1. Introduction
The airway epithelium forms an important barrier at the interface of environment and organism [
1]. AECs consist of heterogenous groups of cells, including pneumocytes, goblet, club, ciliated and basal cells [
2,
3]. Depending on the species and anatomic location in the airways, different proportions of AEC subsets have been observed. The epithelium of large airways in humans forms a pseudostratified layer with ciliated, goblet and club cells at the luminal side, while underlying basal cells are attached to the basement membrane. On the other hand, the pseudostratified epithelium in mice is limited to the trachea, with simpler columnar epithelium present in the bronchi [
1]. It is thought that in mice the majority of airway basal cells are limited to the trachea, with sparse basal cells in bronchi [
1]. Among AECs, basal cells keep the greatest degree of pluripotency allowing them to proliferate and differentiate into various subtypes of epithelial cells. Thus, basal cells play a major role in airway epithelium homeostasis and repair [
2,
3] and they are also essential for
in vitro primary AECs cultures since they can be maintained in a constant proliferative state [
4,
5,
6].
Isolated primary AECs are used in a range of
in vitro experimental systems. Most commonly, when maintained in media containing differentiation inhibitors, primary AECs can be sub-cultured in a monolayer. However, when cultured in well-plate inserts with specialised media, air-liquid interface (ALI) cultures can be established [
4]. ALI cultures recapitulate the epithelium of large airways as these cultures are pseudostratified and AECs differentiate into various subsets including ciliated, club or goblet cells. ALI cultures also exhibit mucociliary motion, produce mucus and develop tight junctions [
7,
8,
9]. Subsets of AECs can also be cultured in a Matrigel matrix to generate tracheospheres, bronchiolospheres or alveolospheres [
10,
11]. All these experimental systems can be used to study AEC biology at homeostasis, after injury e.g. by respiratory pathogens, and during regeneration.
In recent years, it has been recognised that AECs do not only form an inert barrier, but that they are a dynamic and versatile cell population that is involved in maintaining lung homeostasis [
12], mounting initial immune responses [
13] and lung regeneration following injury [
14]. The manifold roles of AECs implicate them in pathogenesis of a number of diseases ranging from asthma [
15], COPD [
16] to lung fibrosis [
17]. An increasing number of research questions focusing on AECs in various contexts creates a demand for improved and well-defined methodologies allowing for their efficient harvest, isolation and culture.
In order to study primary AECs
in vitro, various digestion techniques have been employed that allow for disruption of cell junctions and connective tissue resulting in lung single cell suspension. To isolate and study AECs from a murine lung, researchers commonly utilize a combination of dispase II and DNase I enzymes at 37°C for 30-60min [
18,
19,
20] (hot digestion). However, when we employed this established digestion method, we found that the yield and viability of AECs was unsatisfactory in the context of MACS sorting and
in vitro culture. We therefore investigated alternative workflows. While there are several digestion methods for murine lungs utilizing a range of enzymes including collagenases, dispases, liberase, pronase and trypsin [
21], we investigated the effects of digestion time and temperature rather than the type of enzyme. It has been shown that long, overnight digestion can be beneficial to yield and efficiency when isolating cells from human skin [
22], or human brain tissue [
23]. Following on from a method where murine tracheas were digested for 18h at 4°C using pronase [
24], we assessed a 20h 4°C dispase II/DNase I digestion of murine lungs (cold digestion).
Here, we present a workflow that in comparison to the commonly used hot dispase digestion allows for retrieval of a 3-fold higher number of CD45-CD31-EpCAM+ AECs with a higher proportion of viable cells. This in turn allows for recovery of greater numbers of AECs after MACS sorting and a greater yield of keratin 5 (KRT5)+ p63+ basal cell colonies in vitro after seven-day culture.
2. Materials and Methods
2.1. Animals
C57BL/6 mice were bred and housed at the University of Edinburgh. All procedures approved by the University of Edinburgh Animal Welfare and Ethical Review Board, and performed under UK Home Office licenses with institutional oversight performed by qualified veterinarians. UK Home Office project license to JS, number P4871232F. ARRIVE guidelines were followed.
2.2. Murine Lung Harvest and Digestion
Adult C57BL/6 mice over six weeks of age were anaesthetized with a 1:1 mixture of ketamine and medetomidine through intraperitoneal (IP) injection with a dose dependent on the body weight. Mice were then terminally exsanguinated, and death confirmed by cervical dislocation. The abdominal aorta was cut, and the trachea exposed by removing the salivary glands. The trachea was then cannulated using a blunt needle and bronchoalveolar lavage with ice-cold Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) (Gibco) was performed (0.8ml per mouse). The rib cage was then removed, and cardiovascular system was flushed by injecting 10ml of ice-cold DMEM/F12 into the right ventricle. A volume of 1.5-2ml of enzyme mix was then slowly injected through the tracheal cannula into the lungs, which were then removed (lungs were dissected out of the thoracic cavity by cutting at the primary bronchial bifurcation) and placed in 3ml of digestion mixture (2mg/ml Dispase II (Sigma-Aldrich) + 0.1mg/ml DNaseI (Sigma-Aldrich) re-suspended in DMEM/F12 (Gibco) + 1% v/v Penicillin-Streptomycin (Gibco)). The digestion mixture was previously filtered through 0.22µm filter and frozen. Lungs were then incubated for 1h at 37°C or 20h at 4°C. The digestion solution together with lungs was then poured onto a 70µm strainer, lungs were then dissociated using a 5ml syringe plunger by gently agitating the lungs on the mesh. The strainer was then washed using 10ml of dispase wash (DMEM/F12 (Gibco) + 0.05mg/ml DNaseI (Sigma-Aldrich) + 1% v/v Penicillin-Streptomycin (Gibco)). Each sample was then centrifuged for 15min, 4°C, 130 relative centrifugal force (RCF). The supernatant was then removed and 2ml of ice-cold ACK red blood cell lysis (Gibco) was added to each tube, samples were swirled for 90s, and 5ml of MACS buffer (Phosphate-buffered saline (PBS) - no Mg2+ and Ca2+ + 0.5% bovine serum albumin (BSA) + 2mM ethylenediaminetetraacetic acid (EDTA)) was added. Samples were then passed through a 40µm strainer, washed with 5ml of MACS buffer and centrifuged for 5min, 4°C at 300 RCF. Each sample was then resuspended in 1ml of MACS buffer and blocked with 5µl of 0.5mg/ml anti-mouse CD16/32 antibody (BioLegend) ready for further MACS processing or flow cytometry staining.
2.3. Airway Epithelial Cell Sorting (MACS)
After incubation cells were centrifuged for 5min, 4°C, 300 RCF and resuspended in 85µl of MACS buffer + 5µl of anti-CD31 microbeads and 10µl of anti-CD45 microbeads (Miltenyi Biotec) per 107 total cells. Samples were then incubated for 30 minutes on ice. Each sample was then washed with 1ml/107 cells of MACS buffer and centrifuged for 5min, 4°C at 300 RCF. Cells were then passed through LS MACS columns (Miltenyi Biotec) on a QuadroMACS separator (Miltenyi Biotec) and flow-through was collected. Collected cells were then centrifuged for 5min, 4°C, 300 RCF and resuspended in leftover liquid, and 15µl of anti-EpCAM microbeads (Miltenyi Biotec) was added. Samples were incubated on ice for 30 minutes, washed with 1ml MACS buffer, and centrifuged for 5min, 4°C, 300 RCF. Supernatant was discarded, and cells were resuspended in 500µl of MACS buffer. The cell suspension was then passed through a MS MACS column using an OctoMACS separator (Miltenyi Biotec). EpCAM+ cells were then gently flushed out of the column by applying 2ml of MACS buffer and inserting the plunger into the column. Cells were then centrifuged for 5min at 4°C, 300 RCF, supernatant was removed, and the cell pellet was resuspended in 0.5ml of MACS buffer. 10µl of cell suspension was then aspirated and gently mixed with 10µl of 0.1% trypan blue (Gibco) by pipetting up and down. Immediately afterwards, 10µl of cell suspension was placed in Neubauer improved counting chamber (haemocytometer) and covered with coverslip. Cells were then manually counted and total amount of live (trypan blue negative) and dead (trypan blue positive) cells in suspension was calculated.
2.4. Primary AECs In Vitro Culture
24-well plates were coated with a coating solution that combines 30µg/ml of type I calf skin collagen (Sigma-Aldrich), 10µg/ml human placental fibronectin (Bio-Techne) and 10µg/ml bovine serum albumin (BSA – Sigma-Aldrich) in HBS (Gibco), for at least 4-8h at 37°C. A total of 2x105 MACS sorted AECs were seeded per well and 0.5ml media was changed after 24h, and then every two days thereafter. Media was prepared by combining supplemented PromoCell Epithelial Growth Medium with 1µM A3801 (STEMCELL Technologies) and 0.2µM DMH-1 (STEMCELL Technologies) SMAD signalling inhibitors, 5µM Y27632 ROCK (STEMCELL Technologies) pathway inhibitor, 0.5µM CHIR9902 (STEMCELL Technologies) WNT pathway activator and 1% v/v Penicillin/Streptomycin (Gibco). Cells were cultured for seven days, followed by immunofluorescent microscopy of each well.
2.5. Immunofluorescent Microscopy
Cells in each well were washed with PBS three times and fixed using 4% PFA solution for 10min at room temperature (RT). Fixed cells were then stored in 70% Ethanol at 4°C until staining. Cells were permeabilised with a 1% BSA and 0.2% triton X-100 (Sigma-Aldrich) solution at RT for 10min. Cells were then blocked for 45min at RT using a 5% goat serum, 1% BSA and 0.1% Tween-20 (Sigma-Aldrich) in PBS. Cells were then stained with a mix of primary antibodies (anti-KRT5, anti-p63 and anti-E-Cadherin-eF660), overnight at 4°C, followed by three washes with blocking buffer (each wash for 5min) and staining with secondary antibodies (anti-rabbit-AF488, anti-mouse-AF555) as well as 1:500 10mg/ml Hoechst (Invitrogen) for nuclear counter stain for 1h at RT. Secondary antibodies were then washed away with three washes (each wash for 5min) with blocking buffer followed by filling each well with 0.7ml PBS.
Imaging was performed using EVOS FL Auto 2 (Thermo Fisher Scientific), using 20x objective as well as DAPI, GFP and RFP light cubes. Scans were set to a custom calibrated Corning 24-well plate, where 25% of each well was scanned from the middle using the “more overlap” setting and manually set focus for each channel. Images were analysed using ImageJ, with ICA LUT (look-up table) set for KRT5 for easier basal AECs identification. All micrograph related colony counting was performed while blinded.
Surface area of KRT5 colonies was calculated using ImageJ. At first KRT5 images were converted to 8-bit. Then images were smoothed out using median filter (7px value), followed by thresholding using “mean setting”. Images were then converted to binary format and surface area of black pixels was calculated using “Analyze particles” tool. Obtained pixel numbers were then converted to total surface in mm2 based on image metadata (scale).
Table 1.
List of microscopy antibodies.
Table 1.
List of microscopy antibodies.
| Antibody name |
Supplier |
Host species |
Antibody type |
Clone |
Catalogue number |
Dilution |
| Anti-p63 |
Abcam |
Mouse |
Monoclonal |
4A4 |
ab735 |
1:200 |
| Anti-KRT5 |
BioLegend |
Rabbit |
Polyclonal |
Poly19055 |
905503 |
1:500 |
| Anti-E-cadherin (CD324) eFluor 660 |
Thermo Fisher Scientific |
Rat |
Monoclonal |
DECMA-1 |
50-3249-82 |
1:30 |
| Rat IgG1 kappa Isotype Control (eBRG1) eFluor 660 |
Thermo Fisher Scientific |
Rat |
Isotype control |
eBRG1 |
50-4301-82 |
1:30 |
| Goat Anti-Rabbit IgG (H+L) Alexa Fluor 488 |
Thermo Fisher Scientific |
Goat |
Polyclonal, secondary |
Reactivity - rabbit |
A-11008 |
1:200 |
| Goat Anti-Mouse IgG (H&L) Alexa Fluor 555 |
Abcam |
Goat |
Polyclonal - secondary |
Reactivity - mouse |
ab150114 |
1:200 |
2.6. Flow Cytometry
Following digestion, numbers of cells in each sample were calculated using a haemocytometer. 0.5x106 total cells from each sample were then washed twice with PBS and stained using a LIVE/DEAD fixable near-IR stain (Invitrogen) in PBS for 30min on ice, followed by two washes with PBS and staining with a combination of anti-CD45, anti-CD31 and anti-EpCAM in PBS + 1% BSA for 30min on ice. Stained samples were then washed twice with PBS + 1% BSA. Whole samples were acquired and unmixed using Cytek Aurora with Cytek SpectroFlo 3.0.3 and analysed using De Novo Software FCSexpress 7.
The gating strategy involved debris exclusion (side scatter/forward scatter – SSC-H/FSC-H), followed by selection for singlets (FSC-H/FSC-A) and dead cell exclusion (LIVE/DEAD Fixable Near IR/FSC-H). Dead cell exclusion was performed at the end of gating strategy for viability evaluation in Figure 2b. Leukocytes and endothelial cells were then excluded using CD45/CD31 gate, followed by selection for AECs with EpCAM/SSC-H gate.
Table 2.
List of flow cytometry antibodies.
Table 2.
List of flow cytometry antibodies.
| Antibody name |
Supplier |
Host species |
Antibody type |
Clone |
Catalogue number |
Dilution |
| Anti-CD45 Pacific Blue |
BioLegend |
Rat |
Monoclonal |
S18009F |
157212 |
1:200 |
| Anti-CD45 AF700 |
BioLegend |
Rat |
Monoclonal |
S18009F |
157210 |
1:200 |
| Anti-CD31 BV605 |
BioLegend |
Rat |
Monoclonal |
390 |
102427 |
1:600 |
| Anti-CD31 BV421 |
BioLegend |
Rat |
Monoclonal |
390 |
102423 |
1:300 |
| Anti-EpCAM PE/Dazzle594 |
BioLegend |
Rat |
Monoclonal |
G8.8 |
118236 |
1:300 |
| Anti-EpCAM BV605 |
BioLegend |
Rat |
Monoclonal |
G8.8 |
118227 |
1:300 |
| Anti-CD24 PE/Cyanine7 |
BioLegend |
Rat |
Monoclonal |
M1/69 |
101821 |
1:200 |
| Anti-CD49f BV605 |
BioLegend |
Rat |
Monoclonal |
GoH3 |
313625 |
1:100 |
2.7. RNA Isolation and qPCR (Quantitative Polymerase Chain Reaction)
After CD45-CD31-EpCAM+ MACS sorting cells were centrifuged for 5min, 4°C, 300 RCF and resuspended in 0.5ml of TRizol. Following 5min incubation at RT, samples were stored at -80°C before further processing. Samples were then thawed and 100ul of bromochloropropane (Sigma-Aldrich) was added per 500ul of TRizol. Samples were shaken vigorously for 30s until they acquired a milky-pink shade. Samples were then incubated at RT for 10min, and then cooled down on ice for 5min before centrifugation at 4°C, 16,000 RCF for 20min. 200µl of the top aqueous phase was then transferred to fresh Eppendorf test tubes and 250µl of RNase-free isopropanol (Merck) and 1µl of GlycoBlue Coprecipitant (Invitrogen) were added. Test tubes were inverted ten times and incubated at RT for 10min, followed by centrifugation at 4°C, 16,000 RCF for 10min. Blue RNA pellet could now be observed at the bottom of test tubes. Supernatant was removed and pellet was washed three times using 1ml of RNase-free 75% EtOH (Merck), with 5min 4°C, 16,000 RCF centrifugations between each wash. After final wash, ethanol was discarded, and pellets were air-dried until no liquid could be seen in test tubes. Pellets were then resuspended in 30µl of RNAse-free water (QIAGEN), and amount of RNA was quantified using NanoDrop One/OneC Microvolume UV-Vis Spectrophotometer (Thermo Scientific). The amount of RNA was normalised in each sample using RNase-free water and 300ng of RNA was converted to cDNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCRs were performed in duplicates using Fast SYBR Green Master Mix (Applied Biosystems) and StepOne (Thermo Fisher Scientific) thermocycler. 100nM of forward and reverse primers, as well as 10ng of cDNA, were added to each qPCR reaction. Rpl37 was used as an endogenous control. Genomic DNA contamination was assessed by performing a qPCR reaction on non-reverse transcribed RNA sample.
Table 3.
List of PCR primers.
Table 3.
List of PCR primers.
| Primer |
Sequence |
|
Ldha forward |
CATTGTCAAGTACAGTCCACACT |
|
Ldha reverse |
TTCCAATTACTCGGTTTTTGGGA |
|
Rpl37 forward |
CCAAGCGCAAGAGGAAGTATAAC |
|
Rpl37 reverse |
GAATCCATGTCTGAATCTGCGG |
2.8. Statistical Analysis
Data in the plots are represented as medians with boxes representing interquartile ranges and bars corresponding to minima and maxima. MFI is reported as median fluorescence intensity. Statistical differences were assessed using an unpaired two-tailed t-test. Data in the text is presented as means with standard error. All statistical analysis was performed using GraphPad Prism 8.4.3.
3. Results
3.1. Cold Digestion Provides Greater Yield and Viability of AECs Compared to Hot Digestion
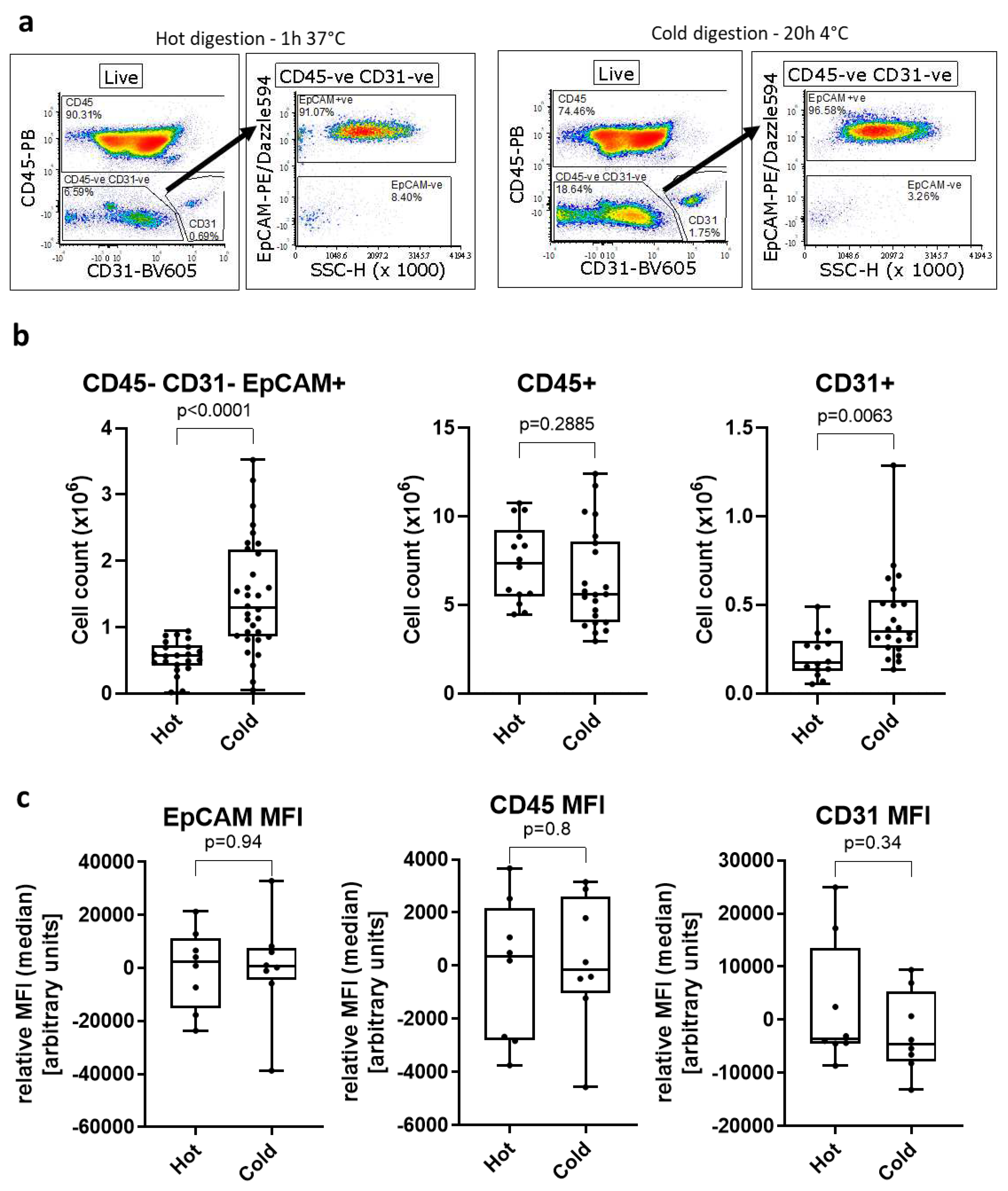
We used flow cytometry (Figure 1a) to assess the yield of three different cell populations after a 1h 37°C dispase II/DNase I digestion of murine lungs. On average, we obtained 5.55 ± 0.05 x105 CD45-CD31-EpCAM+ AECs (7% of total cells), 7.35 ± 0.59 x106 CD45+ immune cells and 2.10 ± 0.03 x105 CD31+ endothelial cells (Figure 1b).
We decided to investigate whether the use of cold digestion of murine lungs would increase the yield and/or viability of AECs. Using the cold digestion approach, the average number of isolated CD45-CD31-EpCAM+ AECs increased to 1.46 ± 0.15 x106, almost 3-fold the number obtained following hot digestion. Likewise, the number of CD31+ endothelial cells obtained with the cold digestion (4.27 ± 0.05 x105) was almost 2-fold higher than after hot digestion. Conversely, the number of CD45+ cells was similar between the two digestion techniques. This suggests that the cold digestion approach is beneficial for recovery of structural cells from the lung, while obtaining similar numbers of lung immune cells.
While several cell surface markers like CD4, CD8 or PD-1L are known to be sensitive to dispase II digestion [
25], we did not find any changes in expression levels of CD45, CD31 and EpCAM between the digestion methods as seen by comparable MFI (median) levels of each marker irrespective of digestion method (
Figure 1c). We also did not observe loss of several AECs-related cell surface markers such as major histocompatibility (MHC) I, MHC-II or CD24 (Supplementary
Figure 1).
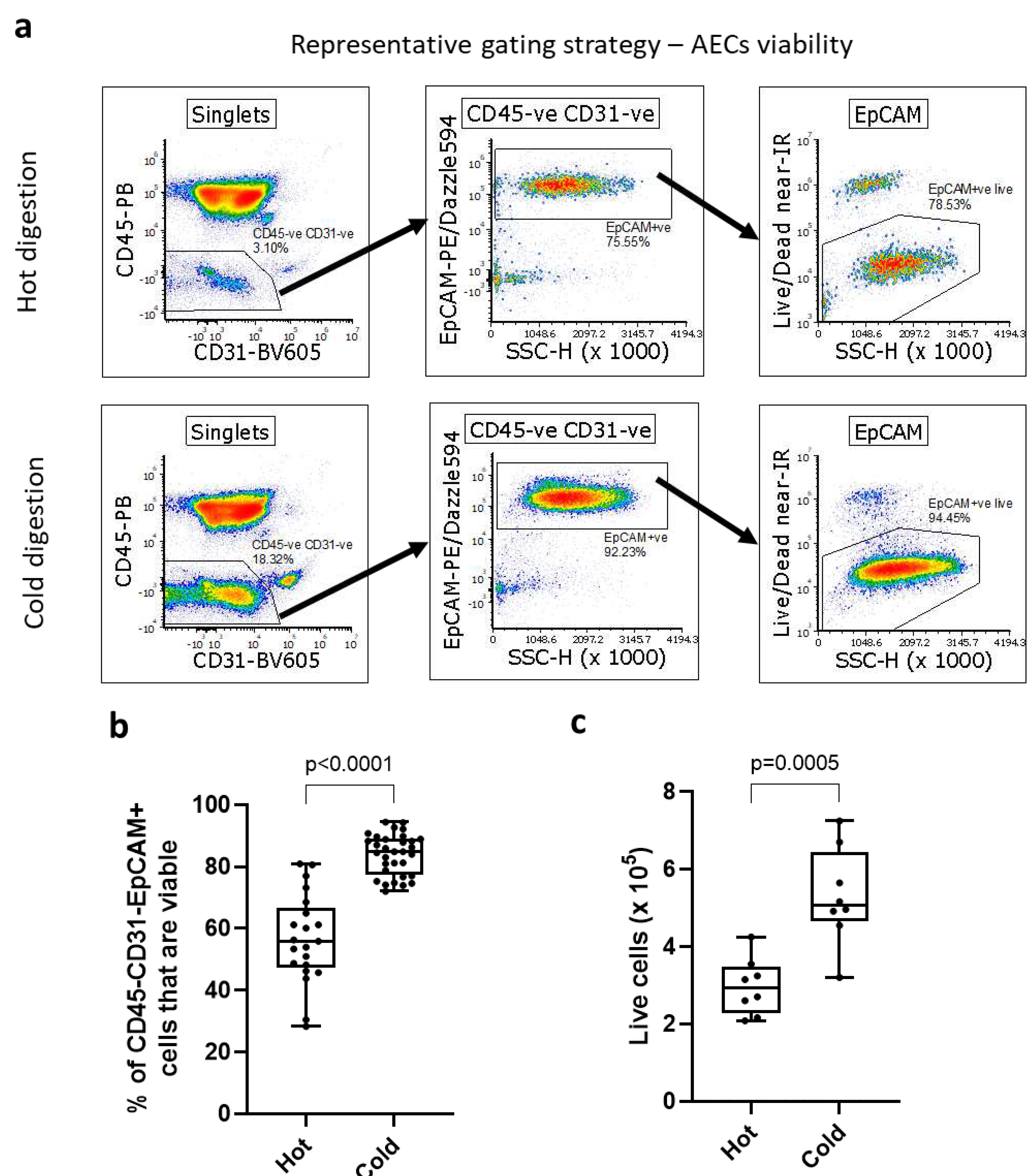
3.2. Following Cold Digestion, More AECs Are Viable Compared to Hot Digestion
Next, we compared the viability of isolated AECs between cold and hot lung digestions by flow cytometry, assessing the proportion of live cells within the CD45
-CD31
-EpCAM
+ population in single cell suspension, with the gating strategy shown in
Figure 2a. We found that on average 83.99 ± 1.16% of CD45
-CD31
-EpCAM
+ cells from cold lung digestions were alive, compared to 56.65 ± 3.16% after hot digestion, a 1.48-fold increase in viable AECs (
Figure 2b). Despite differences in viability of AECs, we did not observe changes in levels of Ldha (a marker of oxidative stress) [
26] between digestion methods (Supplementary
Figure 2).
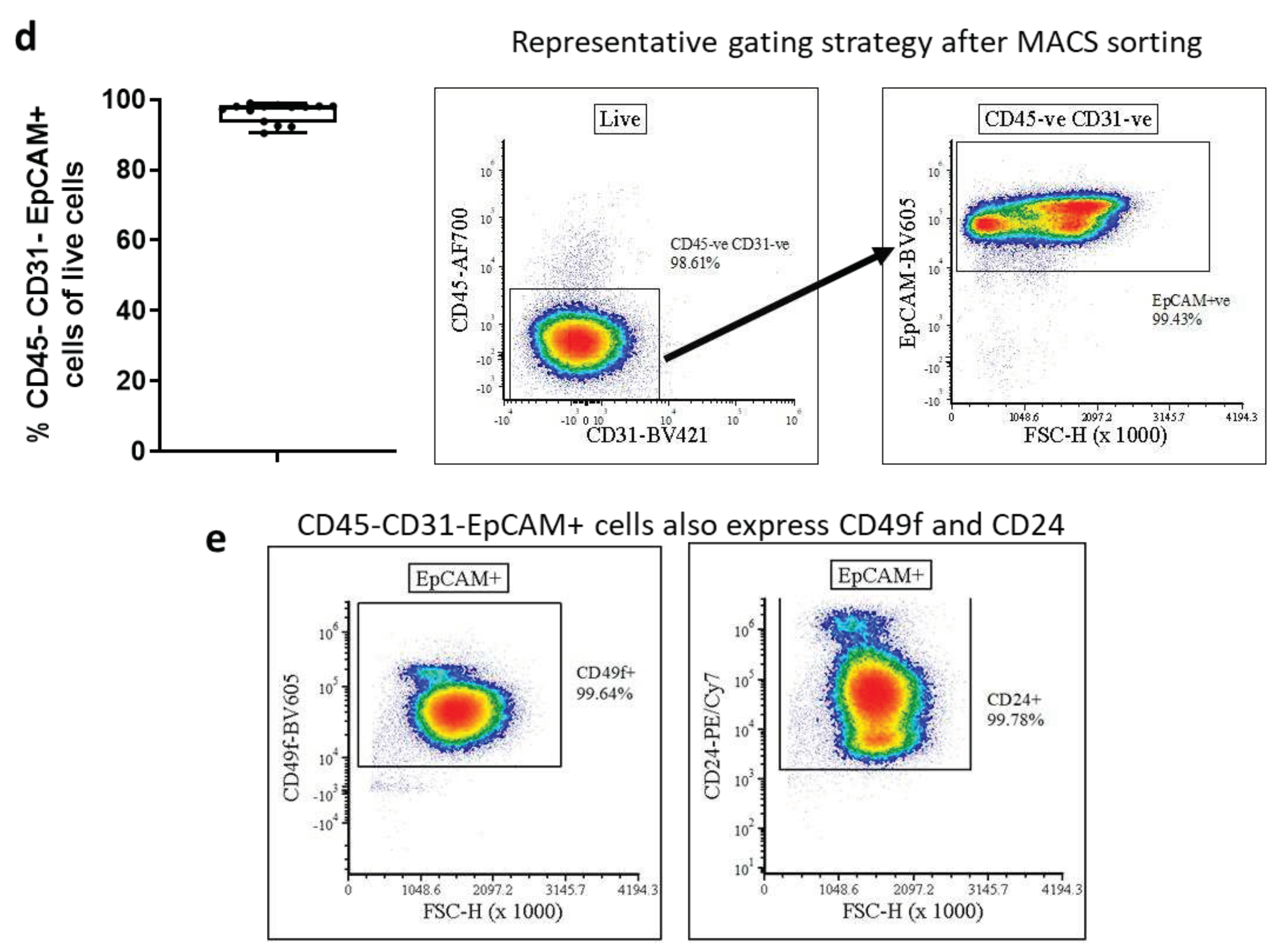
Given this increased viability, we hypothesised that MACS sorting, and subsequent seeding of AECs isolated by cold digestion would result in a greater number of basal AEC colonies in in vitro cultures, comparted to hot digestion. Performing double MACS sorting with initial CD45 and CD31 depletion followed by EpCAM positive selection, we recovered substantially more AECs following cold digestion (5.30 ± 0.45 x10
5 AECs) compared to hot digestion (2.97 ± 0.26 x10
5 AECs) which is equivalent to a 1.78-fold increase in number of viable AECs (
Figure 2c). The AECs recovered after MACS sorting were highly pure (
Figure 2d), with average purity of 96.35 ± 0.75% CD45
-CD31
-EpCAM
+ cells of live cells. To confirm the airway epithelial identity of sorted CD45
-CD31
-EpCAM
+ we used flow cytometry to confirm that these cells also express other epithelial markers. Virtually all CD45
-CD31
-EpCAM
+ cells also expressed CD49f (integrin α6) [
27,
28] and CD24 [
29,
30] (
Figure 2e), albeit as expected, at vastly varying levels.
3.3 Following Cold Digestion and MACS Sorting, More Basal Cell Colonies Proliferate In Vitro
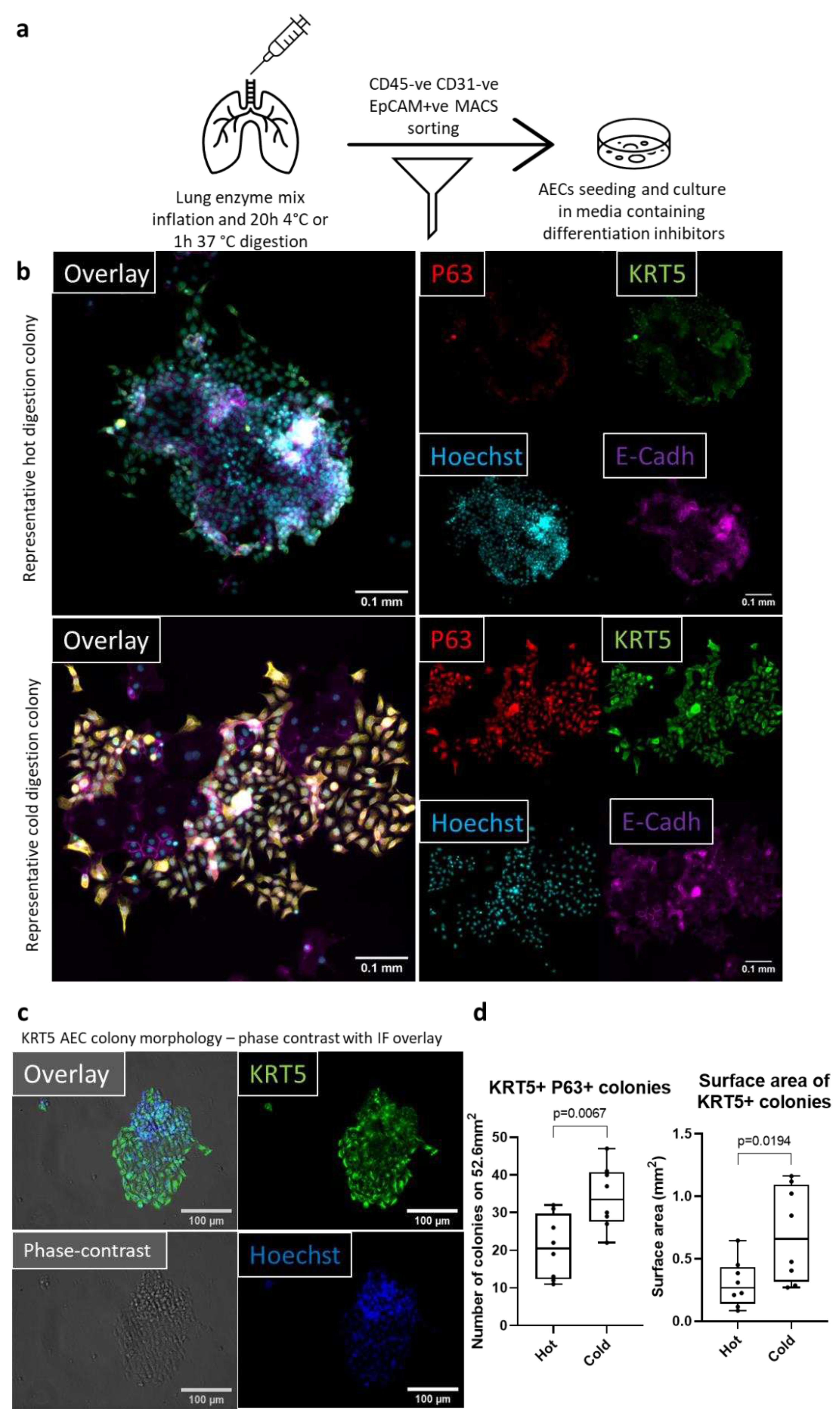
To investigate if the observed increase in AEC viability with cold digestion translated to greater recovery of airway basal cells in vitro, we seeded 2x10
5 MACS sorted CD45
-CD31
-EpCAM
+ cells (98% purity) from hot or cold lung digestions into 24-well plates (workflow indicated in
Figure 3a). Cells were cultured for seven days in Promocell airway epithelial media supplemented with differentiation inhibitors and a Wnt pathway activator[
31,
32,
33]. After seven days the cells were fixed and stained for markers of epithelial (E-cadherin) and basal cells (KRT5 and p63) [
1,
34]. Then 25% of the surface area of each 24-well plate was imaged and the number of basal cell colonies was quantified, as well as the total surface area of KRT5
+ basal cell colonies per well (
Figure 3b). KRT5
+ colony morphology was visualized using phase-contrast microscopy (
Figure 3c). On average there were 21 ± 2.97 basal cell colonies after hot digestion and 34 ± 2.98 colonies after cold digestion per 52.6mm
2, which equals to a 1.62-fold increase in colony counts. When we assessed the total surface area of KRT5
+ colonies between the two digestion methods, we found a 2.3-fold increase in colony surface area with an average surface area occupied by KRT5
+ colonies of 0.66 ± 0.13mm
2 after cold digestions compared to 0.30 ± 0.07mm
2 after hot digestions (
Figure 3d). Furthermore, there were very few E-cadherin negative non-epithelial cells in cultures irrespective of the digestion method employed, suggesting that the combination of cold digestion, MACS sorting, and appropriate media allows for propagating highly pure AECs.
4. Discussion
Here, we demonstrate that by changing standard dispase II/DNase I digestion conditions from 1h 37°C to 20h 4°C we are able to obtain significantly greater numbers of lung structural cells, including AECs. This, together with the observation that a higher proportion of AECs are viable after cold digestion suggests that there is a potential to employ cold dispase II lung digestion for establishing primary murine AECs cultures in vitro. Indeed, in combination with MACS CD45
-CD31
-EpCAM
+ AEC sorting, we demonstrate that culture of AECs after cold dispase digestion results in a significantly greater number of basal AEC colonies with a larger surface area than after hot digestion. Although establishing culture protocols for murine lung basal AECs was not within the scope of this project, by altering media composition we were able to culture CD45-CD31EpCAM+ MACS sorted cells for up to 14 days over two passages (Supplementary
Figure 3a). Additionally, all cells did express KRT5 at the end of passaging period (Supplementary
Figure 3b).
These improvements overcome the need for pooled AEC isolates from several mice [
24,
30,
35] and allow for a more comprehensive analysis of AECs from individual animals. This will contribute to the principles of replacement, reduction, and refinement (3R) [
36] by reducing the number of mice used in AEC research. However, the greater yield and viability are not the only benefits. The fact that after cold digestion the number of isolated AECs increases substantially suggests that overall AEC populations, including rare and difficult-to-isolate populations, may be better represented in comparison to hot digestion. While further analysis is required to formally confirm our prediction, this is likely to have major implications for researchers studying transcriptomics, epigenomics or proteomics of isolated murine AECs, especially at single cell level. It is possible that by employing hot digestion, only the mostly easily dislodged and/or the most robust AECs are released and survive, potentially skewing analysis. When comparing the abundance of cell types in the murine lung between single-cell RNA sequencing (scRNAseq) and single-nuclear RNA sequencing (snRNAseq), it was reported that scRNAseq under-represents epithelial population in comparison to snRNAseq. Indeed, neuroendocrine and basal cells were exclusively seen in the snRNAseq data sets, while type-1 pneumocytes, club cells and ciliated cells were much better represented in scRNAseq. This might be explained by the fact that single-cell suspension for the scRNAseq is obtained through enzymatic digestion, while nuclei for snRNAseq are isolated through physical dissociation of the tissue [
37]. This observation therefore suggests that, in order to isolate representative live epithelial populations through enzymatic digestion, further optimisation is required.
A method commonly used in combination with AEC isolation from tracheas is the removal of fibroblasts by adherence, which requires an additional incubation step, which may further contribute to deteriorating the viability of AECs [
7,
38]. Alternatively, FACS is used to sort AECs from lung digestions, however, FACS may not only be detrimental to viability of sorted cells, but also limits the number of samples that can be sorted in parallel [
39,
40]. Here we suggest combining the cold digestion approach with a two-step MACS sorting of CD45
-CD31
-EpCAM
+ cells, which allows for parallel sorting of multiple highly pure samples from larger animal cohorts.
Historically, researchers used tracheas to isolate basal cells from murine airways[
4,
7], however, the workflow presented here allows for isolation of sufficient numbers of viable basal cells from the lung (without trachea) for successful in vitro cultures. These lung-derived basal cells may well differ from their tracheal counterparts, as has been found in human airways, and culture models based on lung derived basal cells may allow a closer representation of the pulmonary epithelium.
In conclusion, we have established a workflow which is based on cold dispase II/DNase I digestion that allows for recovery of a greater number of highly viable AECs, which together with MACS sorting enables quick and parallel isolation of highly pure AEC populations from murine lungs that are suitable for in vitro culture.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary figure 1: Cold digestion with dispase II does not remove MHC-I, MHC-II or CD24 from the surface of CD45-CD31-EpCAM+ cells. Supplementary figure 2: Digestion type does not affect cellular oxidative stress based on LDHa (lactate dehydrogenase) levels. Supplementary figure 3: Murine AECs can be passaged (P0-2) following cold digestion and CD45-CD31-EpCAM+ MACS sorting.
Author Contributions
PPJ: Conceptualization, Formal analysis, Investigation, Writing - original draft, Writing - review and editing. CC, HJM: Conceptualization, Formal analysis, Investigation, Writing - review and editing; PS, GPW: Formal analysis, Writing - review and editing. JS: Conceptualization, Data curation, Supervision, Funding acquisition, Writing - original draft, Project administration, Writing - review and editing.
Funding
This research was funded by British Lung Foundation and the Breathing Together Consortium, grant number “PHD16-19 BUSH” and Horserace Betting Levy Board, grant number “VET/2020 -1 EPDF 7”.
Institutional Review Board Statement
The animal study protocol was approved by the University of Edinburgh Animal Welfare and Ethical Review Board. UK Home Office project license to JS, number P4871232F.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data underlying this article will be shared upon reasonable request.
Acknowledgments
PPJ and JS thank the British Lung Foundation and the Breathing Together Consortium for support. CC was also supported by a veterinary postdoctoral research fellowship funded by the Horserace Betting Levy Board (VET/2020 -1 EPDF 7). We gratefully acknowledge assistance and expertise from the Flow Core Facility and Microscopy Core Facility at the University of Edinburgh.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Rock, J.R.; Randell, S.H.; Hogan, B.L.M. Airway Basal Stem Cells: A Perspective on Their Roles in Epithelial Homeostasis and Remodeling. Dis Model Mech 2010, 3, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.E.; Brumwell, A.N.; Xi, Y.; Gotts, J.; Brownfield, D.G.; Treutlein, B.; Tan, K.; Tan, V.; Liu, F.; Looney, M.R.; et al. Lineage-Negative Progenitors Mobilize to Regenerate Lung Epithelium after Major Injury. Nature 2015, 517, 621–625. [Google Scholar] [CrossRef]
- Zuo, W.; Zhang, T.; Wu, D.Z.; Guan, S.P.; Liew, A.-A.; Yamamoto, Y.; Wang, X.; Lim, S.J.; Vincent, M.; Lessard, M.; et al. P63+Krt5+ Distal Airway Stem Cells Are Essential for Lung Regeneration. Nature 2015, 517, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Eenjes, E.; Mertens, T.C.J.; Kempen, M.J.B.; Wijck, Y. van; Taube, C.; Rottier, R.J.; Hiemstra, P.S. A Novel Method for Expansion and Differentiation of Mouse Tracheal Epithelial Cells in Culture. Sci Rep-uk 2018, 8, 7349. [Google Scholar] [CrossRef]
- McQualter, J.L.; Yuen, K.; Williams, B.; Bertoncello, I. Evidence of an Epithelial Stem/Progenitor Cell Hierarchy in the Adult Mouse Lung. Proc National Acad Sci 2010, 107, 1414–1419. [Google Scholar] [CrossRef]
- Jiang, D.; Schaefer, N.; Chu, H.W. Lung Innate Immunity and Inflammation, Methods and Protocols. Methods Mol Biology 2018, 1809, 91–109. [Google Scholar] [CrossRef]
- You, Y.; Richer, E.J.; Huang, T.; Brody, S.L. Growth and Differentiation of Mouse Tracheal Epithelial Cells: Selection of a Proliferative Population. Am J Physiol-lung C 2002, 283, L1315–L1321. [Google Scholar] [CrossRef]
- Davidson, D.J.; Kilanowski, F.M.; Randell, S.H.; Sheppard, D.N.; Dorin, J.R. A Primary Culture Model of Differentiated Murine Tracheal Epithelium. Am J Physiol-lung C 2000, 279, L766–L778. [Google Scholar] [CrossRef]
- Broadbent, L.; Villenave, R.; Guo-Parke, H.; Douglas, I.; Shields, M.D.; Power, U.F. Human Respiratory Syncytial Virus, Methods and Protocols. Methods Mol Biology 2016, 1442, 119–139. [Google Scholar] [CrossRef]
- Vazquez-Armendariz, A.I.; Seeger, W.; Herold, S.; Agha, E.E. Protocol for the Generation of Murine Bronchiolospheres. Star Protoc 2021, 2, 100594. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Wen, S.; Cao, W.; Yue, P.; Xu, X.; Zhang, Y.; Luo, L.; Chen, T.; Li, L.; Wang, F.; et al. Lung Organoids, Useful Tools for Investigating Epithelial Repair after Lung Injury. Stem Cell Res Ther 2021, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.H.; Nadarajan, P.; Graham, T.A.; Pipinikas, C.P.; Brown, J.M.; Falzon, M.; Nye, E.; Poulsom, R.; Lawrence, D.; Wright, N.A.; et al. Stochastic Homeostasis in Human Airway Epithelium Is Achieved by Neutral Competition of Basal Cell Progenitors. Elife 2013, 2, e00966. [Google Scholar] [CrossRef] [PubMed]
- Weitnauer, M.; Mijošek, V.; Dalpke, A.H. Control of Local Immunity by Airway Epithelial Cells. Mucosal Immunol 2016, 9, 287–298. [Google Scholar] [CrossRef]
- Crosby, L.M.; Waters, C.M. Epithelial Repair Mechanisms in the Lung. Am J Physiol-lung C 2010, 298, L715–L731. [Google Scholar] [CrossRef]
- Heijink, I.H.; Kuchibhotla, V.; Roffel, M.P.; Maes, T.; Knight, D.A.; Sayers, I.; Nawijn, M.C. Epithelial Cell Dysfunction, a Major Driver of Asthma Development. Allergy 2020, 75, 1902–1917. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Li, L.; Wang, Y.; Zhang, S.; Adcock, I.M.; Barnes, P.J.; Huang, M.; Yao, X. Bronchial Epithelial Cells in COPD. Respirology 2015, 20, 722–729. [Google Scholar] [CrossRef]
- Chakraborty, A.; Mastalerz, M.; Ansari, M.; Schiller, H.B.; Staab-Weijnitz, C.A. Emerging Roles of Airway Epithelial Cells in Idiopathic Pulmonary Fibrosis. Cells 2022, 11, 1050. [Google Scholar] [CrossRef]
- Quantius, J.; Schmoldt, C.; Vazquez-Armendariz, A.I.; Becker, C.; Agha, E.E.; Wilhelm, J.; Morty, R.E.; Vadász, I.; Mayer, K.; Gattenloehner, S.; et al. Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. Plos Pathog 2016, 12, e1005544. [Google Scholar] [CrossRef]
- Raoust, E.; Balloy, V.; Garcia-Verdugo, I.; Touqui, L.; Ramphal, R.; Chignard, M. Pseudomonas Aeruginosa LPS or Flagellin Are Sufficient to Activate TLR-Dependent Signaling in Murine Alveolar Macrophages and Airway Epithelial Cells. Plos One 2009, 4, e7259. [Google Scholar] [CrossRef]
- Naikawadi, R.P.; Disayabutr, S.; Mallavia, B.; Donne, M.L.; Green, G.; La, J.L.; Rock, J.R.; Looney, M.R.; Wolters, P.J. Telomere Dysfunction in Alveolar Epithelial Cells Causes Lung Remodeling and Fibrosis. Jci Insight 2016, 1, e86704. [Google Scholar] [CrossRef]
- Corporation, W.B. Worthington Enzyme Manual. Available online: https://www.worthington-biochem.com/tissuedissociation/Lung.html.
- Supp, D.M.; Hahn, J.M.; Combs, K.A.; McFarland, K.L.; Powell, H.M. Isolation and Feeder-Free Primary Culture of Four Cell Types from a Single Human Skin Sample. Star Protoc 2022, 3, 101172. [Google Scholar] [CrossRef]
- Volovitz, I.; Shapira, N.; Ezer, H.; Gafni, A.; Lustgarten, M.; Alter, T.; Ben-Horin, I.; Barzilai, O.; Shahar, T.; Kanner, A.; et al. A Non-Aggressive, Highly Efficient, Enzymatic Method for Dissociation of Human Brain-Tumors and Brain-Tissues to Viable Single-Cells. Bmc Neurosci 2016, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Richer, E.J.; Huang, T.; Brody, S.L. Growth and Differentiation of Mouse Tracheal Epithelial Cells: Selection of a Proliferative Population. Am J Physiol-lung C 2002, 283, L1315–L1321. [Google Scholar] [CrossRef]
- Autengruber, A.; Gereke, M.; Hansen, G.; Hennig, C.; Bruder, D. Impact of Enzymatic Tissue Disintegration on the Level of Surface Molecule Expression and Immune Cell Function. European J Microbiol Immunol 2012, 2, 112–120. [Google Scholar] [CrossRef]
- Mokwatsi, G.G.; Schutte, A.E.; Kruger, R. A Biomarker of Tissue Damage, Lactate Dehydrogenase, Is Associated with Fibulin-1 and Oxidative Stress in Blacks: The SAfrEIC Study. Biomarkers 2015, 21, 48–55. [Google Scholar] [CrossRef]
- Li, X.; Rossen, N.; Sinn, P.L.; Hornick, A.L.; Steines, B.R.; Karp, P.H.; Ernst, S.E.; Adam, R.J.; Moninger, T.O.; Levasseur, D.N.; et al. Integrin A6β4 Identifies Human Distal Lung Epithelial Progenitor Cells with Potential as a Cell-Based Therapy for Cystic Fibrosis Lung Disease. Plos One 2013, 8, e83624. [Google Scholar] [CrossRef]
- Chernaya, O.; Shinin, V.; Liu, Y.; Minshall, R.D. Behavioral Heterogeneity of Adult Mouse Lung Epithelial Progenitor Cells. Stem Cells Dev 2014, 23, 2744–2757. [Google Scholar] [CrossRef]
- Nakano, H.; Nakano, K.; Cook, D.N. Type 2 Immunity, Methods and Protocols. Methods Mol Biology 2018, 1799, 59–69. [Google Scholar] [CrossRef]
- Chen, H.; Matsumoto, K.; Brockway, B.L.; Rackley, C.R.; Liang, J.; Lee, J.; Jiang, D.; Noble, P.W.; Randell, S.H.; Kim, C.F.; et al. Airway Epithelial Progenitors Are Region Specific and Show Differential Responses to Bleomycin-Induced Lung Injury. Stem Cells 2012, 30, 1948–1960. [Google Scholar] [CrossRef] [PubMed]
- Nichane, M.; Javed, A.; Sivakamasundari, V.; Ganesan, M.; Ang, L.T.; Kraus, P.; Lufkin, T.; Loh, K.M.; Lim, B. Isolation and 3D Expansion of Multipotent Sox9 + Mouse Lung Progenitors. Nat Methods 2017, 14, 1205–1212. [Google Scholar] [CrossRef]
- Eenjes, E.; Mertens, T.C.J.; Kempen, M.J.B.-V.; Wijck, Y.V.; Taube, C.; Rottier, R.J.; Hiemstra, P.S. A Novel Method for Expansion and Differentiation of Mouse Tracheal Epithelial Cells in Culture. Sci Rep-uk 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Levardon, H.; Yonker, L.; Hurley, B.; Mou, H. Expansion of Airway Basal Cells and Generation of Polarized Epithelium. Bio-protocol 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Zhang, T.; Wu, D.Z.A.; Guan, S.P.; Liew, A.A.; Yamamoto, Y.; Wang, X.; Lim, S.J.; Vincent, M.; Lessard, M.; et al. P63 + Krt5 + Distal Airway Stem Cells Are Essential for Lung Regeneration. Nature 2015, 517, 616–620. [Google Scholar] [CrossRef]
- Lam, H.C.; Choi, A.M.K.; Ryter, S.W. Isolation of Mouse Respiratory Epithelial Cells and Exposure to Experimental Cigarette Smoke at Air Liquid Interface. J Vis Exp 2011. [CrossRef]
- Prescott, M.J.; Lidster, K. Improving Quality of Science through Better Animal Welfare: The NC3Rs Strategy. Lab Animal 2017, 46, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Koenitzer, J.R.; Wu, H.; Atkinson, J.J.; Brody, S.L.; Humphreys, B.D. Single-Nucleus RNA-Sequencing Profiling of Mouse Lung. Reduced Dissociation Bias and Improved Rare Cell-Type Detection Compared with Single-Cell RNA Sequencing. Am J Resp Cell Mol 2020, 63, 739–747. [Google Scholar] [CrossRef]
- Lee, D.F.; Salguero, F.J.; Grainger, D.; Francis, R.J.; MacLellan-Gibson, K.; Chambers, M.A. Isolation and Characterisation of Alveolar Type II Pneumocytes from Adult Bovine Lung. Sci Rep-uk 2018, 8, 11927. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wan, J. Methodological Comparison of FACS and MACS Isolation of Enriched Microglia and Astrocytes from Mouse Brain. J Immunol Methods 2020, 486, 112834. [Google Scholar] [CrossRef]
- Sutermaster, B.A.; Darling, E.M. Considerations for High-Yield, High-Throughput Cell Enrichment: Fluorescence versus Magnetic Sorting. Sci Rep-uk 2019, 9, 227. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).