173. [CrossRef]
- Fregel R, Ordóñez AC, Serrano JG. The demography of the Canary Islands from a genetic perspective. Human Molecular Genetics. 2021;30(R1):R64-R71. [CrossRef]
- ázquez JV, Barroso VA, Darias TD, Ben’\itez MM, Lecuyer C, Richardin P. Poblamiento, colonización y primera historia de Canarias: El C14 como paradigma/Settlement, colonization and early history of The Canary Islands: The C14 as a paradigm. Anuario de Estudios Atlánticos. 2020;(66):24-24.
- Orlando L, Allaby R, Skoglund P, et al. Ancient DNA analysis. Nature Reviews Methods Primers. 2021;1(1):14.
- Rodriguez-Varela R, Günther T, Krzewi’nska M, et al. Genomic analyses of pre-European conquest human remains from the Canary Islands reveal close affinity to modern North Africans. Current Biology. 2017;27(21):3396-3402.
- Fregel R, Ordóñez AC, Santana-Cabrera J, et al. Mitogenomes illuminate the origin and migration patterns of the indigenous people of the Canary Islands. PloS one. 2019;14(3):e0209125. [CrossRef]
- Maca-Meyer N, Arnay M, Rando JC, et al. Ancient mtDNA analysis and the origin of the Guanches. European Journal of Human Genetics. 2004;12(2):155-162. [CrossRef]
- Fregel R, Pestano J, Arnay M, Cabrera VM, Larruga JM, González AM. The maternal aborigine colonization of La Palma (Canary Islands). European Journal of Human Genetics. 2009;17(10):1314-1324. 1324. [CrossRef]
- Fregel R, Cabrera VM, Larruga JM, et al. Isolation and prominent aboriginal maternal legacy in the present-day population of La Gomera (Canary Islands). European Journal of Human Genetics. 2015;23(9):1236-1243. [CrossRef]
- Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. The American Journal of Human Genetics. 2002;71(5):1150-1160. [CrossRef]
- Garcia-Olivares V, Rubio-Rodr’\iguez LA, Muñoz-Barrera A, et al. Digging into the admixture strata of current-day Canary Islanders based on mitogenomes. iScience. 2022:105907. [CrossRef]
- Soares P, Ermini L, Thomson N, et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. The American Journal of Human Genetics. 2009;84(6):740-759. [CrossRef]
- Cabrera VM. Human molecular evolutionary rate, time dependency and transient polymorphism effects viewed through ancient and modern mitochondrial DNA genomes. Scientific Reports. 2021;11(1):1-8. [CrossRef]
- Cabrera VM. Counterbalancing the time-dependent effect on the human mitochondrial DNA molecular clock. BMC Evolutionary Biology. 2020;20(1):1-9. [CrossRef]
- MISSING:berbersgenomic. MISSING:berbersgenomic. 2023.
- Ordóñez AC, Fregel R, Trujillo-Mederos A, Hervella M, de-la-Rúa C, Arnay-de-la-Rosa M. Genetic studies on the prehispanic population buried in Punta Azul cave (El Hierro, Canary Islands). Journal of Archaeological Science. 2017;78:20-28.
- Maca-Meyer N, Cabrera VM, Arnay M, et al. Mitochondrial DNA diversity in 17th-18th century remains from Tenerife (Canary Islands). American Journal of Physical Anthropology: The Official Publication of the American Association of Physical Anthropologists. 2005;127(4):418-426.
- Santana J, Fregel R, Lightfoot E, et al. The early colonial Atlantic world: New insights on the African diaspora from isotopic and ancient DNA analyses of a multiethnic 15th-17th century burial population from the Canary Islands, Spain. American Journal of Physical Anthropology. 2016;159(2):300-312. [CrossRef]
- Santos C, Fregel R, Cabrera VM, González AM, Larruga JM, Lima M. Mitochondrial DNA patterns in the Macaronesia islands: Variation within and among archipelagos. American Journal of Physical Anthropology: The Official Publication of the American Association of Physical Anthropologists. 2010;141(4):610-619.
- Rando J, Cabrera V, Larruga J, et al. Phylogeographic patterns of mtDNA reflecting the colonization of the Canary Islands. Annals of human genetics. 1999;63(5):413-428. [CrossRef]
- Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: a human mitochondrial genome database. Nucleic acids research. 1996;24(1):177-179. [CrossRef]
- Huber N, Parson W, Dür A. Next generation database search algorithm for forensic mitogenome analyses. Forensic Science International: Genetics. 2018;37:204-214. [CrossRef]
- Weissensteiner H, Pacher D, Kloss-Brandstätter A, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic acids research. 2016;44(W1):W58-W63. [CrossRef]
- Van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Human mutation. 2009;30(2):E386-E394. [CrossRef]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature genetics. 1999;23(2):147-147. [CrossRef]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular ecology notes. 2006;6(1):288-295. [CrossRef]
- Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59(4):935-45.
- Coudray C, Olivieri A, Achilli A, et al. The complex and diversified mitochondrial gene pool of Berber populations. Annals of Human Genetics. 2009;73(2):196-214. [CrossRef]
- Secher B, Fregel R, Larruga JM, et al. The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC evolutionary biology. 2014;14(1):1-17. [CrossRef]
- Pennarun E, Kivisild T, Metspalu E, et al. Divorcing the Late Upper Palaeolithic demographic histories of mtDNA haplogroups M1 and U6 in Africa. BMC evolutionary biology. 2012;12(1):1-12. [CrossRef]
- Pierron D, Heiske M, Razafindrazaka H, et al. Genomic landscape of human diversity across Madagascar. Proceedings of the National Academy of Sciences. 2017;114(32):E6498-E6506. 6498. [CrossRef]
- Pino-Yanes M, Corrales A, Basaldúa S, et al. North African influences and potential bias in case-control association studies in the Spanish population. PLoS One. 2011;6(3):e18389. 1838. [CrossRef]
- Flores C, Maca-Meyer N, Perez J, Gonzalez A, Larruga J, Cabrera V. A predominant European ancestry of paternal lineages from Canary Islanders. Annals of human genetics. 2003;67(2):138-152. [CrossRef]
- Van de Loosdrecht M, Bouzouggar A, Humphrey L, et al. Pleistocene North African genomes link near Eastern and sub-Saharan African human populations. Science. 2018;360(6388):548-552. [CrossRef]
- Kefi R, Hechmi M, Naouali C, et al. On the origin of Iberomaurusians: new data based on ancient mitochondrial DNA and phylogenetic analysis of Afalou and Taforalt populations. Mitochondrial DNA Part A. 2018;29(1):147-157. [CrossRef]
- Drosou K, Collin TC, Freeman PJ, Loynes R, Freemont T. The first reported case of the rare mitochondrial haplotype H4a1 in ancient Egypt. Scientific Reports. 2020;10(1):1-8. [CrossRef]
- Plaza S, Calafell F, Helal A, et al. Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean. Annals of human genetics. 2003;67(4):312-328. [CrossRef]
- Aboukhalid R, Sturk-Andreaggi K, Bouabdellah M, Squalli D, Irwin JA, Amzazi S. Mitochondrial DNA control region variation from samples of the Moroccan population. International journal of legal medicine. 2013;127:757-759. [CrossRef]
- Rando JC, Pinto F, González AM, et al. Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with European, near-eastern, and sub-Saharan populations. Annals of human genetics. 1998;62(6):531-550. [CrossRef]
- González AM, Garc’\ia O, Larruga JM, Cabrera VM. The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country. BMC genomics. 2006;7(1):1-7. [CrossRef]
- Martiniano R, Cassidy LM, Ó’Maoldúin R, et al. The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS genetics. 2017;13(7):e1006852. [CrossRef]
- Richards M, Macaulay V, Hickey E, et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67(5):1251-76.
- Sampietro ML, Lao O, Caramelli D, et al. Palaeogenetic evidence supports a dual model of Neolithic spreading into Europe. Proceedings of the Royal Society B: Biological Sciences. 2007;274(1622):2161-2167. [CrossRef]
- Krings M, Bauer K, Geisert H, et al. mtDNA analysis of Nile River Valley populations: A genetic corridor or a barrier to migration? The American Journal of Human Genetics. 1999;64(4):1166-1176.
- Del Arco M, Del Arco M, Atiénzar E, et al. Dataciones absolutas en la prehistoria de Tenerife. 1997.
- Maca-Meyer N, González AM, Pestano J, Flores C, Larruga JM, Cabrera VM. Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC genetics. 2003;4(1):1-11. [CrossRef]
- Pereira L, Silva NM, Franco-Duarte R, et al. Population expansion in the North African late Pleistocene signalled by mitochondrial DNA haplogroup U6. BMC evolutionary biology. 2010;10(1):1-10. [CrossRef]
- Henn BM, Gignoux CR, Feldman MW, Mountain JL. Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Molecular biology and evolution. 2009;26(1):217-230. [CrossRef]
- Fregel R, Gomes V, Gusmão L, et al. Demographic history of Canary Islands male gene-pool: replacement of native lineages by European. BMC evolutionary Biology. 2009;9(1):1-14. [CrossRef]
- Velasco-Vázquez J, Alberto-Barroso V, Delgado-Darias T, Moreno-Ben’\itez M. A propósito del poblamiento aborigen en Gran Canaria. Demograf’\ia, dinámica social y ocupación del territorio. Complutum. 2021.
- Navarro Mederos JF. Arqueolog’\ia de las islas Canarias. 1997.
- Hooton EA. The Ancient Inhabitants of the Canary Islands, Harvard. African Studies, Cambridge. 1925;7:261-267.
- Fuste M. Physical anthropology of the Canary Islands: old and new views. American Journal of Physical Anthropology. 1965;23(3):285-291. [CrossRef]
- Schwidetzky I. La población prehispánica de las Islas Canarias: investigaciones antropológicas. Museo arqueológico; 1963.
- Guatelli-Steinberg D, Irish J, Lukacs J. Canary islands-north African population affinities: measures of divergence based on dental morphology. Homo. 2001;52(2):173-188. [CrossRef]
- Atoche Peña P, Ram’\irez Rodr’\iguez MÁ. El yacimiento de Buenavista, un asentamiento fenicio púnico en Lanzarote, Islas Canarias (circa 960-360 ane). 2019.
- Fidel DR, del Arco Aguilar M del C. Desde el taller de púrpura romano de Lobos 1, una mirada a las actividades haliéuticas/From the roman purple workshop of Lobos 1, a look at halieutic activities. Coloquios de Historia Canario Americana. 2020.
- Mart’\in AM, Cobo GE. Mare purpureum. Producción y comercio de la púrpura en el litoral atlántico norteafricano. Rivista di studi fenici. 2006;34:71-96.
- Mercer J. The Canary Islanders: their prehistory, conquest, and survival. London: Collings; 1980. [CrossRef]
- Padilla AP. El poblamiento prehistórico de las islas Canarias. El Museo Canario. 2009;64:79-97.
- Hagenblad J, Morales J, Leino MW, Rodr’\iguez-Rodr’\iguez AC. Farmer fidelity in the Canary Islands revealed by ancient DNA from prehistoric seeds. Journal of Archaeological Science. 2017;78:78-87. [CrossRef]
- Amills M, Capote J, Tomàs A, et al. Strong phylogeographic relationships among three goat breeds from the Canary Islands. Journal of Dairy Research. 2004;71(3):257-262. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




