1. Introduction
The virus responsible for the COVID-19 pandemic, SARSCoV-2, is a betacoronavirus like severe acute respiratory syndrome coronavirus (SARSCoV) and the Middle East respiratory syndrome coronavirus (MERSCoV) [
1]. Several patients with SARSCoV-2 infection develop acute respiratory distress syndrome likely due to excessive pro-inflammatory host response, including aberrant induction of inflammatory cytokines, and is associated with severe lung pathology with a fatal outcome in a non-negligible number of cases [
23].
The global clinical experience with ribavirin for the treatment of COVID-19 pneumonia started with SARSCoV, for which RBV was initially indicated based on the pathological similarity of SARSCoV with the acute respiratory syndrome, which requires a typical administration of RBV and corticosteroid [
4,
5].
Ribavirin for inhalation solution is FDA-approved for the treatment of infants and young children with severe lower respiratory tract infections due to respiratory syncytial virus [
6] and has been shown to be effective against different influenza viruses [
7,
8]. Recently, the administration of RBV through aerosol has been made available in Italy for patients with COVID-19 as part of a compassionate use program [
9].
From a molecular point of view, RBV can limit viral replication basing on multimodal antiviral proprieties: first, RBV is a guanosine analogue, and the most straightforward mechanism of action is the inhibition of viral RNA synthesis. A second mechanism of action involves the competitive inhibition of the inosine monophosphate dehydrogenase (IMPDH) enzyme, through specific binding to the IMPDH substrate (inosine-5-monophosphate) ultimately leading to decreased synthesis and lower levels of guanosine triphosphate (GTP). A third mechanism of action is inferred from data demonstrating that RBV triphosphate can be incorporated into the viral genome resulting in ‘lethal RNA mutagenesis’. Ribavirin can interact with viral genome by both being incorporated as a substrate into the RNA molecule and by causing transition mutations [
10,
11,
12]. Since RBV can base pair with both cytosine and uracil, once incorporated in the genome, it would drive an increasing number of mutations [
13,
14].
The SARSCoV-2 genome consists of a positive-sense single stranded RNA ranging from 26.0 kilobase (kb) to 32.0 kb, having several open reading frames (ORFs), rendering its genomic organization like other known SARSCoVs [
15]. Two-thirds of the viral genomic region located downstream to 5′-end, involves replicase gene referred to as open reading frame 1a and ab (ORF1ab), which encode the nonstructural proteins (nsps). The remaining 10 kb region preceding 3′-end encodes various structural proteins involving surface (S), envelope (E), membrane (M), and nucleocapsid (N). Additionally, the structural genes encode nine accessory proteins, encoded by ORF3a, ORF3d, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF14, and ORF10 genes. Genomic organization of SARSCoV-2 is described in Figure 1.
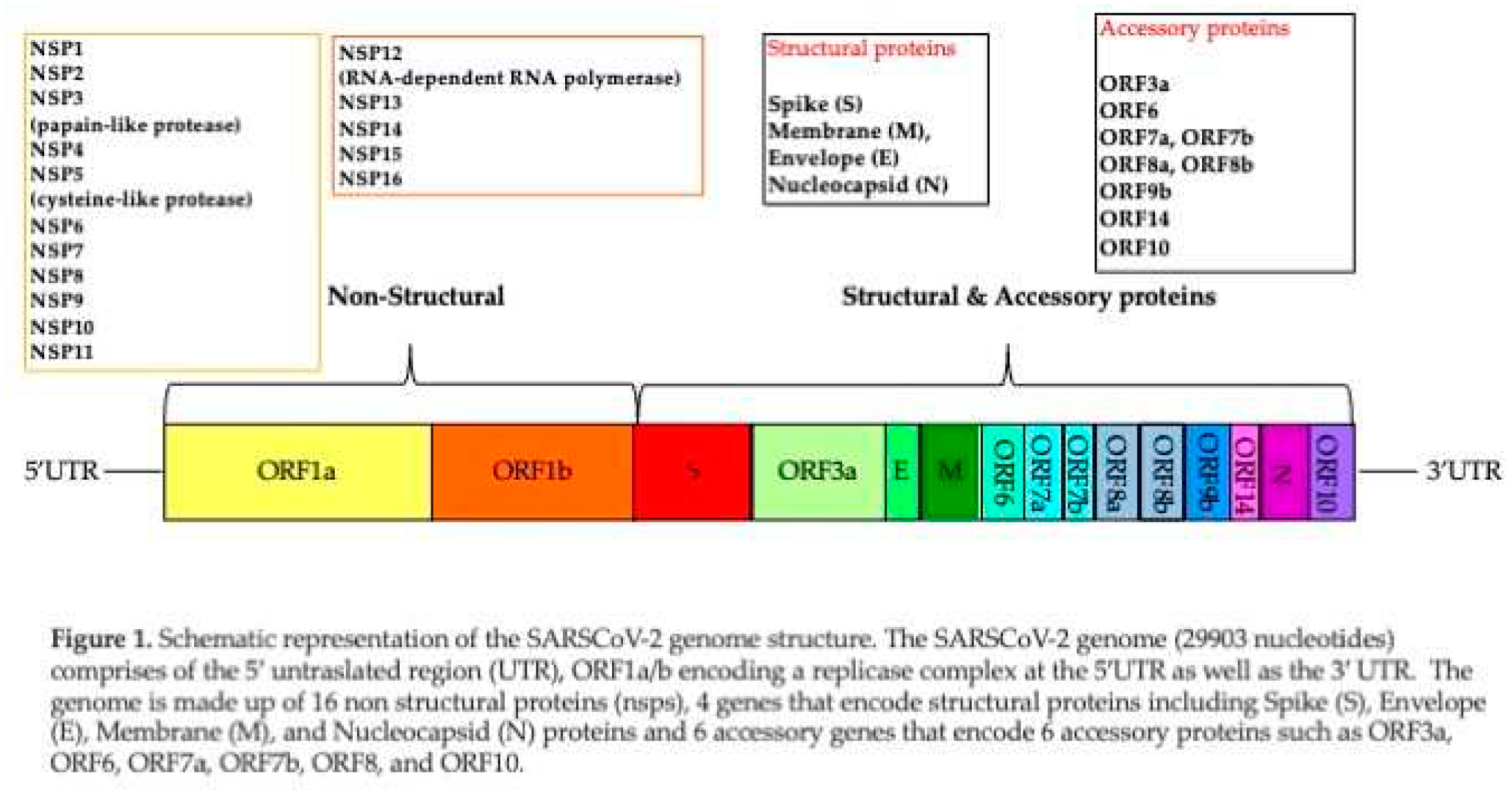
As other RNA viruses, SARSCoV-2 may present high intra-host adaptability.
It is well known how during the COVID-19 pandemic a higher incidence of SARSCoV-2 infection in poorest countries resulted in the emergence of more transmissible viral variants often showing improved immune-escape compared to previously circulating variants. Therefore, virus adaptability represents a key molecular aspect that drives evolution of variants of concern (VOC) with higher or lower pathogenicity. Last, the presence of mutations can affect vaccine efficacy, laboratory diagnosis and therapies already available [
15]. Antivirals, target viral enzymes involved in virus intracellular replication or hamper virus entry process. However, due to adaptive selection of certain mutations, several drugs can become less efficient or, in the worst scenarios, ineffective.
As detailed above, RBV, may limit SARSCoV-2 replication by multimodal mechanisms, possibly limiting the emergence of novel virus variants.
Here we describe possible effect of RBV administered via aerosol on viral load and clinical outcomes in hospitalized participants with COVID-19 pneumonia.
2. Materials and Methods
This retrospective study included data collected between July 2020 and August 2022, within the compassionate use study allowing the treatment with RBV for inhalation solution, USP, in adult participants (PTs) with a laboratory confirmed SARSCoV-2 infection, diagnosed with primary mild-moderate COVID-19 pneumonia (
https://www.aifa.gov.it/programmi-di-uso-compassionevole-covid-19).
Computed tomography-Scan was performed to assess the presence of pneumonia and evaluate the extent of parenchyma inflammation. From a descriptive point of view, our radiologists defined the degree of pulmonary involvement using a visual assessment expressed on a scale (<10%, 10-25%, 25- 50%, 50-75%, > 75%) [
16]. All PTs underwent standard of care treatment according to local guidelines. The primary endpoint was to assess the effect of RBV aerosol on SARSCoV-2 viral load in nasopharyngeal samples, expressed as the proportion of participants achieving a “virus-negative” status by means of real-time polymerase chain reaction (PCR) at the time of discharge.The secondary endpoints were to measure the dynamic of SARSCoV-2 in nasopharyngeal samples by using a semi quantitative real-time polymerase chain reaction (PCR) assay threshold cycle (Ct) at baseline (BL), end of treatment (EOT) and discharge. In some PTs who were discharged with a SARSCoV-2 positive PCR, a subsequent specimen was investigated within 14 days from discharge. Another secondary end point was the evaluation of clinical efficacy expressed through the variation of clinical status severity scale (CSS), calculated on the 7-point ordinal scale from the first dose date (BL) up to 6 days (EOT) or date of RBV aerosol discontinuation and discharge or death. The clinical status severity (CSS) was rated on a 7-point scale recommended by the World Health Organization (WHO) R&D Blueprint expert group [
17]. The change in clinical/respiratory parameters was evaluated according to the compassionate use study protocol: PaO2/FiO2 ≥300, peripheral capillary oxygen saturation (SpO2), and change in pulmonary imaging by CT-Scan. Laboratory blood exams considered at baseline and discharge included: white blood cells (WBC) count, lymphocytes, C-reactive protein, lactic dehydrogenase (LDH), ferritin, fibrinogen, D-dimer, interleukin-6 (IL-6) in plasma samples. Was also considered the proportion of participants with drug-related adverse events leading or not to RBV discontinuation.The study was conducted in accordance with the ethical principles that have their origin in the current Declaration of Helsinki and will be consistent with International Conference on Harmonization Good Clinical Practice (ICH GCP), Good Epidemiology Practices (GEP), and applicable regulatory requirements. Samples were collected upon informed consent in accordance with the Helsinki Declaration and with local ethical committee approvals: Covid-BioB, ClinicalTrials.gov NCT04318366; Ethical Committee approval number 34/int/2020.
Sequential nasopharyngeal samples (taken at least at day1, BL evaluation and day6, EOT were aliquoted and stored at -80°C until testing. Identification and semiquantitative analysis of SARSCoV-2 were performed through the Cobas® SARSCoV-2Test (Roche Diagnostics) using the fully automated Cobas® 6800/8800 Systems under FDA Emergency Use Authorization (EUA). The assay is a single-well dual target assay, which includes specific detection of SARSCoV-2 by targeting conserved regions within the ORF 1a/b and E genes, a full-process negative control, positive control, and internal control. Results were recorded both as positive/negative and semi-quantitatively based on amplification cycle threshold (Ct) values for one of the two target genes. A decrease in viral load corresponds to an increase in Ct value; a CT>40 was considered as negative. The mutational profile was assessed by sequence analysis of SARSCoV-2 whole genome (WGS) of nasopharyngeal samples, as previously described [
18]. Briefly, RNA extracts were processed with the CleanPlex
® SARSCoV-2 Panel (Paragon Genomics, Hayward, CA, USA) and sequenced with MiSeq Reagent Kit v2 (300-cycles) (Illumina, San Diego, CA, USA) on the Illumina
® MiSeq platform. Genomic reconstruction was performed using the SOPHiA DDM
™ platform (SOPHiA Genetics, Lausanne, Switzerland). The mutational spectrum was inferred on genome consensus sequences by using the Stanford Coronavirus Resistance Database (CoV-RDB;
https://covdb.stanford.edu). A maximum-likelihood phylogenetic tree containing the SARSCoV-2 sequences was constructed by using Clustal_Omega of EMBL site: Bioinformatics Tools for Multiple Sequence Alignment < EMBL-EBI. The nucleotide distances were calculated by generating a distance matrix using the maximum-likelihood model in the DNADIST program (PHYLIP 3.5c package). The phylogenetic tree was drawn using TreeViewPPC version 1.5.3.
3. Results
3.1. Clinical features in the study group
Characteristics of PTs at BL at EOT and at discharge are summarized in Table 1.
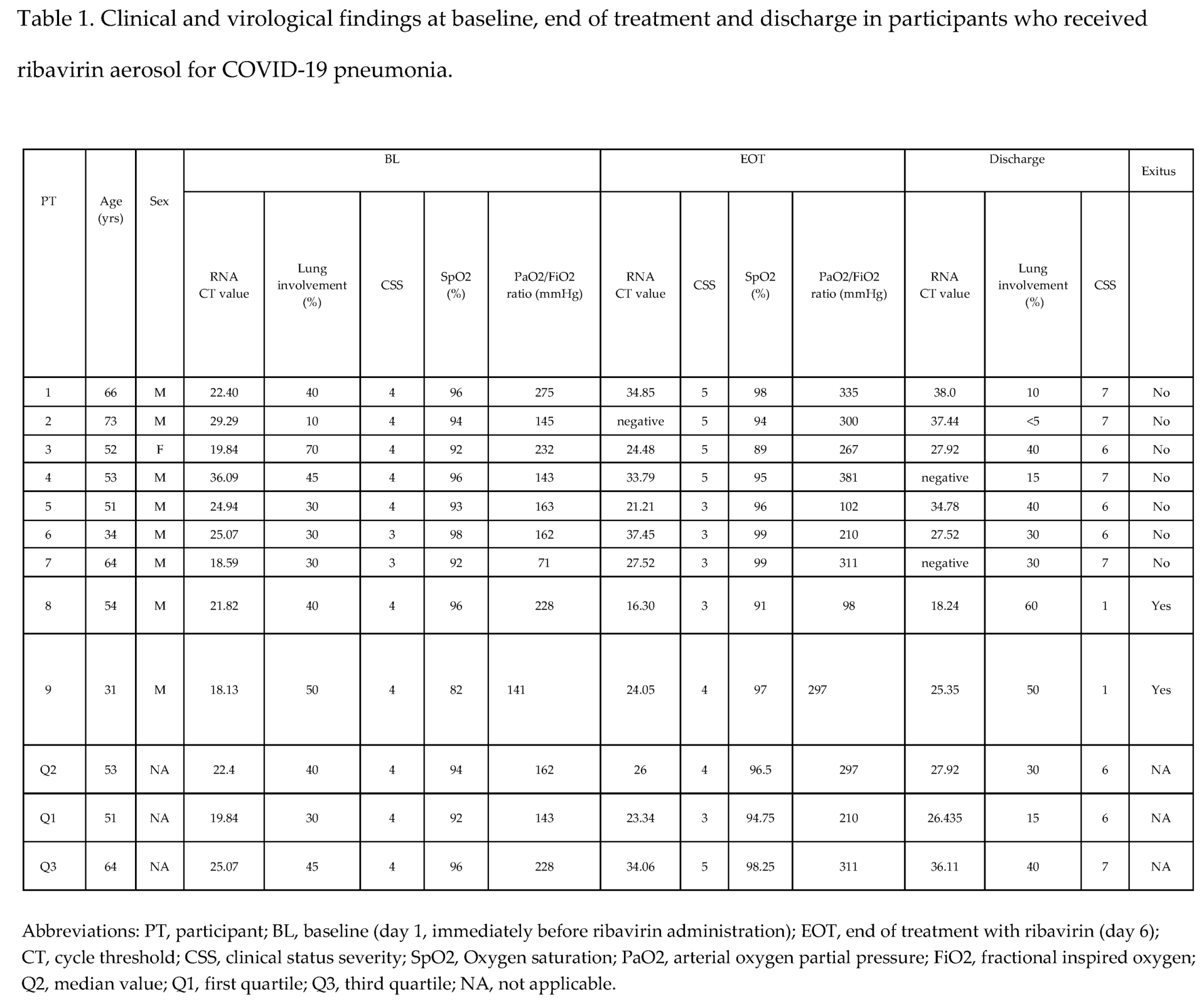
In total were included 9 PTs with mild-moderate COVID-19 pneumonia: 8 males and one female. The median age was 53 years (IQR 51-64). The most common pre-existing comorbidities were cardiovascular/metabolic diseases: PT1, PT3, PT4 and PT7 suffered hypertension controlled by optimal antihypertensive therapy, PT2 and PT4 had dyslipidemia. PTs 2 and 3 also suffered type 2 diabetes mellitus. Two PTs (PT8 and PT9) had hematologic malignancy, while PT5 and PT6 had no comorbidities. The median percentage of lung involvement by CT-Scan was 40% (IQR 30-45 %).
Seven PTs (PT1-PT5 and PT8, PT9) had a CSS value of 4. Two PTs (PT6, PT7) had a CSS value of 3.
The median SpO2 value at BL under different oxygen flow, was 94 (IQR 92-96) with a median PaO2/FiO2 ratio of 162 mmHg (IQR 143-228).
At EOT, 4/9 PTs (PT 1-4) experienced an improvement of their CSS, 3 (PT6, PT7, PT9) did not have CSS change and the remaining 2 (PT8 and PT5) had a worsening of their CSS scale-point. SpO2 ameliorated in 5 PTs, was unchanged in 2 and slightly worsened in two other PTs, while PaO2/FiO2 increased in 7/9 PTs (Table 1).
At discharge all these parameters reflecting clinical status were improved in 7 PTs with a favorable outcome, while worsened in two PTs (PT8, PT9) who deceased during hospitalization (Table 1).
Among the 7 PTs with a favorable outcome was observed a decrease of lung involvement in 4, (PT1-4) while lung damage was unchanged in two PTs (PT6,7) and slightly increased in the remaining one (PT5). In the 2 PTs (PT8 and PT9) who deceased a progressive worsening of clinical status was observed because of superinfections or a progression of the underlying hematological malignancy (Table 1).
3.2. Biochemistry in the study group
Biochemistry at BL and discharge are described in Table 2. At BL evaluation most participants had lymphopenia with median total lymphocytes of 0.9 x10
9/L, (IQR 0.6-1.4) a median ferritin levels of 2266 ng/mL (IQR 1038-3391). Other markers of inflammation or cytokine activation like C-reactive protein, fibrinogen, lactate dehydrogenase and interleukin-6 (IL-6) values were found abnormally high (Table 2).
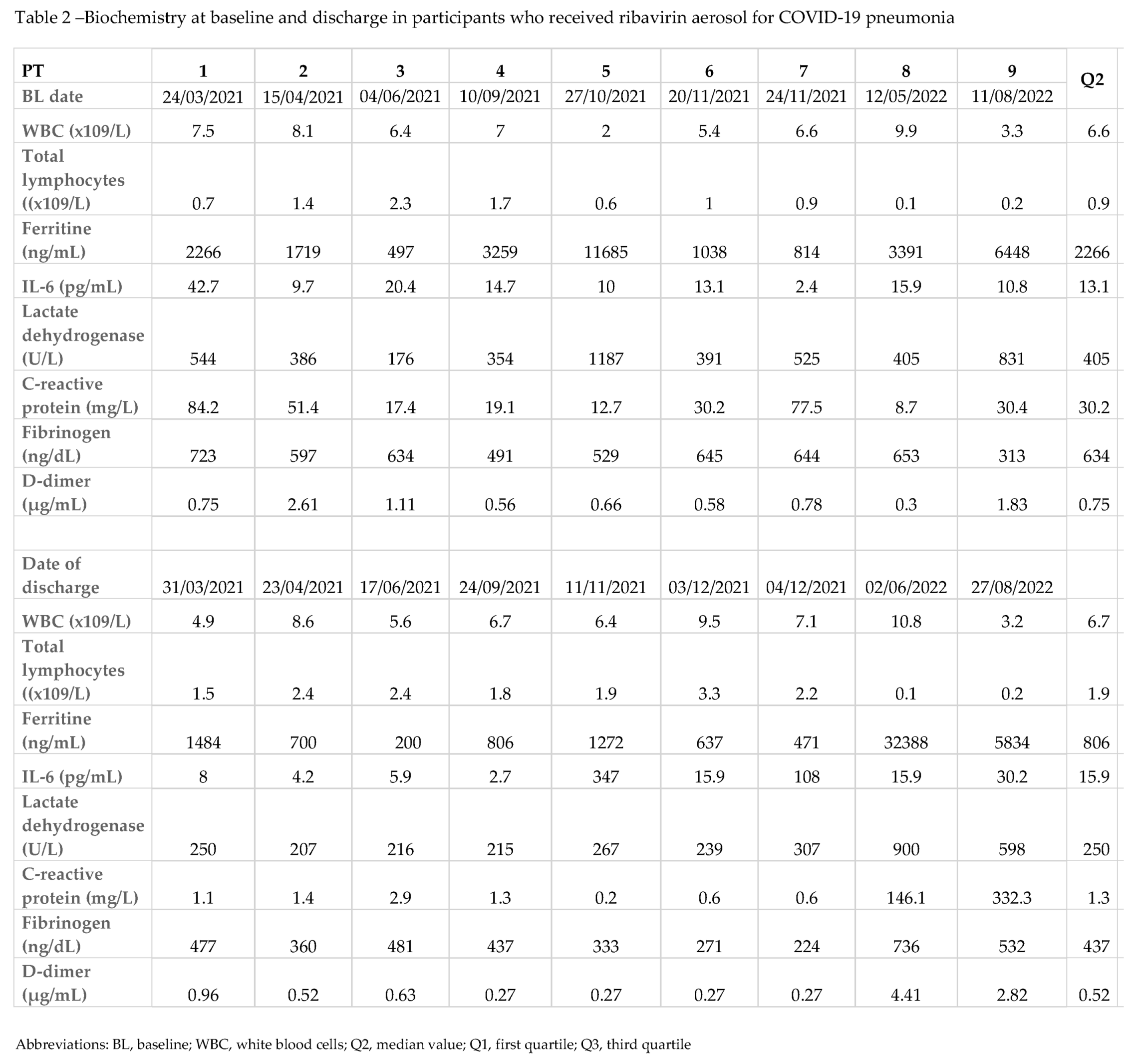
At discharge, total lymphocytes were increased, and biomarkers of inflammation (ferritin, C-reactive protein, fibrinogen, and lactate dehydrogenase) decreased, while IL-6 value persisted increased with a median value of 15.9 picogram/mL (IQR 5.9-30.2). The median time from BL to discharge or death was 13 days (IQR 10-16). None of PTs had adverse events potentially associated with RBV therapy.
3.3. Concomitant treatment
During hospitalization, all the 9 PTs received low molecular weight heparin and, with exclusion of PT8, received intravenous glucocorticoid treatment. Two (PT1 and PT2) received, according to local protocols/guidelines, the anti-interleukin 1 agent, anakinra at the dose of 5 mg/Kg bid for a total duration of 10 days; 3 PTs (PT6, PT7 and PT8) received the anti-IL6 agent, tocilizumab at dose of 8 mg/Kg according to local therapy protocols and AIFA (Italian drug agency) approvals.
Targeted antibiotic therapy for the onset of bacterial superinfection was administered to 3/9 PTs (PT3, PT8, PT9).
One participant (PT3) received remdesivir for 5 days and hyperimmune plasma infusion, before RBV therapy.
3.4. Virological findings
Overall, the median Ct values progressively decreased during the period of observation, including the two PTs (PT8 and PT9) who dead during hospitalization: BL, median value Ct 22.4, IQR 19.84-5.07; EOT, median value Ct 26, IQR 23.34-34.06; discharge, median value Ct 27.92, IQR 26.43-36.11. Only one PT (PT2) had undetectable viral load at EOT with reappearance of viral RNA at discharge, but with a high Ct, indicating a low viral replication (Table 1).
Considering the PTs with a favorable outcome, the clearance of the virus was observed in 2/7 (28.6%, PT4 and PT7) at discharge, and the cumulative clearance rate was 71.4% within day 14th from discharge, because three other PTs had a negative RNA at a follow-up visit after discharge (Table 1).
Consensus sequence of SARSCoV-2 whole genomes (WGS) was available in 5/9 PTs at BL and EOT. In three out of five PTs (PT3, PT8, PT9) WGS was also available in at least one other time point before BL evaluation. In total were analyzed 15 SARSCoV-2 genomes. The sequences from sequential nasopharyngeal samples of these five PTs were analyzed by inferring Stanford Coronavirus Resistance Database (CoV-RDB;
https://covdb.stanford.edu) for the definition of VOCs and viral lineages.
In detail, nasopharyngeal samples from PT3 belonged to Alfa variant, lineage B.1.1., samples from PT6 and PT7 clustered with Delta variant, lineage AY75, while PT8 and PT9 were infected by the Omicron variant, lineage BA.2 and BA.1.17, respectively. The phylogenetic tree construction showed that each sequence clustered within the respective variant and was related to each other of the same PT (Figure 2).
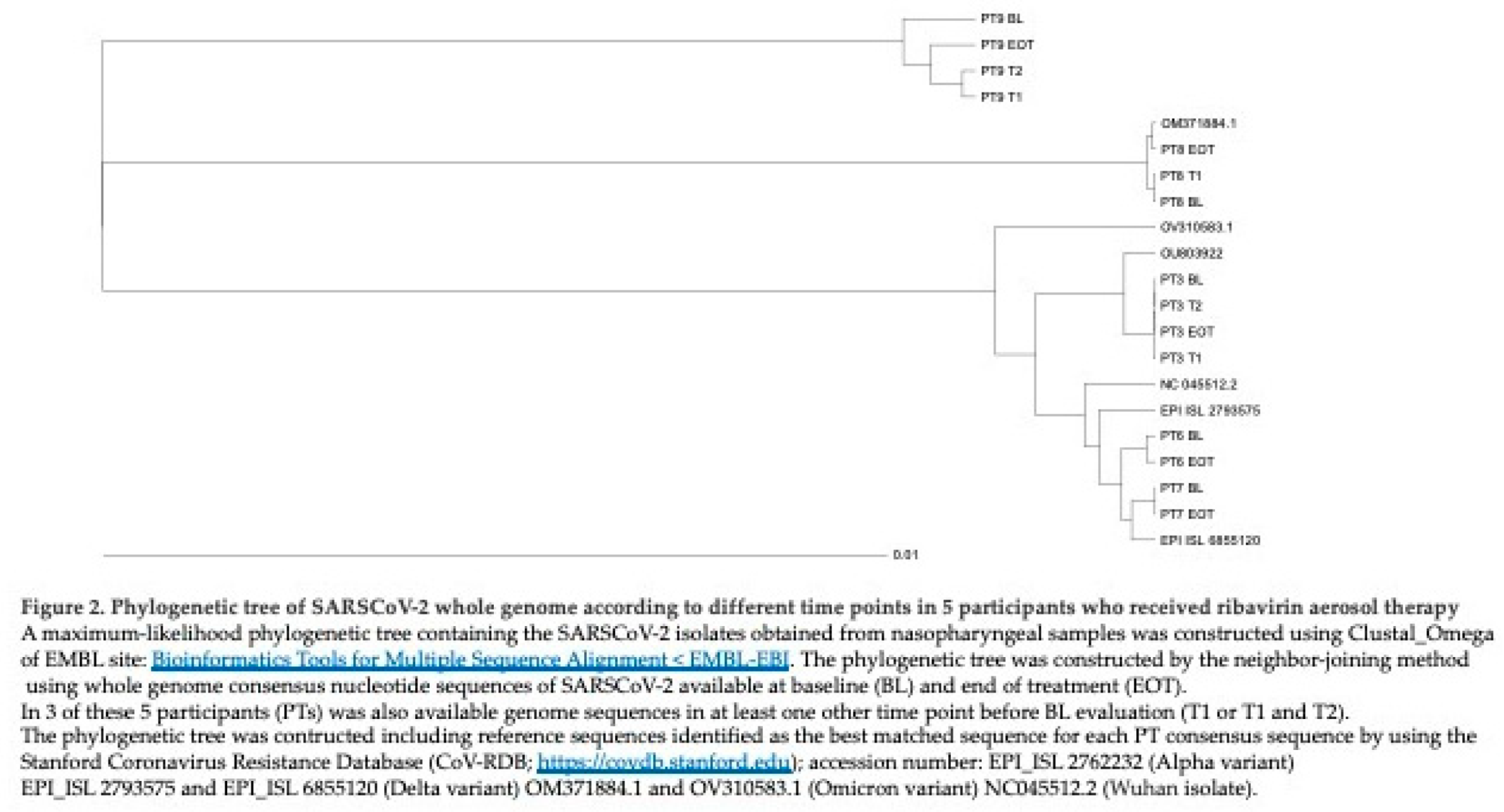
Four out of five PTs (PT3, PT6, PT7, PT8) had very closely related sequences at different time points investigated, while the remaining PT, (PT9) had an interesting profile: EOT sequence was more closely related to those obtained at T1 and T2 (3 and 2 months before RBV administration, respectively) than to sequence obtained at BL. Notably, this PT had a long lasting SARSCoV-2 infection with hospitalization in April 2022. Sequences of the five PTs were also analyzed for their amino acid (aa) mutational profile along different time points. Data on aa change at different time points are summarized in Table 3.

All aa substitutions were considered in each PT’s sample respect to the best matched isolate that was used for comparison.
Participant-3 and PT7 showed several aa changes compared with the reference sequence described in Table 3. However, their sequence pattern remained unchanged in sequential samples (Table 3). Regarding the spike protein targeting-mAbs, PT3 showed the deletion at position 144 (Δ144) [
19]. In addition, PT7 displayed L452R, already described for its association with reduced susceptibility to bamlanivimab, with also 5.7 fold-reduction in susceptibility to cilgavimab.
An interesting mutational spectrum was revealed along different time points in PT6, PT8 and PT9. In detail, PT6 showed G446V substitution within the spike protein associated with reduced susceptibility to several neutralizing mAbs. This mutation was found at EOT but not at BL. In PT8 was detected an aa substitution in the Spike region (K182N) only at BL evaluation, while this substitution was not present at EOT. At EOT emerged substitution (G96V) in the Nucleoprotein. The sample obtained from this PT (PT6) two days before RBV therapy (T1) did not exhibit any change. Several aa changes commonly present in the Omicron variant and associated with various degree of reduced susceptibility to neutralizing mAbs were invariably detected in the samples belonging to PT8 [G142D, S371F, D405N, K417N, N440K, E484A Q493R (Table 3)].
The longitudinal analysis of sequences belonging to PT9, considering also the first two sequences (T1 and T2), obtained from nasopharyngeal samples three and two months before RBV administration, respectively, showed an aa substitution, E155K within the nsp8, and two mutations within the Spike protein: V67A and L455S. These mutations were revealed only at EOT. Furthermore, the substitution C464F within nsp12 (RNA-dependent RNA polymerase) was present at the BL but not in the other time points analyzed (T1, T2, EOT). The ORF7a showed a higher complexity in its mutational profile with the disappearance of aa mutations and the appearance of other aa substitution during the period of observation (Table 3). In detail, at BL was revealed the aa substitution T39I, while, at EOT emerged the aa change T111I. Also in this case, several aa changes commonly present in the Omicron variant and associated with various degree of resistance to neutralizing mAbs were detected in all the samples investigated (Δ142-144, R346K, S371L, K417N, N440K, G446S, E484A, Q493R). This pattern of resistance to neutralizing mAbs was similar to that detected in PT8 with the exception of the aa substitution G446S that was located within the binding site of mAbs tixagevimab and cilgavimab. Notably, PT9 received, about one month before RBV administration, a specific treatment with both monoclonals, with no benefit.
4. Discussion
We retrospectively evaluated virological and clinical findings in hospitalized adults with COVID-19 pneumonia with the primary objective being the effects of RBV aerosol on SARS-CoV-2 viral load.
At discharge, we found the clearance of the virus in 2/7 (28.6%) PTs with a favorable clinical course. The other 5 PTs with a good clinical outcome had very low viral replication at discharge, and 3/3 PTs with an available follow-up sample achieved a virus negative status within 14 days from discharge. We had a cumulative viral clearance rate of 71% at last follow-up visit. Additionally, several PTs (including the two PTs deceased during hospitalization) showed a decline of viral load in nasopharyngeal samples measured from BL to discharge.
A recent trial on RBV aerosol administration in hospitalized adults with respiratory distress for COVID-19 showed SARSCoV-2 test negative in 24/28 (85.7%) patients who completed the trial (30 days assessment) [
20]. One other study from China on critically ill COVID-19 patients who received RBV orally, showed virus clearance in 63% of patients at day 21 of treatment with also a significant decrease in viral load when comparing day 21 to the baseline for RBV therapy [
21].
Our previous report within a compassionate study with RBV aerosol in patients with SARSCoV-2 infection showed a negative SARSCoV-2 test in participants at the end of quarantine period (day 14
th after hospital discharge) [
9]. Altogether, these previous studies and the present report suggest that RBV aerosol limits SARSCoV-2 replication, which may play a role in the prevention and/or attenuation of damages exerted by local and systemic inflammation. Of note, 89% of PTs in this our study were on treatment with corticosteroids which might reduce the antiviral effect of RBV, but also the inflammatory response.
Among PTs with a favorable outcome CSS scale at discharge was 6 or 7. Respiratory function parameters were ameliorated and markers of inflammation, in particular ferritin, fibrinogen and C-reactive protein showed a declining trend at discharge.
The study by Poulakou et al., investigating the potential of RBV inhalation solution to reduce COVID-19 disease severity in adults with a diagnosis of respiratory distress, showed an improvement of clinical severity status rating in 31.4% (16/51) of patients at the end of treatment and 78.4% (40/51) of patients on day 30 assessment [
20].
One other study from China including severe and critical COVID-19 patients who received RBV orally, showed 100% survival [
21]. Since the viral load of SARSCoV-2 in lower respiratory tract was declining with the use of RBV, the authors suggested that the decrease of viral load positively influenced survival rate.
Eslami G. et al., compared oral RBV vs. sofosbuvir/daclatasvir combination treatment showed a better outcome in the sofosbuvir/daclatasvir arm vs. RBV arm [
22]. However, the comparison was performed with a combination of two potential antivirals vs. RBV alone, this latter compound administered at high dosage.
Notably, high doses of oral RBV have many adverse effects like anemia and may impair renal function, which might complicate advanced case of COVID-19. In this regard, the authors suggested that the relative advantage of the combination treatment with sofosbuvir daclatasvir could be the consequence of an excess of adverse events in the RBV arm. Of note, the severity of the disease increased from Alpha to Delta variant and subsequently decreased with the spread of Omicron sub lineages [
23,
24]. The most pronounced differences at mutational level were noted for the last two variants, Delta and Omicron. It is also well known that this latter variant presents higher transmissibility and reduced susceptibility to neutralization by most mAbs rendering of pivotal importance the approach with antivirals, especially in the early phase of infection and in participants with comorbidities that may negatively influence the outcome. We therefore investigated the mutational profile of SARSCoV-2 in 5/9 PTs before and immediately after the last dose of RBV, also because
in vitro studies indicated an inhibitory and mutagenic activity of RBV on SARSCoV-2 [
25]. Of note, our PTs were infected during different waves, therefore harbored different mutational pattern: one PT (PT3) was infected by Alpha variant lineage B.1.1.7. Two (PT6 and PT7) by Delta (lineage AY75) and the remaining two PTs (PT8 and PT9) by the most recent Omicron variants (lineage BA.2, and BA.1.17, respectively) (Table 3).
Regarding the mutational profile of SARSCoV-2 during RBV treatment, we found that in 2 PTs (PT3 and PT7) the genome detected in the first sample remained unchanged in the subsequent samples. This finding suggests that in these two cases, even in the presence of RBV, no novel mutations were fixed during 6 days of treatment. Interestingly, PT3 received remdesivir 5 days before RBV (Table 3). Sequence analysis did not show a clear effect of remdesivir on viral evolution by comparison of mutational pattern at T1 (before remdesivir administration) T2 (corresponding to first remdesivir administration) and BL administration of RBV (corresponding to EOT for remdesivir).
Interestingly, PT6, PT8 and PT9 showed aa mutation at EOT respect to BL evaluation. Of note, these mutations were not located in the nsp12 corresponding to RNA-dependent RNA polymerase (RdRp) codifying region.
It is well known that the nsp8 acts as a cofactor of RdRp playing a role together with nsp7 on the stabilization of the replication/transcription complex [
26]. Two different conformations of nsp8 and one conformation of nsp7 interact with nsp12. Together, the four proteins form the minimal core polymerase complex. It has been described that without these co-factors, the nsp12 may have a low efficiency in polymerase activity [
27,
28]. Interestingly, the mutation E155K within the nsp8 was detected only at EOT in PT9.
However, to the best of our knowledge, no data are available on the possible impact on the polymerase activity of mutations within nsp8 domain.
Sequences from PT8 showed that were closely related (apart 2 aa changes, one at BL and one at EOT) to the isolate used for comparison, while sequences from PT9 were more divergent respect to the respective reference isolate, probably because of long covid infection in this PT, and immune deficiency status (he had hematologic malignancy and was previously under immunosuppressive drugs). In this PT, was found the aa substitution G446S in all sequences analyzed within the binding sites of tixagevimab and cilgavimab, which may result in reduction activity of these two mAbs on Omicron variants [
29]. Notably, he received these two mAbs about one month before RBV with no benefit.
The main limitation of this study is its retrospective nature including a small group of PTs with similar baseline pulmonary feature but in the context of heterogeneous clinical background, which in two cases influenced an unfavorable clinical outcome.
One other important limitation is that we cannot firmly assess a role of RBV on the clinical improvement nor on virologic outcome because we did not include a placebo control group.
Finally, our PTs, were infected by different SARSCoV-2 variants. Therefore, clinical and virological outcomes could be influenced by differences in the severity of disease, over than BL mutational pattern.
In conclusion, our finding of clinical improvement in a relative short period in 7/9 PTs who received RBV aerosol could be, at least in part, related to the decline and clearance of RNA in a relatively short time.
Ribavirin may also have contributed to the mutational spectrum of SARSCoV-2.
Our results encourage extending clinical trials to consider the use of RBV aerosol in combination therapies in view of new emerging SARSCoV-2 variants that could severely affect current vaccination strategies, diagnostics and therapeutics.









