1. Structural Basis of ESCRT
The Endosomal Sorting Complexes Required for Transport (ESCRT) system is a peripheral membrane protein complex composed of ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, VPS4-VTA1, and several accessory proteins 1, 2. They are recruited to distinct cellular locations to perform key steps in essential processes such as protein degradation, cell division, and membrane sealing 3. The ESCRT system was initially discovered in yeast cells 4 and subsequently found to exist in insects, mammalian cells, and even archaea. It is evolutionarily conserved in all eukaryotes and plays important roles in cytoplasmic separation 5, cellular autophagy 6, 7, membrane fission and remodeling 8, as well as replication and budding processes of certain enveloped viruses 9, 10, among other biological activities. Although proteins and complexes involved in the ESCRT pathway must work collaboratively, they are recruited sequentially and carry out distinct functions. Early-acting factors such as ESCRT-I and ALIX recognize and assist in remodeling the appropriate membrane, concentrating vesicular cargo, and recruiting late-acting factors 11.
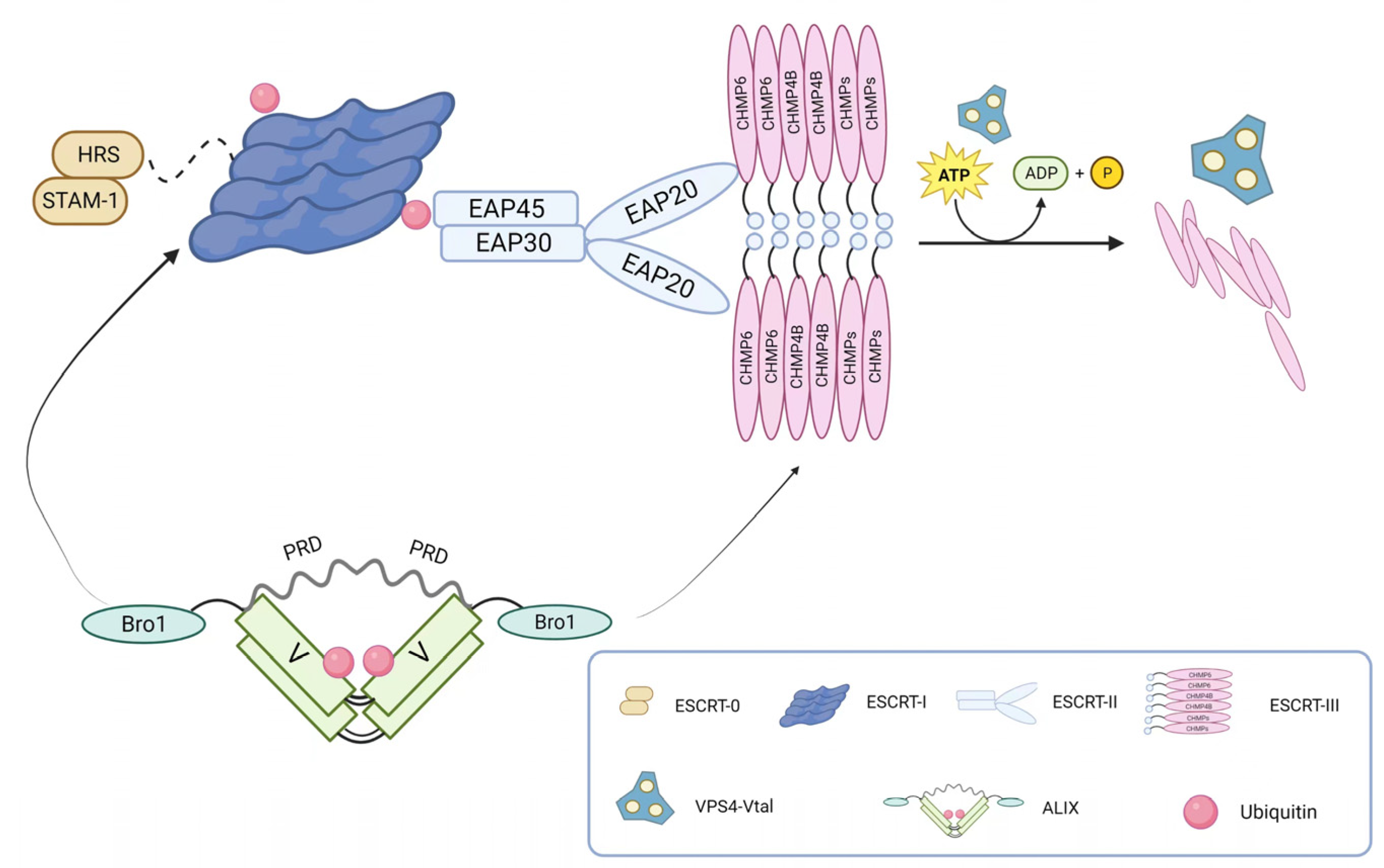
The core structure of ESCRT is illustrated in
Figure 1. The ESCRT-0 complex, a heterodimer, primarily recognizes ubiquitinated substrates. In eukaryotes, it consists of hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) and signals transducing adaptor molecule (STAM1/2), whereas in yeast cells, it is composed of VPS27 and HSE1. These subunits form a composite helix through the GAT (GGA and Tom1) domain in a 1:1 ratio
12, 13. As a regulatory factor in one of the essential intracellular vesicular transport pathways, ESCRT-0 recognizes, recruits, and binds to membrane proteins on the cell membrane. It then associates with ESCRT-I and ESCRT-II complexes to form vesicles for intracellular transport and degradation of substances. Therefore, ESCRT-0 plays a critical role in regulating vesicular transport pathways in the cytoplasm.
ESCRT-I, structurally forming an elongated heterotetramer, was the first discovered ESCRT structure. It consists of VPS23/TSG101, VPS28, VPS37, and MVB12/UBAP1 (Ubiquitin Associated Protein 1) in a 1:1:1:1 ratio. On one hand, the UEV domain of VPS23/TSG101 interacts with ESCRT-0 and ubiquitinated membrane proteins. ESCRT-I can bind two ubiquitin moieties on the endosome due to the association of UBAP1 and yeast Mvb12 with ubiquitin. On the other hand, VPS28 interacts with the GLUE domain of ESCRT-II protein EAP45, thereby interacting with the ESCRT-II complex. Additionally, ESCRT-I and Alix can recruit ESCRT-III components, which mediate the final scission step during cell division. Moreover, ESCRT-I is involved in regulating various cellular processes such as viral infection 14 and apoptosis 14, 15.
As a key factor in intracellular membrane transport and degradation pathways, ESCRT-II participates in forming and dissociating intracellular vesicles in the endoplasmic reticulum system. In yeast cells, this complex consists of VPS36, VPS25, and VPS22, while in mammals, it is composed of the endosomal sorting complex required for transport-associated proteins (EAP). The Y-shaped structure of ESCRT-II is formed by EAP30, EAP45, and EA20 in a 1:1:2 ratio 16. Similar to ESCRT-I, ESCRT-II has only one ubiquitin-binding domain, which limits its ability to recruit ubiquitinated proteins. However, it contains domains that interact with ubiquitin 17, ESCRT-I 18, and specific membrane components. Therefore, it does not directly participate in sorting ubiquitinated proteins but functions after cargo selection by ESCRT-0. Additionally, the SNF8 protein (VPS22) of ESCRT-II interacts with calcium transporters, thereby participating in receptor-mediated calcium signaling in various cells and tissues 19.
In contrast to early ESCRT complexes (ESCRT-0, -I, and -II) that form stable protein complexes in the cytoplasm, the ESCRT-III complex assembles only transiently on endosomes. ESCRT-III belongs to the CHMP family, soluble monomers that are widely present in eukaryotes and participate in intracellular membrane transport and degradation pathways. It also acts as a membrane remodeling machinery, providing the driving force for membrane constriction at the neck and subsequent scission 20, 21. The ESCRT-III complex consists of 12 proteins, including CHMP1-6 and IST1, and their recruitment and assembly are regulated by early ESCRT complexes (ESCRT-0, -I, and -II). In most cases, ESCRT-III exists as filaments in cells, assembling on membranes to form tight helical structures. It mediates the rupture and sealing of various cellular membranes, including those between the nuclear envelope and plasma membrane, and is released from the membrane in the final stage through the transient ATP-dependent reaction with other ESCRT proteins, facilitated by Vps4. Furthermore, several studies have shown that the ESCRT-III complex plays important roles in various biological processes, including cell division 22, 23, immune regulation 24, and neurodegenerative diseases 25, 26.
The AAA-ATPase complex Vacuolar protein sorting 4 (VPS4) plays a crucial role in intracellular protein transport, cell division, and retroviral budding. It consists of the type I AAA-ATPase Vps4 and its cofactor Vta1. By harnessing the energy provided by ATP hydrolysis, it disassembles the ESCRT-III complex on endosomal membranes, enabling recycling. Upon completion of ESCRT-III disassembly, the Vps4 complex dissociates into its inactive protomers. Therefore, the Vps4 complex terminates each round of MVB cargo sorting and vesicle formation 27, 28. Two types of VPS4 complexes, VPS4A and VPS4B, have been identified in mammals, and they play important roles in cell biology and disease, including cell division 29, membrane fusion and fission 30, 31, and viral infection 32, 33 among other biological processes. Nearly all viruses that utilize ESCRT budding require the recruitment of VPS4, indicating its critical role in viral budding 34.
ALIX protein is a multifunctional cellular protein with a structure depicted in
Figure 1. It is primarily composed of three domains: Bro1, V, and PRD (proline-rich domain). Each domain mediates interactions with different cellular and protein partners
35. The interaction between AAA-ATPase complex Vacuolar protein sorting 4 (VPS4) and apoptosis-linked gene 2 (ALG-2)-interacting protein X (ALIX) is a potential determinant of ESCRT-I and ESCRT-II function, promoting certain ESCRT-mediated activities through direct interactions with ESCRT-I and ESCRT-III. ALIX can also associate with various effector and adaptor proteins, such as TSG101
36, 37, tyrosine kinases
38, and CEP55 (centrosomal protein 55)
39, and these interactions are crucial for many viral budding processes. Furthermore, ALIX exhibits diverse functions, including promoting exocytosis
40, cell migration
41, and apoptosis
42.
2. Functions and Mechanisms of the ESCRT Complex
The ESCRT complex is a group of peripheral membrane protein complexes that play important roles in various biological processes, including the formation of multivesicular bodies (MVBs), retroviral budding (such as HIV), cytokinesis, autophagy, and other cellular activities
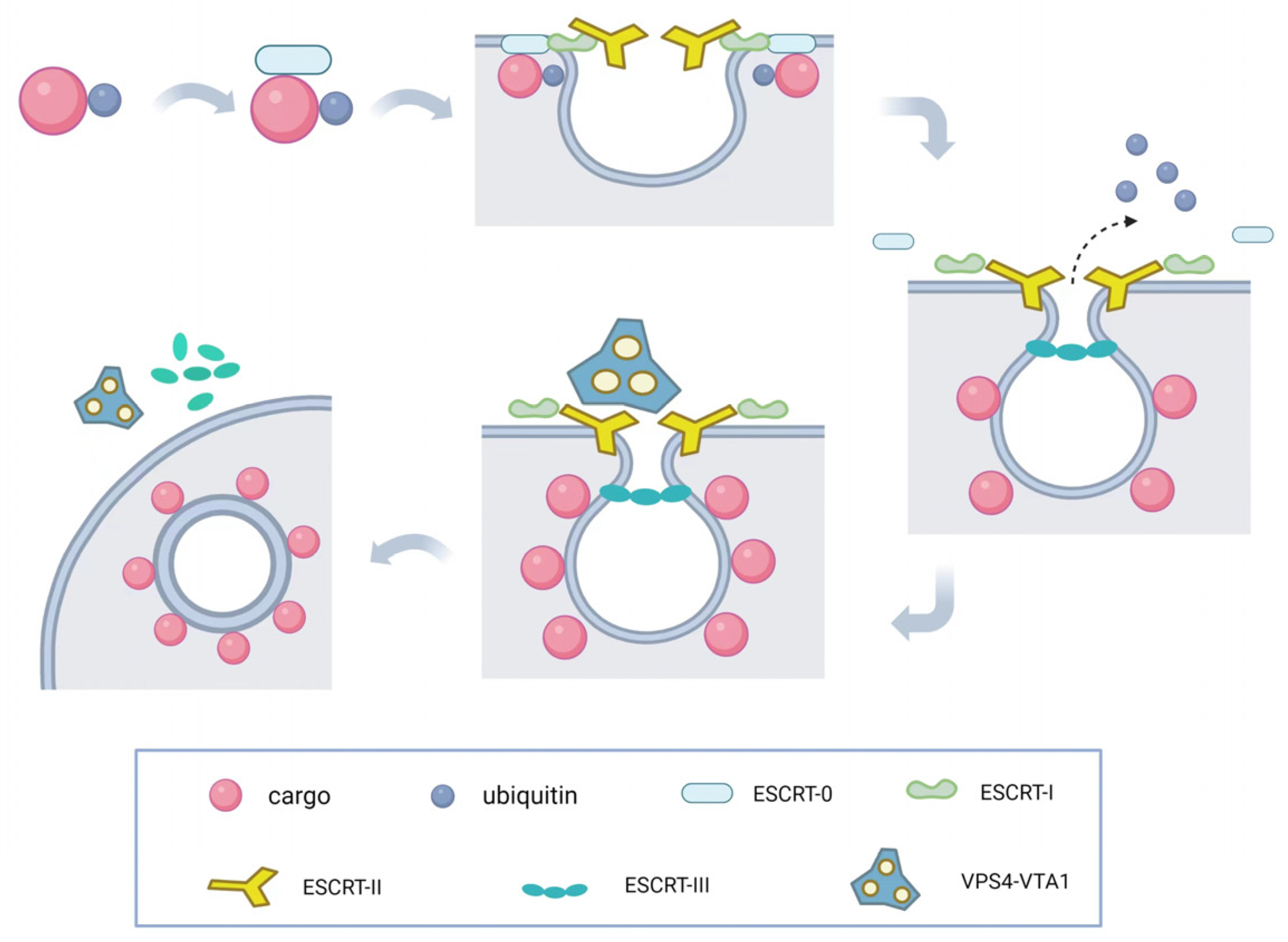
5-7, 9, 10, 43-45. The molecular mechanism underlying membrane invagination is illustrated in
Figure 2. Initially, ESCRT-0 recognizes and enriches ubiquitinated substrates to recruit ESCRT-I and initiate the budding process. Ubiquitination serves as a critical entry point for the ESCRT pathway and plays a crucial role in retroviral budding
46. Ubiquitinated cargo molecules are sorted into MVBs, where proteins such as TSG101, UBAP1, EAP45, and ALIX contain ubiquitin-binding sites. ESCRT-I and ESCRT-II then interact with each other to induce inward membrane invagination and form initial buds. Subsequently, ESCRT-III cuts the neck of the buds, releasing small vesicles into the lumen of the endosome, completing the budding process. Finally, the VPS4-VTA1 complex, powered by ATP hydrolysis, dissociates the ESCRT-III complexes from the endosomal membrane, separating the ESCRT structures and allowing for recycling. Once MVBs fuse with lysosomes, the intraluminal vesicles (ILVs) are degraded within the lysosomal lumen. Therefore, the ESCRT system plays a crucial role in sorting substances and downregulating surface receptors, while also participating in the precise regulation of cell signaling
47, 48. During this process, upstream ESCRT mechanisms or other recruiting factors, such as viral Gag proteins, initiate membrane deformation and allow downstream ESCRT machinery to aggregate in the absence of pre-existing membrane necks or pores. ESCRT-III components are recruited by ESCRT-I/-II and/or Bro domain proteins
49 and preferentially aggregate on curved membranes
50-53. The following sections will describe several important functions of the ESCRT complex.
2.1. ESCRT and Autophagy
Degradation and recycling of intracellular substances are fundamental mechanisms for maintaining cellular homeostasis and responding to adverse external conditions. Most cells employ three distinct degradation systems: the ubiquitin-proteasome degradation system, endocytosis, and autophagy 54. Among these, autophagy is a conserved cellular mechanism in all eukaryotes, through which organelles, proteins, and invading pathogens can be sequestered and degraded within vesicles 55. Within cells, various types of autophagic pathways coexist, relying on different combinations of cellular and molecular mechanisms, but ultimately delivering cargo for degradation in lysosomes 6. The ESCRT machinery is capable of participating in different types of autophagic processes to degrade cytoplasmic components, such as macroautophagy and microautophagy. The distinction between the two lies in the fact that, in macroautophagy, autophagic substrates are transported to vesicles called autophagosomes, while in microautophagy, substrates are directly engulfed by invaginations of the vacuolar membrane.
The formation of autophagosomes requires the induction, transport, and fusion of membrane-bound organelles. Previous studies have shown that disruption of the ESCRT machinery in mammals leads to autophagosome accumulation, indicating the important role of ESCRT in regulating autophagy 25, 56. The relationship between ESCRT and autophagy is linked through shared molecular mechanisms involved in autophagosome formation and fusion. The ESCRT machinery rescues mildly damaged lysosomes and interacts with autophagy-related proteins (ATGs) to form a bridge between endocytic and macroautophagy processes, facilitating the formation of the endo-lysosomal system and cell death. Closure of autophagosomes requires membrane scission, which is topologically similar to ESCRT-III-mediated intraluminal vesicle (ILV) formation on multivesicular endosomes (MVEs), suggesting a similar or identical membrane scission machinery is involved. Upon maturation, autophagosomes fuse with late endosomes/lysosomes for degradation, and this process involves ESCRT proteins such as ALIX 57 and VPS4. Additionally, certain components of the ESCRT complex participate in regulating different steps of the autophagic pathway. For instance, the ESCRT-III protein SNF7 plays a role in autophagosome formation and membrane sealing 58, while VPS4 promotes the fusion of autophagosomes with late endosomes, facilitating the progression of autophagy 59, 60. Therefore, the ESCRT complex plays a crucial role in the autophagic pathway and is closely associated with the mechanisms of autophagy.
2.2. ESCRT-Dependent Membrane Remodeling and Fission
ESCRT is an evolutionarily conserved membrane remodeling complex with the unique ability to catalyze membrane constriction within necks. Among them, ESCRT-III, in collaboration with the AAA ATPase VPS4, plays a central role in the main membrane remodeling and scission functions of the ESCRT machinery 61. ESCRT-III exerts its membrane fission catalytic ability on almost all cellular membranes. It catalyzes the formation of intraluminal vesicles (ILVs) within endosomal membranes, drives membrane scission from the cell surface, facilitates nuclear envelope reformation, and mediates autophagosome closure 8. It is recruited to various distinct sites of action through interactions with multiple adaptor proteins. VPS4, on the other hand, is recruited to the membrane to facilitate the disassembly of ESCRT-III components 62. Cells utilize this system for reverse membrane fission during processes such as protein degradation, cell division, and retroviruses release. Retroviruses recruit ESCRT-III to bud sites to ensure cell exit. In this process, the ESCRT complex, through interaction with viral particle proteins, forms a protein membrane fission machinery that promotes viral particle release and aids in the evolution of the L domain 63.
Not all organisms require the complete ESCRT complex, but ESCRT-III and VPS4 appear to be indispensable, with CHMP2, CHMP3, CHMP4, and VPS4 considered essential components in membrane remodeling. However, even in the presence of ESCRT-III proteins, membrane fission is halted when AAA ATPase VPS4 is absent. Furthermore, ESCRT demonstrates critical activity in many other cellular membranes remodeling events, such as enveloped virus budding, cytokinesis abscission, exosome biogenesis 64, plasma membrane repair, immune synapse formation, and nuclear membrane homeostasis 65.
2.3. ESCRT and Endosomal Sorting
The ESCRT machinery was initially discovered through genetic studies in yeast, where mutations led to improper sorting of proteins into vesicles, functionally equivalent to lysosomes 66. These mutant strains, known as "E-vps mutants," exhibited enlarged pre-vacuolar endosome-like compartments containing undegraded proteins 67. Subsequently, it was found that most E-vps genes act sequentially, concentrating transport cargoes and incorporating them into late endosomes (also known as multivesicular bodies or MVBs), which eventually fuse with lysosomes for degradation 68. During the sorting process, the various complexes of ESCRT are sequentially recruited from the cytoplasm to the endosomal membrane through specific subunits, packaging cellular waste, proteins, and other molecules into MVBs via the formation of internal vesicles, followed by their transport to lysosomes for degradation.
ESCRT complexes play crucial roles in this process, with the ubiquitination of cargoes providing a critical signal for initial cargo binding by ESCRT-0 69. The ESCRT-0 subunits HRS and STAM, ESCRT-I subunit TSG101, and ESCRT-II subunit Vps36 all contain ubiquitin-binding domains that interact with ubiquitinated cargoes. Specifically, ESCRT-0, ESCRT-I, and ESCRT-II subunits facilitate the aggregation and recognition of membrane receptors by interacting with specific signaling molecules on the membrane. Subsequently, ESCRT-III and VPS4-VTA1 subunits act on the outer membrane of the MVB, forming a cone-shaped structure that ultimately drives MVB formation and separation of internal vesicles 70. With the assistance of ESCRT complexes, the VPS4 complex dissociates ESCRT structures from the limiting membrane, and after fusion of the MVB with lysosomes, intraluminal vesicles (ILVs) are degraded within the lysosomal lumen. Additionally, several accessory proteins participate in the regulation of this process, such as ALIX and VPS27, which interact with different subunits of the ESCRT complex, thereby modulating endosomal sorting 71, 72. Thus, the ESCRT pathway also plays a critical role in the sorting and downregulation of cell surface receptors, and it is involved in fine-tuning cell signaling pathways 73.
2.4. ESCRT and Cell Division: Key Players in Membrane Dynamics
ESCRT was initially discovered as a regulator of intracellular cargo sorting 68, 74. Recent studies have shown that ESCRT is also involved in a series of critical events during cell division, such as gametogenesis, chromosome segregation, and cytokinesis75. The final stage of cell division is abscission, which involves the severing of the narrow membrane bridge between the two daughter cells 76. ESCRT is essential for cell division, and its role in cytokinesis can be traced back to ancient bacteria 77, suggesting that the ancestral function of these proteins was to promote membrane remodeling for division and was selected throughout evolution to perform various other cellular biological functions. During this process, ESCRT-associated components are sequentially recruited to the membrane to mediate the scission event 77 and exhibit membrane-severing activity in vitro 78. The charged multivesicular body protein 1B (CHMP1B) of ESCRT-III interacts with the microtubule-severing enzyme spastin, which is crucial for cell division 79. The ESCRT-I subunit tumor susceptibility gene 101 (TSG101) and the ESCRT-III subunit charged multivesicular body protein 4B (CHMP4B) are sequentially recruited to the central region of the intercellular bridge, forming a series of cortical rings. In the late stage of cell division, CHMP4B is acutely recruited to the constricted site where abscission occurs, and the ESCRT disassembly factor vacuolar protein sorting 4 (VPS4) follows CHMP4B to that location, triggering immediate cell separation 80. ESCRT-III and VPS4 are spatially and temporally associated with the abscission event, indicating their direct involvement in membrane scission during cytokinesis.
During cell division, ESCRT interacts with structures such as the centrosome, chromosomes, and the cytoskeletal framework, participating in the regulation of cytokinesis, chromosome segregation, and recombination, as well as gametogenesis and embryonic development. Moreover, ESCRT may be closely associated with cell fate determination during cell division, including cell fate transitions, stem cell differentiation, and tumor development 81, 82. In conclusion, the role of ESCRT in cell division is gradually being unraveled, and it holds significant implications for a comprehensive understanding of the mechanisms underlying cell division regulation and related diseases.
2.5. ESCRT and cell apoptosis
Cell apoptosis is a programmed cell death and the most extensively studied form of regulated cell death. It is characterized by sequential activation of the cysteine-aspartic protease caspases 83. This process involves chromatin fragmentation and marginalization, as well as the generation and membrane budding of apoptotic bodies 84. During cell apoptosis, cells release various cell factors and vesicles containing cellular components and nuclear fragments referred to as apoptotic bodies. The ESCRT complex can participate in the formation and clearance of apoptotic bodies. Specifically, the ESCRT-0 and ESCRT-III complexes interact with vesicle-associated proteins on the cell membrane, inducing membrane curvature and eventually leading to vesicle formation and release. Subsequently, the ESCRT-III complex assembles within the cell, forming a contractile structure that can engulf and release the apoptotic bodies to the extracellular space. Under normal circumstances, the binding of TSG101 with ALIX prevents cell apoptosis, but disruption of this process may occur due to dysregulation of cellular solute Ca2+ and subsequent upregulation of ALG-2 (apoptosis-linked gene 2) 85. Additionally, the ESCRT-III complex can also form mitochondrial exosomes, participating in apoptosis by segregating and releasing mitochondria. ESCRT-III plays a crucial role in repairing damaged membranes involved in various types of regulated cell death, such as necroptosis, pyroptosis, and ferroptosis. Inhibition of the ESCRT-III mechanism through genetic depletion of its core components increases susceptibility to cell death induced by anticancer drugs, indicating that ESCRT-III could be a potential target to overcome drug resistance during cancer treatment. Therefore, the ESCRT complex plays a significant role in regulating the release and clearance of cellular components during cell apoptosis, maintaining normal physiological processes within the organism 85.
Furthermore, cells undergoing necroptosis do not always lead to cell death. The known repair mechanisms of ESCRT involve vesiculation or shedding of damaged membranes and play a critical balancing role in sorting and downregulating activated cell surface receptors 73 and repairing damaged membranes to maintain membrane integrity 65. This delays cell death pathway, including cell apoptosis, necroptosis, pyroptosis, and ferroptosis 86-88. Cell apoptosis, necroptosis, and pyroptosis are all passive "suicidal" forms of molecular processes that result in the formation of several na-nanometer-sized membrane pores and catastrophic cell rupture 89. In summary, ESCRT provides time for dying cells, and ESCRT-dependent membrane repair negatively regulates cell death: ESCRT-I and -III are involved in cell apoptosis, while ESCRT-III mediates necroptosis, pyroptosis, and fer- roptosis.
3. ESCRT and Enveloped Viruses
The ESCRT machinery is not only essential for cellular processes such as membrane repair but is also hijacked by certain viruses during different stages of replication, particularly during the budding phase. Late budding (L) domains have been widely identified in the structural proteins of these viruses through three conserved tetrapeptide motifs (Y(L)XXL, PT/SAP, and PPXY). These motifs mediate the recruitment and interaction of class E proteins to facilitate virus budding, envelopment of retroviruses (e.g., HIV), and RNA viruses (e.g., filoviruses, orthomyxoviruses, bunyaviruses, and paramyxoviruses), redirecting cellular ESCRT proteins to the cytoplasmic membrane for viral particle release and egress from infected cells.
Many enveloped viruses utilize the ESCRT pathway to catalyze membrane scission, leading to the release or budding of viral particles 90. This process is dependent on specific late-domain structures of the respective viruses. All viruses that bud in an ESCRT-dependent manner require the membrane-remodeling functions provided by ESCRT-III and VPS4, but their requirements for upstream factors may vary. For instance, HIV-1 recruits the ESCRT pathway through specific motifs on the viral Gag protein 91, while the influenza A virus utilizes an ESCRT-independent budding pathway 92. Enveloped virus infection begins with the attachment of the virus to the host cell's plasma membrane, followed by viral entry, replication, and expression within the host cell. Finally, mature virus particles are released from the host cell, initiating a new infectious cycle. Studies have revealed the hijacking of the ESCRT system by HIV for its budding, and subsequent research has demonstrated the hijacking of the ESCRT system by various viruses, mediating different steps of the virus life cycle in different cells, thereby facilitating virus proliferation, budding, and dissemination. The ESCRT system has become an integral component of enveloped virus infections 90, 93. In the following, we summarize the reported interactions between various types of enveloped viruses and different ESCRT components, aiming to enhance our understanding of the interplay between ESCRT complexes and viral infections. Additionally, we aim to provide new research directions and contribute to the development of therapeutic strategies for future studies.
Table 1.
Interactions between different types of enveloped viruses and various ESCRT components.
Table 1.
Interactions between different types of enveloped viruses and various ESCRT components.
| Virus Category |
Virus Name |
ESCRT Components |
References |
| Enveloped RNA Viruses |
NDV |
ESCRT-III |
97、98
|
| SeV |
ALIX |
101 |
| HPIV1 |
ALIX |
109 |
| NIV |
ESCRT-I |
113 |
| HIV-1 |
ESCRT-I、-III、VPS4、ALIX |
115-117;122-124
|
| HCV |
ESCRT-0、-III、VPS4 |
134、136-137
|
| PRRSV |
ESCRT-I |
145 |
| EBOV |
ESCRT-I、ALIX |
149、157
|
| Enveloped DNA Viruses |
HBV |
ESCRT-0、-I、-II、-III、VPS4 |
158、163、164
|
| HSV-1 |
ESCRT-III |
169-171 |
3.1. ESCRT Pathway and Enveloped RNA Viruses
Enveloped RNA viruses utilize the ESCRT pathway to facilitate budding and release from host cells. By interacting with ESCRT proteins, these viruses can redirect intracellular proteins to the cell membrane, promoting viral particle budding and separation. The involvement of ESCRT plays a critical regulatory role in the infection, replication, and spread of enveloped RNA viruses.
3.1.1. The Role of ESCRT Machinery in Retroviral Budding
Avian paramyxoviruses, including Newcastle disease virus (NDV), are single-stranded RNA viruses that can cause highly contagious diseases in various avian species. The M protein of paramyxoviruses has been identified as a nucleocapsid-associated protein that plays a crucial role in the virus life cycle 94. During early infection, the M protein typically enters the cell nucleus through its nuclear localization signal and transiently accumulates in the nucleolus. Previous studies have shown that this M protein aggregation, observed in respiratory syncytial virus (RSV), NDV, and measles virus (MeV), can affect the transcription, synthesis, and post-translational modifications of host proteins 95. The M protein contains conserved peptides known as the L domain, which can utilize the ESCRT complex to facilitate paramyxovirus release. However, single amino acid mutations in this domain can result in severe budding defects in paramyxoviruses 96. While the FPIV-type L domain has been identified in some paramyxoviruses, its functional role in the virus life cycle and the mechanisms by which it recruits the ESCRT machinery remain poorly understood. As a core component of the ESCRT-III complex, CHMP4B has been identified as a key factor linking the FPIV L domain and the ESCRT machinery. Previous studies by Li et al.97 confirmed that the binding between NDV M protein and charged multivesicular body protein (CHMP4) promotes virus replication. Similarly, the study by Pei et al. 98 demonstrated that the ESCRT-III complex CHMP4s directly interacts with NDV M protein through the FPIV L domain, independently of TSG101, ESCRT-II, or ALIX as assembly factors. In summary, critical amino acid mutations in the FPIV L domain prevent effective interaction between NDV M protein and CHMP4B, resulting in the stalling of viral sub-particles during membrane budding and leading to defects in NDV release and growth. However, further research is needed to elucidate the detailed mechanisms underlying the interaction between NDV and the ESCRT system, as well as whether other ESCRT components are utilized to facilitate virus propagation.
Sendai virus (SeV), belonging to the Paramyxoviridae family and Respirovirus genus, is a pneumotropic virus in rodents and one of the most extensively studied members of the Respirovirus genus 99. Its structure contains a multifunctional accessory protein called the C protein, which enhances the virulence of the virus by stimulating viral replication 100. The C protein is crucial for the in vivo proliferation and pathogenesis of the virus, and the absence of C protein in SeV leads to the production of more infectious viral particles 101. Oda et al. conducted virus infection experiments using SeV generated with the Y3:Bro1 complex crystal structure and analyzed the impact of the interaction between SeV's C protein and Alix, a component of the ESCRT complex, on SeV budding. Their study revealed that a decrease in the affinity between the C protein and Alix significantly reduced the infectivity of the virus, indicating that the binding of the C protein to Alix enhances SeV budding 101. Furthermore, some studies have suggested that Alix also regulates cellular immune responses and inflammatory reactions, which may be associated with the immune response and inflammation triggered by SeV infection, although the specific mechanisms remain unclear 102-104. While the role of the ESCRT system in other viruses has been extensively studied, there is currently a lack of comprehensive research on the interaction between the Sendai virus and the ESCRT system.
Human parainfluenza virus type 1 (HPIV1) is a common respiratory pathogen and a single-stranded, negative-sense, non-segmented RNA virus belonging to the Paramyxoviridae family. The viral genome consists of 15,600 nucleotides and encodes six proteins from six genes. Among them, the C protein serves as an important virulence factor in HPIV1. It can inhibit apoptosis, influence host gene transcription, and regulate the production and signaling of type I interferon 105-107. The C protein of HPIV1 interacts with the Bro1 domain of Alix at a specific site, which is also essential for the interaction between Alix and CHMP4B. The C protein is ubiquitinated and subject to proteasome-mediated degradation, but its interaction with Alix's Bro1 domain protects it from degradation. Studies have shown that the Sendai virus (SeV) shares a high degree of homology with the C protein of HPIV1 108. However, the major difference between them is that SeV also expresses another accessory protein called the V protein, which has inhibitory effects on the host's innate antiviral response. In contrast, HPIV1 does not express the V protein, and the C protein is the only known viral antagonist of innate immune responses. Boonyaratanakornkit et al.109 proposed that during HPIV1 infection, Alix binds to the C protein and recruits it to the cytoplasmic surface of late endosomes. Subsequently, CHMP4 may recruit the Alix-C core-shell to a common site on the cytoplasmic surface of late endosomes, which serves as the source for virus assembly and budding.
Nipah virus (NIV) is a deadly zoonotic virus belonging to the Paramyxoviridae family, capable of causing severe respiratory and neurological diseases in humans and animals. The M protein of NIV is considered to play a central role in virus assembly and budding 110, 111. When expressed alone, the M protein is released from the cells as virus-like particles, and its budding is independent of the ESCRT pathway 112. The C protein of NIV acts as an accessory factor to counteract innate immunity, enhancing the budding of the M protein through the ESCRT pathway. Research by Park et al. 113 indicates that NIV-C directly interacts with the ESCRT factor TSG101, which is essential for NIV-C to enhance budding and efficiently release infectious Nipah virus. Further studies are being conducted to gain a better understanding of the interaction mechanisms between NIV and the ESCRT system.
3.1.2. The ESCRT machinery is essential for the proper assembly and budding of retroviral particles
HIV (Human Immunodeficiency Virus) is a single-stranded RNA virus belonging to the Retroviridae family. It primarily infects human immune cells, leading to immune system impairment. HIV-1 Gag is synthesized in the cytoplasm and subsequently directed to the plasma membrane, where virus assembly takes place 114. The formation of HIV viral particles occurs through the multimerization of the Gag precursor on the plasma membrane, and the budding of the virus is mediated by the interaction between Gag and various ESCRT (Endosomal Sorting Complex Required for Transport) proteins. Early components of the cellular ESCRT system, ESCRT-I (TSG101/VPS23) and ALIX mediate HIV-1 budding, both of which interact with the P6 domain of Gag. Within this domain, there are PS/TAP and YPXnL motifs that simultaneously promote virus budding, with the PS/TAP motif binding to ESCRT-I's TSG101 and the YPXnL motif binding to ESCRT-III-associated ALIX protein. Both pathways require downstream ESCRT-III and VPS4 ATPase 115-117. Additionally, this domain can interact with ESCRT-II's EAP20, directing HIV particles to the cell membrane and facilitating particle release 118-120. However, Langelier et al. suggested that HIV-1 release is ESCRT-II-independent since siRNA depletion of ESCRT-II subunit EAP20 did not significantly impact virus release and infectivity. Nevertheless, they did report detrimental effects on virus production at later time points following ESCRT-II knockdown 121. Furthermore, the Gag protein interacts with multiple members of the ESCRT-III complex, including CHMP2, CHMP3, CHMP4, and CHMP6, leading to the formation of membrane helical structures within the cell and aiding in virus particle formation and release 122-124.
Following this, Janvier et al. 125 investigated the additional role of the ESCRT machinery in HIV-1 release. Their research demonstrated that BST-2/tetherin, acting as a restriction factor, hinders HIV release by tethering mature virus particles to the plasma membrane. As a crucial component of ESCRT-0, HRS facilitates Vpu protein-induced downregulation and degradation of BST-2, thereby aiding in efficient HIV release. Physiologically, HRS is believed to initiate ESCRT-mediated multivesicular body (MVB) formation, with the PSAP motif in HRS recruiting the ESCRT machinery by interacting with ESCRT-I component TSG101 126. The assembly and efficient release of HIV-1 requires highly coordinated interactions between virus-encoded proteins and cellular key components. In summary, we propose that ESCRT-0 and ESCRT-II may be dispensable for HIV budding, while Alix is indispensable. Among the ESCRT-III subunits, only CHMP4 and CHMP2 appear to be essential. Loss of ESCRT function inhibits virus release, resulting in accumulated arrested buds on the surface of infected cells.
3.1.3. Interactions between HCV and the ESCRT System Promote Viral Envelopment through Ubiquitination
The Flaviviridae family comprises enveloped, single-stranded positive-sense RNA viruses, including the Hepatitis C virus (HCV), which is the causative agent of chronic hepatitis. It encodes a large polyprotein precursor of approximately 3,000 amino acid residues, which is processed by viral proteases and cellular signalises to generate three structural proteins (core, E1, and E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) 127. The non-structural proteins NS3, NS4A, NS4B, NS5A, and NS5B form the viral replication machinery, while p7 and NS2 are crucial for the production of infectious viruses 128, 129. NS5A is a membrane-associated RNA-binding phosphoprotein that also participates in the assembly and maturation of infectious HCV particles 130, 131.
Enveloped viruses typically hijack the ESCRT system through late-domain motifs or ubiquitination to bud from the plasma membrane. However, some viruses, such as HCV, lack identifiable late-domain motifs despite lacking late structural domains, suggesting that their association with ESCRT proteins may have diverse and complex mechanisms. On one hand, HCV proteins involved in viral assembly, such as core and NS2, are ubiquitinated 132, 133, providing a potential mechanism for the recruitment of HCV proteins into ESCRT complexes. Subsequently, Ariumi et al. 134 demonstrated the binding of the HCV core to CHMP4B, indicating that the core possesses a novel motif required for HCV production. In accordance with this, Blanchard et al. reported that an aspartic acid residue at position 111 in the core is critical for virus assembly, as the conversion of aspartic acid to proline at amino acid 111 (PTDP to PTAP) generates a PTAP late-domain motif, enhancing the release of HCV core in cell culture media 135.
HRS serves as a critical factor in HCV envelopment, while ESCRT-III and VPS4/VTA1 factors are hijacked by the HCV core and NS5A, respectively, to mediate viral assembly. The essential role of VPS4 and the ESCRT-III complex in HCV production has been demonstrated by Corless et al. 136. Furthermore, integrated approaches combining proteomics, RNA interference, viral genetics, and biophysics have validated the recruitment of ESCRT components by HCV proteins to facilitate viral envelopment 134, 137. ESCRT-0 component HRS has been identified as crucial for the release of HCV via exosome secretion by Tamai et al. 138. Subsequently, the study by Barouch-Bentov et al. 137 revealed that HCV hijacks HRS as an entry point into the ESCRT pathway to promote viral envelopment, highlighting the role of ubiquitinated NS2 residues in HRS recruitment and HCV assembly. The ubiquitination of viral proteins was found to serve as a signal for HRS binding and HCV assembly, thereby compensating for the absence of late structural domains. Additionally, HRS interacts with the viral core protein and envelope glycoproteins, facilitating their incorporation into newly formed viral particles. This interaction is crucial for the generation of infectious viral particles and the dissemination of HCV infection.
3.1.4. ESCRT Interacts with N Protein and Participates in Early Secretory Pathway in PRRSV Assembly
Porcine Reproductive and Respiratory Syndrome (PRRS) is an acute, contact-transmitted infectious disease that can cause abortion, stillbirth, weak-born piglets, mummified fetuses, and acute respiratory distress in pigs of all ages. The causative agent, PRRSV, is an enveloped, non-segmented, positive-sense RNA virus belonging to the Arteriviridae family and the genus Arterivirus 139. PRRSV infection is initiated by interaction between the virus and host cell receptors/factors attached to the plasma membrane 140. Upon internalization, PRRSV utilizes host machinery for its translation, transcription, and replication 141. Subsequently, the PRRSV nucleocapsid enters the endoplasmic reticulum (ER) or Golgi apparatus (GA), leading to the formation of enveloped viral particles. Finally, PRRSV viral particles are released through exocytosis, establishing a new cycle of infection. The PRRSV N protein is involved in the formation of the viral envelope by covalent and non-covalent interactions, enveloping the viral genome 142. Additionally, it interacts with various host cell factors to modulate viral infection.
Among the ESCRT proteins that are crucial for PRRSV infection, TSG101 has been identified as the most upstream factor. It has been reported that TSG101 is involved in the replication of classical swine fever virus and the assembly and release of various viruses 143. As a novel host factor interacting with PRRSV N protein, TSG101 facilitates the formation of PRRSV viral particles and is involved in viral particle assembly and budding, rather than an attachment, internalization, RNA replication, and N protein translation during the infection process 144, 145. The study by Zhang et al. 145 confirmed that inhibiting TSG101 expression or function significantly affects PRRSV replication and infectivity. Furthermore, PRRSV infection leads to the aberrant relocation and dysregulation of TSG101, affecting the balance of the ESCRT pathway and membrane trafficking within the cells.
Although significant progress has been made in elucidating the PRRSV lifecycle, a comprehensive understanding of PRRSV assembly is still lacking. The mechanisms of other identified ESCRT components involved in PRRSV infection remain unclear. Further research is needed to explore the interaction mechanisms between PRRSV and the ESCRT system to better understand the PRRSV lifecycle and viral proliferation process. Moreover, considering the importance of TSG101 in various viral infections, it holds promise as a broad-spectrum antiviral target for development.
3.1.5. The Role of ESCRT in the EBOV Budding Process
Ebola virus (EBOV) is a single-stranded negative-sense enveloped RNA virus belonging to the family Filoviridae, which can cause a severe hemorrhagic fever syndrome with a high fatality rate. The viral matrix protein VP40 is a major component of EBOV virions and is essential for filamentous particle formation 146, 147. Oligomerization of VP40 on the plasma membrane is required for particle assembly and virus budding 148. The late (L) budding domain of VP40 recruits host ESCRT (endosomal sorting complex required for transport) and ESCRT-related proteins to facilitate membrane scission, thereby mediating efficient virus-cell separation. This structural protein plays a crucial role in the late stages of virus particle assembly and release. Additionally, ESCRT-III is also critical for EBOV release.
Previous studies have indicated the involvement of TSG101, a component of the ESCRT-I complex, in EBOV budding 149. TSG101 recruitment to the plasma membrane, away from its normal functional site in endosomes, has been shown to occur through binding with EBOV VP40 148, 149. The interaction between the two requires specific amino acid motifs or the late (L) domain 150, which is responsible for the binding of TSG101 to the PTAP motif of VP40. In the final step of the vacuolar protein sorting (VPS) pathway, the ATPase activity of VPS4 generates the energy necessary for the disassembly of protein complexes, allowing for subsequent rounds of sorting. Dominant-negative VPS4 mutants lacking ATPase activity have been shown to inhibit EBOV release 151, 152. EBOV VP40 also redirects other ESCRT-I proteins from endosomes to the plasma membrane, although these interactions have not been fully characterized.
Furthermore, the host ESCRT protein ALIX has been extensively implicated in the budding of retroviruses such as HIV-1 and EIAV 153-155, as well as in the egress of other RNA viruses 156. Han et al. 157 first proposed a role for ALIX in EBOV particle budding and identified the domains of ALIX that mediate its interaction with the required VP40. Specifically, EBOV VP40 recruits host ALIX through the YPx(n)L/I motif, which can act as an alternative to the L domain in promoting virus release. These findings are of significant importance for a comprehensive understanding of the intricate virus-host interactions required for efficient EBOV egress and dissemination.
3.2. ESCRT and Enveloped DNA Viruses
In addition to enveloped RNA viruses, the ESCRT complex is also involved in the assembly and release processes of enveloped DNA viruses. For instance, Hepatitis B virus (HBV) and Herpes simplex virus type 1 (HSV-1).
3.2.1. Involvement of ESCRT in the HBV Infection Process
Hepatitis B virus (HBV) is the major pathogen responsible for human liver disease. It is an enveloped, DNA-containing retrovirus that replicates primarily through reverse transcription of its pregenomic RNA(pgRNA)158. Despite the success of global HBV vaccination programs, a definitive cure for HBV infection has yet to be found 159, 160. Persistent HBV infection can lead to the development of liver cirrhosis, liver damage, and hepatocellular carcinoma 161. When the nucleocapsid of HBV reaches genomic maturation in the cytoplasm, two distinct outcomes can occur: one involves encapsidation of the viral particles and their release through the ESCRT system, while the other involves the return of the viral RC DNA genome to the cell nucleus for further amplification of covalently closed circular DNA 162.
During HBV infection, all components of ESCRT-0 are essential for HBV replication. The core antigen of HBV (HBcAg) interacts with ESCRT-0, facilitating assembly and release of HBV particles. Specifically, the HBcAg protein forms a complex with the HRS subunit of ESCRT-0, which promotes the intracellular aggregation and assembly of HBV core proteins, ultimately leading to the release of HBV particles. Additionally, the surface antigen of HBV (HBsAg) can interact with the ESCRT complex, facilitating the release of HBV particles. Studies by Chou et al. 163 have shown that both downregulation and overexpression of HRS result in the inhibition of HBV RNA transcription, DNA replication, and virus particle production, partly through promoting naked capsid secretion.
Due to the absence of the P(S/T)AP-like late-domain motif in HBV structural proteins, which is known to interact with TSG101, it was previously believed that TSG101 was dispensable for HBV export. However, recent research findings have demonstrated that knockdown of TSG101 reduces extracellular HBV DNA levels and results in the accumulation of viral capsids within cells. This supports the essential role of TSG101 in the process of HBV extracellular capsid release164. Furthermore, it has been observed that ESCRT-II colocalizes and interacts with viral capsid proteins 162. Although the absence of ESCRT-II does not impair the capsid assembly reaction, it selectively impairs the formation and/or stability of nuclear capsids. In cells depleted of ESCRT-II, the levels of enveloped pgRNA are significantly reduced, indicating that ESCRT-II guides steps in capsid formation that accompany replicative capacities, such as facilitating RNA transport and capsid envelopment. Stieler et al. performed knockdown experiments targeting ESCRT-II components EAP20, EAP30, and EAP45, and found that depletion of ESCRT-II components dramatically reduced virus budding. Therefore, depletion of ESCRT-II components EAP20, EAP30, and EAP45 not only inhibits the production and/or release of enveloped virus particles but also impairs the formation of intracellular nuclear capsids. HBV budding on cellular membranes also requires the involvement of ESCRT-III and the VPS4 complex in membrane scission and separation 158.
3.2.2. The Link between HSV-1 and the ESCRT System
Herpes simplex virus type 1 (HSV-1) is a structurally complex enveloped double-stranded DNA virus belonging to the Herpesviridae family. It is a human pathogen that can cause diseases such as oral herpes, keratitis, and encephalitis. Its capsid can package together with the DNA genome in the cell nucleus 165. The viral capsid undergoes primary packaging at the inner nuclear membrane 166 and then undergoes final secondary packaging within the lumen of cellular compartments in the cytoplasm 167. During this process, it utilizes its viral-encoded proteins and the cellular ESCRT mechanism to drive its envelope assembly. ESCRT-III and VPS4 are essential for HSV-1 to accomplish cytoplasmic secondary packaging and play important roles in the wrapping process of HSV-1 at the inner nuclear membrane168, 169.
Generally, viruses recruit one or more early ESCRT components to access ESCRT-III, with the most common being ALIX and the ESCRT-I complex 90. However, unlike many other members of the enveloped virus family, HSV-1 appears to not require the ESCRT-I subunit TSG101 or the protein ALIX containing the Bro1 domain for recruitment and activation of ESCRT-III. Both of them are dispensable for HSV-1 envelopment even when simultaneously depleted 169. Its structural components may directly interact with ESCRT-II to control ESCRT-III assembly. Following this, Jenna Barnes and colleagues proposed a hypothesis that due to the lack of ALIX and ESCRT-I involvement, the most likely remaining cellular proteins used by HSV-1 to assemble the ESCRT-III machinery are the ESCRT-II complex and Bro1 family members other than ALIX. Knockdown experiments targeting the critical EAP25/VPS1 subunit of ESCRT-II and the remaining known ESCRT-related Bro1 domain proteins, HD-PTP and BROX, revealed no impact on HSV-1 replication 170.
Considering the aforementioned findings, it is reasonable to hypothesize that ESCRT-III may be directly recruited by proteins present in the capsid or envelope of HSV-1. HSV-1 appears to redundantly utilize multiple cellular ESCRT components for ESCRT-III recruitment, or the virus may completely bypass the requirement for Bro1 domain proteins, ESCRT-I, and ESCRT-II. In the latter scenario, the structural proteins within HSV-1 particles, whether internal or external to the capsid, might have evolved to directly recruit and trigger assembly of ESCRT-III subunits at sites of envelope constriction and scission 171, 172. The interaction between HSV-1 and the ESCRT system may involve the participation of other components, and further investigation is needed to elucidate the specific mechanisms and their implications.
4. Conclusions
ESCRT plays a crucial role in various cellular processes, including the budding of enveloped viruses, regulation of innate immune responses, and degradation of damaged or unwanted cellular components. Although significant progress has been made in understanding the molecular mechanisms of ESCRT-mediated virus budding, much remains to be explored. For instance, the molecular mechanisms underlying the regulation of ESCRT components' expression and activity require further investigation. Moreover, the precise roles of ESCRT in virus replication and pathogenesis need to be elucidated. An enhanced understanding of how the ESCRT system is hijacked by viruses can greatly contribute to our knowledge of normal cellular physiological functions.
In the future, research on ESCRT function in virus-host interactions will provide new insights into the development of antiviral therapies. A better understanding of the ESCRT complex's role in innate immune response regulation will lead to the development of novel approaches to treat viral infections. Moreover, the identification and characterization of novel ESCRT-interacting viral proteins and their interaction mechanisms will expand our knowledge of virus-host interactions. Finally, the development of new technologies, such as single-particle cryo-electron microscopy, will allow us to determine the three-dimensional structure of ESCRT and provide a more detailed understanding of its molecular mechanisms. Overall, further research on ESCRT and its interactions with viruses will help to develop new strategies to combat viral infections.
Author Contributions
Conceptualization, S.L.H. and X.F.L.; methodology, Y.C. and C.X.W.; resources, Y.C., S.L.H. and X.F.L.; writing—original draft preparation, C.X.W. and Y.C.; writing—review and editing, S.L.H. and X.F.L.; supervision, S.L.H. and X.F.L.; funding acquisition, S.L.H. and Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation (32202767), the Earmarked Fund for China Agriculture Research System (CARS-40), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
This review did not require ethical approval.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data was created.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Johnson, D. S.; Bleck, M.; Simon, S. M. Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. Elife 2018, 7, e36221. [Google Scholar] [CrossRef]
- Zhang, L.; Ju, Y.; Chen, S.; Ren, L. Recent Progress on Exosomes in RNA Virus Infection. Viruses-Basel 2021, 13(2), 256. [Google Scholar] [CrossRef] [PubMed]
- Migliano, S. M.; Wenzel, E. M.; Stenmark, H. Biophysical and molecular mechanisms of ESCRT functions, and their implications for disease. Current Opinion in Cell Biology 2022, 75, 102062. [Google Scholar] [CrossRef]
- Rothman, J. H.; Stevens, T. H. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell 1986, 47(6), 1041–51. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, C.; Schellmann, S.; Sabovljevic, A.; Shahriari, M.; Keshavaiah, C.; Bechtold, N.; Herzog, M.; Mueller, S.; Hanisch, F.-G.; Huelskamp, M. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 2006, 133(23), 4679–4689. [Google Scholar] [CrossRef]
- Rusten, T. E.; Stenmark, H. How do ESCRT proteins control autophagy? Journal of Cell Science 2009, 122(13), 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Isono, E. ESCRT Is a Great Sealer: Non-Endosomal Function of the ESCRT Machinery in Membrane Repair and Autophagy. Plant and Cell Physiology 2021, 62(5), 766–774. [Google Scholar] [CrossRef]
- Schoneberg, J.; Lee, I.-H.; Iwasa, J. H.; Hurley, J. H. Reverse-topology membrane scission by the ESCRT proteins. Nature Reviews Molecular Cell Biology 2017, 18(1), 5–17. [Google Scholar] [CrossRef]
- Ahmed, I.; Akram, Z.; Iqbal, H. M. N.; Munn, A. L. The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview. International Journal of Biological Macromolecules 2019, 127, 1–11. [Google Scholar] [CrossRef]
- Rose, K. M.; Hirsch, V. M.; Bouamr, F. Budding of a Retrovirus: Some Assemblies Required. Viruses-Basel 2020, 12(10). [Google Scholar] [CrossRef]
- Hurley, J. H. The ESCRT complexes. Critical Reviews in Biochemistry and Molecular Biology 2010, 45(6), 463–487. [Google Scholar] [CrossRef] [PubMed]
- Asao, H.; Sasaki, Y.; Arita, T.; Tanaka, N.; Endo, K.; Kasai, H.; Takeshita, T.; Endo, Y.; Fujita, T.; Sugamura, K. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. Journal of Biological Chemistry 1997, 27(52), 32785–32791. [Google Scholar] [CrossRef]
- Prag, G.; Watson, H.; Kim, Y. C.; Beach, B. M.; Ghirlando, R.; Hummer, G.; Bonifacino, J. S.; Hurley, J. H. The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Developmental Cell 2007, 12(6), 973–986. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Sandrin, V.; Alam, S. L.; Eckert, D. M.; Gygi, S. P.; Sundquist, W. I. Identification of human MVB12 proteins as ESCRT-I Subunits that function in HIV budding. Cell Host & Microbe 2007, 2(1), 41–53. [Google Scholar] [CrossRef]
- Kaul, Z.; Chakrabarti, O. Tumor susceptibility gene 101 regulates predisposition to apoptosis via ESCRT machinery accessory proteins. Molecular Biology of the Cell 2017, 28(15), 2106–2122. [Google Scholar] [CrossRef] [PubMed]
- Alam, S. L.; Langelier, C.; Whitby, F. G.; Koirala, S.; Robinson, H.; Hill, C. P.; Sundquist, W. I. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nature Structural & Molecular Biology 2006, 13(11), 1029–1030. [Google Scholar]
- Slagsvold, T.; Aasland, R.; Hirano, S.; Bache, K. G.; Raiborg, C.; Trambaiolo, D.; Wakatsuki, S.; Stenmark, H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. Journal of Biological Chemistry 2005, 280(20), 19600–19606. [Google Scholar] [CrossRef]
- Teo, H. L.; Gill, D. J.; Sun, J.; Perisic, O.; Veprintsev, D. B.; Vallis, Y.; Emr, S. D.; Williams, R. L. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 2006, 125(1), 99–111. [Google Scholar] [CrossRef]
- Carrasquillo, R.; Tian, D.; Krishna, S.; Pollak, M. R.; Greka, A.; Schloendorff, J. SNF8, a member of the ESCRT-II complex, interacts with TRPC6 and enhances its channel activity. Bmc Cell Biology 2012, 13. [Google Scholar] [CrossRef]
- Fabrikant, G.; Lata, S.; Riches, J. D.; Briggs, J. A. G.; Weissenhorn, W.; Kozlov, M. M. Computational Model of Membrane Fission Catalyzed by ESCRT-III. Plos Computational Biology 2009, 5(11), e1000575. [Google Scholar] [CrossRef]
- Wollert, T.; Hurley, J. H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464(7290), 864–U73. [Google Scholar] [CrossRef] [PubMed]
- Harker-Kirschneck, L.; Hafner, A. E.; Yao, T.; Vanhille-Campos, C.; Jiang, X.; Pulschen, A.; Hurtig, F.; Hryniuk, D.; Culley, S.; Henriques, R.; Baum, B.; Saric, A. Physical mechanisms of ESCRT-III-driven cell division. Proceedings of the National Academy of Sciences of the United States of America 2022, 119(1), e2107763119. [Google Scholar] [CrossRef] [PubMed]
- Hurtig, F.; Burgers, T. C. Q.; Cezanne, A.; Jiang, X.; Mol, F. N.; Traparic, J.; Pulschen, A. A.; Nierhaus, T.; Tarrason-Risa, G.; Harker-Kirschneck, L.; Lowe, J.; Saric, A.; Vlijm, R.; Baum, B. The patterned assembly and stepwise Vps4-mediated disassembly of composite ESCRT-III polymers drives archaeal cell division. Science advances 2023, 9(11), eade5224–eade5224. [Google Scholar] [CrossRef]
- Li, Z.; Mo, F.; Wang, Y.; Li, W.; Chen, Y.; Liu, J.; Chen-Mayfield, T.-J.; Hu, Q. Enhancing Gasdermin-induced tumor pyroptosis through preventing ESCRT-dependent cell membrane repair augments antitumor immune response. Nature Communications 2022, 13(1), 6321. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-A.; Beigneux, A.; Ahmad, S. T.; Young, S. G.; Gao, F.-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Current Biology 2007, 17(18), 1561–1567. [Google Scholar] [CrossRef]
- Han, J.-H.; Ryu, H.-H.; Jun, M.-H.; Jang, D.-J.; Lee, J.-A. The functional analysis of the CHMP2B missense mutation associated with neurodegenerative diseases in the endo-lysosomal pathway. Biochemical and Biophysical Research Communications 2012, 421(3), 544–549. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Xia, H.; Yoshino-Koh, K.; Zhou, J.; Xu, Z. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. Journal of Molecular Biology 2007, 374(3), 655–670. [Google Scholar] [CrossRef]
- Shestakova, A.; Hanono, A.; Drosner, S.; Curtiss, M.; Davies, B. A.; Katzmann, D. J.; Babst, M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Molecular Biology of the Cell 2010, 21(6), 1059–1071. [Google Scholar] [CrossRef]
- Morita, E.; Colf, L. A.; Karren, M. A.; Sandrin, V.; Rodesch, C. K.; Sundquist, W. I. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proceedings of the National Academy of Sciences of the United States of America 2010, 107(29), 12889–12894. [Google Scholar] [CrossRef]
- Finken-Eigen, M.; Roehricht, R. A.; Koehrer, K. The VPS4 gene is involved in protein transport out of a yeast pre-vacuolar endosome-like compartment. Current Genetics 1997, 31(6), 469–480. [Google Scholar] [CrossRef]
- Migliano, S. M.; Teis, D. ESCRT and Membrane Protein Ubiquitination. In Endocytosis and Signaling; Lamaze, C., Prior, I., Eds.; 2018; Vol. 57, pp. 107–135. [Google Scholar]
- Kieffer, C.; Skalicky, J. J.; Morita, E.; De Domenico, I.; Ward, D. M.; Kaplan, J.; Sundquist, W. I. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Developmental Cell 2008, 15(1), 62–73. [Google Scholar] [CrossRef]
- Broniarczyk, J.; Pim, D.; Massimi, P.; Bergant, M.; Gozdzicka-Jozefiak, A.; Crump, C.; Banks, L. The VPS4 component of the ESCRT machinery plays an essential role in HPV infectious entry and capsid disassembly. Scientific Reports 2017, 7, 45159. [Google Scholar] [CrossRef]
- Hurley, J. H.; Hanson, P. I. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nature Reviews Molecular Cell Biology 2010, 11(8), 556–566. [Google Scholar] [CrossRef] [PubMed]
- Bissig, C.; Gruenberg, J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends in Cell Biology 2014, 24(1), 19–25. [Google Scholar] [CrossRef] [PubMed]
- Strack, B.; Calistri, A.; Craig, S.; Popova, E.; Gottlinger, H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 2003, 114(6), 689–699. [Google Scholar] [CrossRef]
- von Schwedler, U. K.; Stuchell, M.; Muller, B.; Ward, D. M.; Chung, H. Y.; Morita, E.; Wang, H. E.; Davis, T.; He, G. P.; Cimbora, D. M.; Scott, A.; Krausslich, H. G.; Kaplan, J.; Morham, S. G.; Sundquist, W. I. The protein network of HIV budding. Cell 2003, 114(6), 701–713. [Google Scholar] [CrossRef]
- Schmidt, M. H. H.; Dikic, I.; Bogler, O. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. Journal of Biological Chemistry 2005, 280(5), 3414–3425. [Google Scholar] [CrossRef] [PubMed]
- Carlton, J. G.; Agromayor, M.; Martin-Serrano, J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proceedings of the National Academy of Sciences of the United States of America 2008, 105(30), 10541–10546. [Google Scholar] [CrossRef]
- Fan, W.; Guo, J.; Gao, B.; Zhang, W.; Ling, L.; Xu, T.; Pan, C.; Li, L.; Chen, S.; Wang, H.; Zhang, J.; Wang, X. Flotillin-mediated endocytosis and ALIX-syntenin-1-mediated exocytosis protect the cell membrane from damage caused by necroptosis. Science Signaling 2019, 12(583), eaaw3423. [Google Scholar] [CrossRef]
- Xie, Q.-H.; Wang, W.-M.; Yang, J.-G.; Xia, H.-F.; Xiao, B.-L.; Chen, G.-H.; Huang, J.; Li, R.-F.; Chen, G. ALIX promotes cell migration and invasion of head and neck squamous cell carcinoma by regulating the expression of MMP9, MMP14, VEGF-C. Archives of oral biology 2023, 151, 105696–105696. [Google Scholar] [CrossRef]
- Strappazzon, F.; Torch, S.; Chatellard-Causse, C.; Petiot, A.; Thibert, C.; Blot, B.; Verna, J.-M.; Sadoul, R. Alix is involved in caspase 9 activation during calcium-induced apoptosis. Biochemical and Biophysical Research Communications 2010, 397(1), 64–69. [Google Scholar] [CrossRef] [PubMed]
- Henne, W. M.; Buchkovich, N. J.; Emr, S. D. The ESCRT Pathway. Developmental Cell 2011, 21(1), 77–91. [Google Scholar] [CrossRef]
- Moyano, S.; Musso, J.; Feliziani, C.; Zamponi, N.; Frontera, L. S.; Ropolo, A. S.; Lanfredi-Rangel, A.; Lalle, M.; Touz, M. Exosome Biogenesis in the Protozoa Parasite Giardia lamblia: A Model of Reduced Interorganellar Crosstalk. Cells 2019, 8(12), 1600. [Google Scholar] [CrossRef]
- Hoban, K.; Lux, S. Y.; Poprawski, J.; Zhang, Y.; Shepherdson, J.; Castineira, P. G.; Pesari, S.; Yao, T.; Prosser, D. C.; Norris, C.; Wendland, B. ESCRT-dependent protein sorting is required for the viability of yeast clathrin-mediated endocytosis mutants. Traffic 2020, 21(6), 430–450. [Google Scholar] [CrossRef]
- Strack, B.; Calistri, A.; Accola, M. A.; Palu, G.; Gottlinger, H. G. A role for ubiquitin ligase recruitment in retrovirus release. Proceedings of the National Academy of Sciences of the United States of America 2000, 97(24), 13063–13068. [Google Scholar] [CrossRef] [PubMed]
- Taelman, V. F.; Dobrowolski, R.; Plouhinec, J.-L.; Fuentealba, L. C.; Vorwald, P. P.; Gumper, I.; Sabatini, D. D.; De Robertis, E. M. Wnt Signaling Requires Sequestration of Glycogen Synthase Kinase 3 inside Multivesicular Endosomes. Cell 2010, 143(7), 1136–1148. [Google Scholar] [CrossRef]
- Sigismund, S.; Confalonieri, S.; Ciliberto, A.; Polo, S.; Scita, G.; Di Fiore, P. P. ENDOCYTOSIS AND SIGNALING: CELL LOGISTICS SHAPE THE EUKARYOTIC CELL PLAN. Physiological Reviews 2012, 92(1), 273–366. [Google Scholar] [CrossRef]
- Christ, L.; Wenzel, E. M.; Liestol, K.; Raiborg, C.; Campsteijn, C.; Stenmark, H. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. Journal of Cell Biology 2016, 212(5), 499–513. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-H.; Kai, H.; Carlson, L.-A.; Groves, J. T.; Hurley, J. H. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proceedings of the National Academy of Sciences of the United States of America 2015, 112(52), 15892–15897. [Google Scholar] [CrossRef] [PubMed]
- de Franceschi, N.; Alqabandi, M.; Weissenhorn, W.; Bassereau, P. Dynamic and Sequential Protein Reconstitution on Negatively Curved Membranes by Giant Vesicles Fusion. Bio-Protocol 2019, 9(13), e3294. [Google Scholar] [CrossRef]
- Bertin, A.; de Franceschi, N.; de la Mora, E.; Maity, S.; Alqabandi, M.; Miguet, N.; di Cicco, A.; Roos, W. H.; Mangenot, S.; Weissenhorn, W.; Bassereau, P. Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nature Communications 2020, 11(1), 2663. [Google Scholar] [CrossRef] [PubMed]
- Nepal, B.; Sepehri, A.; Lazaridis, T. Mechanisms of negative membrane curvature sensing and generation by ESCRT III subunit Snf7. Protein Science 2020, 29(6), 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, C.; Legouis, R.; Culetto, E. ESCRT and autophagies: Endosomal functions and beyond. Seminars in Cell & Developmental Biology 2018, 74, 21–28. [Google Scholar]
- Marshall, R. S.; Vierstra, R. D. Autophagy: The Master of Bulk and Selective Recycling. In Annual Review of Plant Biology, Vol 69; Merchant, S. S., Ed.; 2018; Vol. 69, pp. 173–208. [Google Scholar]
- Filimonenko, M.; Stuffers, S.; Raiborg, C.; Yamamoto, A.; Malerod, L.; Fisher, E. M. C.; Isaacs, A.; Brech, A.; Stenmark, H.; Simonsen, A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. The Journal of cell biology 2007, 179(3), 485–500. [Google Scholar] [CrossRef] [PubMed]
- Petiot, A.; Sadoul, R. Autophagy discriminates between Alix and ESCRTs. Autophagy 2009, 5(1), 106–107. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J. A.; Schessner, J. P.; Bircham, P. W.; Tsuji, T.; Funaya, C.; Pajonk, O.; Schaeff, K.; Ruffini, G.; Papagiannidis, D.; Knop, M.; Fujimoto, T.; Schuck, S. ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. Embo Journal 2020, 39(2), e102586. [Google Scholar] [CrossRef]
- Teis, D.; Saksena, S.; Judson, B. L.; Emr, S. D. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. Embo Journal 2010, 29(5), 871–883. [Google Scholar] [CrossRef]
- Tang, S.; Henne, W. M.; Borbat, P. P.; Buchkovich, N. J.; Freed, J. H.; Mao, Y.; Fromme, J. C.; Emr, S. D. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. Elife 2015, 4, e12548. [Google Scholar] [CrossRef]
- Stoten, C. L.; Carlton, J. G. ESCRT-dependent control of membrane remodelling during cell division. Seminars in Cell & Developmental Biology 2018, 74, 50–65. [Google Scholar]
- Yang, B.; Stjepanovic, G.; Shen, Q.; Martin, A.; Hurley, J. H. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nature Structural & Molecular Biology 2015, 22(6), 492-U88. [Google Scholar]
- Scheffer, L. L.; Sreetama, S. C.; Sharma, N.; Medikayala, S.; Brown, K. J.; Defour, A.; Jaiswal, J. K. Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nature Communications 2014, 5, 5646. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. Journal of Cell Biology 2020, 219(3), e201904113. [Google Scholar] [CrossRef]
- Christ, L.; Raiborg, C.; Wenzel, E. M.; Campsteijn, C.; Stenmark, H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends in Biochemical Sciences 2017, 42(1), 42–56. [Google Scholar] [CrossRef] [PubMed]
- Alfred, V.; Vaccari, T. When membranes need an ESCRT: endosomal sorting and membrane remodelling in health and disease. Swiss Medical Weekly 2016, 146, w14347. [Google Scholar] [CrossRef] [PubMed]
- Haguenauer-Tsapis, R. An MBoC Favorite: Morphological classification of the yeast vacuolar protein-sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Molecular Biology of the Cell 2012, 23(14), 2622–2622. [Google Scholar] [CrossRef] [PubMed]
- Katzmann, D. J.; Babst, M.; Emr, S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106(2), 145–155. [Google Scholar] [CrossRef]
- Urbe, S. Ubiquitin and endocytic protein sorting. In Essays in Biochemistry, Vol 41: The Ubiquitin-Proteasome System; Mayer, R. J., Layfield, R., Eds.; 2005; Vol. 41, pp. 81–98. [Google Scholar]
- Huotari, J.; Helenius, A. Endosome maturation. Embo Journal 2011, 30(17), 3481–3500. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, P. S.; Winistorfer, S. C.; Kearney, W. R.; Robertson, A. D.; Piper, R. C. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. Journal of Cell Biology 2003, 163(2), 237–243. [Google Scholar] [CrossRef]
- Murrow, L.; Malhotra, R.; Debnath, J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nature Cell Biology 2015, 17(3), 300–10. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458(7237), 445–452. [Google Scholar] [CrossRef]
- Babst, M.; Katzmann, D. J.; Snyder, W. B.; Wendland, B.; Emr, S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Developmental Cell 2002, 3(2), 283–289. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Michel-Hissier, P.; Boucherit, V.; Huynh, J.-R. The deubiquitinase USP8 targets ESCRT-III to promote incomplete cell division. Science 2022, 376(6595), 818–823. [Google Scholar] [CrossRef] [PubMed]
- Steigemann, P.; Gerlich, D. W. Cytokinetic abscission: cellular dynamics at the midbody. Trends in Cell Biology 2009, 19(11), 606–616. [Google Scholar] [CrossRef] [PubMed]
- Wollert, T.; Yang, D.; Ren, X.; Lee, H. H.; Im, Y. J.; Hurley, J. H. The ESCRT machinery at a glance. Journal of Cell Science 2009, 122(13), 2163–2166. [Google Scholar] [CrossRef] [PubMed]
- Wollert, T.; Wunder, C.; Lippincott-Schwartz, J.; Hurley, J. H. Membrane scission by the ESCRT-III complex. Nature 2009, 458(7235), 172–7. [Google Scholar] [CrossRef]
- Yang, D.; Rismanchi, N.; Renvoise, B.; Lippincott-Schwartz, J.; Blackstone, C.; Hurley, J. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Faseb Journal 2009, 23. [Google Scholar] [CrossRef]
- Carlton, J. G.; Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science 2007, 316(5833), 1908–1912. [Google Scholar] [CrossRef]
- Valentine, M.; Hogan, J.; Collier, S. The Drosophila Chmp1 Protein Determines Wing Cell Fate Through Regulation of Epidermal Growth Factor Receptor Signaling. Developmental Dynamics 2014, 243(8), 977–987. [Google Scholar] [CrossRef]
- Wilson, C.; Kavaler, J.; Ahmad, S. T. Expression of a human variant of CHMP2B linked to neurodegeneration in Drosophila external sensory organs leads to cell fate transformations associated with increased Notch activity. Developmental Neurobiology 2020, 80(3–4), 85–97. [Google Scholar] [CrossRef]
- Green, D. R. The Coming Decade of Cell Death Research: Five Riddles. Cell 2019, 177(5), 1094–1107. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicologic Pathology 2007, 35(4), 495–516. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, M.; Zhang, Y.-Y.; Zhao, S.-Z.; Gu, S. The endosomal sorting complex required for transport repairs the membrane to delay cell death. Frontiers in Oncology 2022, 12, 1007446. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A. J.; Maiuri, P.; Lafaurie-Janvore, J.; Divoux, S.; Piel, M.; Perez, F. ESCRT Machinery Is Required for Plasma Membrane Repair. Science 2014, 343(6174), 1247136. [Google Scholar] [CrossRef]
- Gong, Y.-N.; Guy, C.; Olauson, H.; Becker, J. U.; Yang, M.; Fitzgerald, P.; Linkermann, A.; Green, D. R. ESCRT-III Acts Downstream of MLKL to Regulate Necroptotic Cell Death and Its Consequences. Cell 2017, 169(2), 286–300. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, S.; Shkarina, K.; Demarco, B.; Heilig, R.; Santos, J. C.; Broz, P. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science 2018, 362(6417), 956–960. [Google Scholar] [CrossRef] [PubMed]
- Espiritu, R. A. Repairing plasma membrane damage in regulated necrotic cell death. Molecular Biology Reports 2021, 48(3), 2751–2759. [Google Scholar] [CrossRef]
- Votteler, J.; Sundquist, W. I. Virus Budding and the ESCRT Pathway. Cell Host & Microbe 2013, 14(3), 232–241. [Google Scholar]
- Morita, E.; Sundquist, W. I. Retrovirus budding. Annual Review of Cell and Developmental Biology 2004, 20, 395–425. [Google Scholar] [CrossRef]
- Rossman, J. S.; Jing, X.; Leser, G. P.; Lamb, R. A. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell 2010, 142(6), 902–913. [Google Scholar] [CrossRef]
- Meng, B.; Lever, A. M. L. The Interplay between ESCRT and Viral Factors in the Enveloped Virus Life Cycle. Viruses-Basel 2021, 13(2), 324. [Google Scholar] [CrossRef]
- Pentecost, M.; Vashisht, A. A.; Lester, T.; Voros, T.; Beaty, S. M.; Park, A.; Wang, Y. E.; Yun, T. E.; Freiberg, A. N.; Wohlschlegel, J. A.; Lee, B. Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Paramyxovirinae Matrix Proteins. Plos Pathogens 2015, 11(3), e1004739. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Shahriari, S.; Li, H.-M.; Ghildyal, R. Measles Virus Matrix Protein Inhibits Host Cell Transcription. Plos One 2016, 11(8), e0161360. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Hu, Z.; Zhu, J.; Xu, H.; Chen, J.; Liu, H.; Hu, S.; Liu, X. Mutations in the FPIV motif of Newcastle disease virus matrix protein attenuate virus replication and reduce virus budding. Archives of Virology 2014, 159(7), 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, X.; Cao, H.; Wang, Y.; Zheng, S. J. Engagement of new castle disease virus (NDV) matrix (M) protein with charged multivesicular body protein (CHMP) 4 facilitates viral replication. Virus Research 2013, 171(1), 80–88. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Xue, J.; Teng, Q.; Feng, D.; Huang, M.; Liang, R.; Li, X.; Zhao, Y.; Zhao, J.; Zhang, G. Mutation of Phenylalanine 23 of Newcastle Disease Virus Matrix Protein Inhibits Virus Release by Disrupting the Interaction between the FPIV L-Domain and Charged Multivesicular Body Protein 4B. Microbiology Spectrum 2023, e0411622. [Google Scholar] [CrossRef]
- Henrickson, K. J. Parainfluenza viruses. Clinical Microbiology Reviews 2003, 16(2), 242–+. [Google Scholar] [CrossRef]
- Oda, K.; Matoba, Y.; Sugiyama, M.; Sakaguchi, T. Structural Insight into the Interaction of Sendai Virus C Protein with Alix To Stimulate Viral Budding. Journal of Virology 2021, 95(19). [Google Scholar] [CrossRef]
- Kurotani, A.; Kiyotani, K.; Kato, A.; Shioda, T.; Sakai, Y.; Mizumoto, K.; Yoshida, T.; Nagai, Y. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes To Cells 1998, 3(2), 111–124. [Google Scholar] [CrossRef]
- Yan, H.; Yi, H.; Xia, L.; Zhan, Z.; He, W.; Cao, J.; Yang, P.-C.; Liu, Z. Staphylococcal enterotoxin B suppresses Alix and compromises intestinal epithelial barrier functions. Journal of Biomedical Science 2014, 21, 29. [Google Scholar] [CrossRef]
- Chen, J.-L.; Yan, H. Dicaine represses apoptosis-linked gene 2-interacting protein X expression to induce airway epithelial barrier dysfunction. Molecular Medicine Reports 2015, 12(1), 238–242. [Google Scholar] [CrossRef]
- Chen, X.; Wei, Q.; Sun, H.; Zhang, X.; Yang, C.; Tao, Y.; Nong, G. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Regulate Macrophage Polarization to Attenuate Systemic Lupus Erythematosus-Associated Diffuse Alveolar Hemorrhage in Mice. International Journal of Stem Cells 2021, 14(3), 331–340. [Google Scholar] [CrossRef] [PubMed]
- Van Cleve, W.; Amaro-Carambot, E.; Surman, S. R.; Bekisz, J.; Collins, P. L.; Zoon, K. C.; Murphy, B. R.; Skiadopoulos, M. H.; Bartlett, E. J. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology 2006, 352(1), 61–73. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, E. J.; Cruz, A.-M.; Esker, J.; Castano, A.; Schomacker, H.; Surman, S. R.; Hennessey, M.; Boonyaratanakornkit, J.; Pickles, R. J.; Collins, P. L.; Murphy, B. R.; Schmidt, A. C. Human parainfluenza virus type 1 C proteins are nonessential proteins that inhibit the host interferon and apoptotic responses and are required for efficient replication in nonhuman primates. Journal of Virology 2008, 82(18), 8965–8977. [Google Scholar] [CrossRef] [PubMed]
- Boonyaratanakornkit, J. B.; Bartlett, E. J.; Amaro-Carambot, E.; Collins, P. L.; Murphy, B. R.; Schmidt, A. C. The C Proteins of Human Parainfluenza Virus Type 1 (HPIV1) Control the Transcription of a Broad Array of Cellular Genes That Would Otherwise Respond to HPIV1 Infection. Journal of Virology 2009, 83(4), 1892–1910. [Google Scholar] [CrossRef] [PubMed]
- Andrejeva, J.; Childs, K. S.; Young, D. F.; Carlos, T. S.; Stock, N.; Goodbourn, S.; Randall, R. E. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proceedings of the National Academy of Sciences of the United States of America 2004, 101(49), 17264–17269. [Google Scholar] [CrossRef] [PubMed]
- Boonyaratanakornkit, J.; Schomacker, H.; Collins, P.; Schmidt, A. Alix Serves as an Adaptor That Allows Human Parainfluenza Virus Type 1 to Interact with the Host Cell ESCRT System. Plos One 2013, 8(3), e59462. [Google Scholar] [CrossRef]
- Patch, J. R.; Crameri, G.; Wang, L.-F.; Eaton, B. T.; Broder, C. C. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virology Journal 2007, 4, 1. [Google Scholar] [CrossRef]
- Harrison, M. S.; Sakaguchi, T.; Schmitt, A. P. Paramyxovirus assembly and budding: Building particles that transmit infections. International Journal of Biochemistry & Cell Biology 2010, 42(9), 1416–1429. [Google Scholar]
- Patch, J. R.; Han, Z.; McCarthy, S. E.; Yan, L.; Wang, L.-F.; Harty, R. N.; Broder, C. C. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virology Journal 2008, 5, 137. [Google Scholar] [CrossRef]
- Park, A.; Yun, T.; Vigant, F.; Pernet, O.; Won, S. T.; Dawes, B. E.; Bartkowski, W.; Freiberg, A. N.; Lee, B. Nipah Virus C Protein Recruits Tsg101 to Promote the Efficient Release of Virus in an ESCRT-Dependent Pathway. Plos Pathogens 2016, 12(5), e1005659. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Freed, E. O.; van Engelenburg, S. B. A Consensus View of ESCRT-Mediated Human Immunodeficiency Virus Type 1 Abscission. In Annual Review of Virology, Vol 4; Enquist, L., Ed.; 2017; Vol. 4, pp. 309–325. [Google Scholar] [CrossRef]
- Zavitz, K.; Morham, S.; Wettstein, D. A. TSG101-GAG interaction and use thereof. US 0733 5468, Feb 26 2008, 2008. [Google Scholar]
- Meng, B.; Lever, A. M. L. Wrapping up the bad news - HIV assembly and release. Retrovirology 2013, 10, 5. [Google Scholar] [CrossRef]
- Sette, P.; Nagashima, K.; Piper, R. C.; Bouamr, F. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 2013, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Pincetic, A.; Medina, G.; Carter, C.; Leis, J. Avian Sarcoma Virus and Human Immunodeficiency Virus, Type 1 Use Different Subsets of ESCRT Proteins to Facilitate the Budding Process. Journal of Biological Chemistry 2008, 283(44), 29822–29830. [Google Scholar] [CrossRef] [PubMed]
- Medina, G. N.; Ehrlich, L. S.; Chen, M. H.; Khan, M. B.; Powell, M. D.; Carter, C. A. Sprouty 2 Binds ESCRT-II Factor Eap20 and Facilitates HIV-1 Gag Release. Journal of Virology 2011, 85(14), 7353–7362. [Google Scholar] [CrossRef]
- Ghoujal, B.; Milev, M. P.; Ajamian, L.; Abel, K.; Mouland, A. J. ESCRT-II's involvement in HIV-1 genomic RNA trafficking and assembly. Biology of the Cell 2012, 104(12), 706–721. [Google Scholar] [CrossRef] [PubMed]
- Langelier, C.; von Schwedler, U. K.; Fisher, R. D.; De Domenico, I.; White, P. L.; Hill, C. P.; Kaplan, J.; Ward, D.; Sundquist, W. I. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. Journal of Virology 2006, 80(19), 9465–9480. [Google Scholar] [CrossRef]
- Baumgaertel, V.; Ivanchenko, S.; Dupont, A.; Sergeev, M.; Wiseman, P. W.; Kraeusslich, H.-G.; Braeuchle, C.; Mueller, B.; Lamb, D. C. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nature Cell Biology 2011, 13(4), 469–U277. [Google Scholar] [CrossRef]
- Cashikar, A. G.; Shim, S.; Roth, R.; Maldazys, M. R.; Heuser, J. E.; Hanson, P. I. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife 2014, 3, e02184. [Google Scholar] [CrossRef]
- Johnson, D. S.; Bleck, M.; Simon, S. M. Recruitment Dynamics of Escrt-III Proteins during HIV-1 Gag Assembly and Plasma Membrane Scission. Biophysical Journal 2019, 116(3), 373A–373A. [Google Scholar] [CrossRef]
- Janvier, K.; Pelchen-Matthews, A.; Renaud, J.-B.; Caillet, M.; Marsh, M.; Berlioz-Torrent, C. The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation. Plos Pathogens 2011, 7(2), e1001265. [Google Scholar] [CrossRef] [PubMed]
- Bache, K. G.; Brech, A.; Mehlum, A.; Stenmark, H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. Journal of Cell Biology 2003, 162(3), 435–442. [Google Scholar] [CrossRef]
- Scheel, T. K. H.; Rice, C. M. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nature Medicine 2013, 19(7), 837–849. [Google Scholar] [CrossRef] [PubMed]
- Jones, C. T.; Murray, C. L.; Eastman, D. K.; Tassello, J.; Rice, C. M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. Journal of Virology 2007, 81(16), 8374–8383. [Google Scholar] [CrossRef] [PubMed]
- Jirasko, V.; Montserret, R.; Lee, J. Y.; Gouttenoire, J.; Moradpour, D.; Penin, F.; Bartenschlager, R. Structural and Functional Studies of Nonstructural Protein 2 of the Hepatitis C Virus Reveal Its Key Role as Organizer of Virion Assembly. Plos Pathogens 2010, 6(12), e1001233. [Google Scholar] [CrossRef] [PubMed]
- Appel, N.; Zayas, M.; Miller, S.; Krijnse-Locker, J.; Schaller, T.; Friebe, P.; Kallis, S.; Engel, U.; Bartenschlager, R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. Plos Pathogens 2008, 4(3), e1000035. [Google Scholar] [CrossRef]
- Tellinghuisen, T. L.; Foss, K. L. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. Plos Pathogens 2008, 4(3), e1000032. [Google Scholar] [CrossRef]
- Shirakura, M.; Murakami, K.; Ichimura, T.; Suzuki, R.; Shimoji, T.; Fukuda, K.; Abe, K.; Sato, S.; Fukasawa, M.; Yamakawa, Y.; Nishijima, M.; Moriishi, K.; Matsuura, Y.; Wakita, T.; Suzuki, T.; Howley, P. M.; Miyamura, T.; Shoji, I. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. Journal of Virology 2007, 81(3), 1174–1185. [Google Scholar] [CrossRef]
- Welbourn, S.; Jirasko, V.; Breton, V.; Reiss, S.; Penin, F.; Bartenschlager, R.; Pause, A. Investigation of a role for lysine residues in non-structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. Journal of General Virology 2009, 90, 1071–1080. [Google Scholar] [CrossRef]
- Ariumi, Y.; Kuroki, M.; Maki, M.; Ikeda, M.; Dansako, H.; Wakita, T.; Kato, N. The ESCRT system is required for hepatitis C virus production. PloS one 2011, 6(1), e14517–e14517. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, E.; Hourioux, C.; Brand, D.; Ait-Goughoulte, M.; Moreau, A.; Trassard, S.; Sizaret, P. Y.; Dubois, F.; Roingeard, P. Hepatitis C virus-like particle budding: Role of the core protein and importance of its Asp(111). Journal of Virology 2003, 77(18), 10131–10138. [Google Scholar] [CrossRef] [PubMed]
- Corless, L.; Crump, C. M.; Griffin, S. D. C.; Harris, M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. Journal of General Virology 2010, 91, 362–372. [Google Scholar] [CrossRef]
- Barouch-Bentov, R.; Neveu, G.; Xiao, F.; Beer, M.; Bekerman, E.; Schor, S.; Campbell, J.; Boonyaratanakornkit, J.; Lindenbach, B.; Lu, A.; Jacob, Y.; Einav, S. Hepatitis C Virus Proteins Interact with the Endosomal Sorting Complex Required for Transport (ESCRT) Machinery via Ubiquitination To Facilitate Viral Envelopment. Mbio 2016, 7(6), e01456–16. [Google Scholar] [CrossRef] [PubMed]
- Tamai, K.; Shiina, M.; Tanaka, N.; Nakano, T.; Yamamoto, A.; Kondo, Y.; Kakazu, E.; Inoue, J.; Fukushima, K.; Sano, K.; Ueno, Y.; Shimosegawa, T.; Sugamura, K. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology 2012, 422(2), 377–385. [Google Scholar] [CrossRef]
- Lefkowitz, E. J.; Dempsey, D. M.; Hendrickson, R. C.; Orton, R. J.; Siddell, S. G.; Smith, D. B. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Research 2018, 46 (D1), D708–D717. [Google Scholar] [CrossRef]
- Su, C.-M.; Rowland, R. R. R.; Yoo, D. Recent Advances in PRRS Virus Receptors and the Targeting of Receptor-Ligand for Control. Vaccines 2021, 9(4), 354. [Google Scholar] [CrossRef]
- Pasternak, A. O.; Spaan, W. J. M.; Snijder, E. J. Nidovirus transcription: how to make sense... ? Journal of General Virology 2006, 87, 1403–1421. [Google Scholar] [CrossRef]
- Dokland, T. The structural biology of PRRSV. Virus Research 2010, 154(1–2), 86–97. [Google Scholar] [CrossRef]
- Liu, C.-c.; Liu, Y.-y.; Cheng, Y.; Zhang, Y.-N.; Zhang, J.; Liang, X.-D.; Gao, Y.; Chen, H.; Baloch, A. S.; Yang, Q.; Go, Y. Y.; Zhou, B. The ESCRT-I Subunit Tsg101 Plays Novel Dual Roles in Entry and Replication of Classical Swine Fever Virus. Journal of Virology 2021, 95(6), e01928–20. [Google Scholar] [CrossRef]
- Sang, Y.; Shi, J.; Sang, W.; Rowland, R. R. R.; Blecha, F. Replication-Competent Recombinant Porcine Reproductive and Respiratory Syndrome (PRRS) Viruses Expressing Indicator Proteins and Antiviral Cytokines. Viruses-Basel 2012, 4(1), 102–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, R.; Geng, R.; Wang, L.; Chen, X.-X.; Qiao, S.; Zhang, G. Tumor Susceptibility Gene 101 (TSG101) Contributes to Virion Formation of Porcine Reproductive and Respiratory Syndrome Virus via Interaction with the Nucleocapsid (N) Protein along with the Early Secretory Pathway. Journal of Virology 2022, 96(6), e0000522. [Google Scholar] [CrossRef] [PubMed]
- Timmins, J.; Scianimanico, S.; Schoehn, G.; Weissenhorn, W. Vesicular release of Ebola virus matrix protein VP40. Virology 2001, 283(1), 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bavari, S.; Bosio, C. M.; Wiegand, E.; Ruthel, G.; Will, A. B.; Geisbert, T. W.; Hevey, M.; Schmaljohn, C.; Schmaljohn, A.; Aman, M. J. Lipid raft microdomains: A gateway for compartmentalized trafficking of Ebola and Marburg viruses. Journal of Experimental Medicine 2002, 195(5), 593–602. [Google Scholar] [CrossRef]
- Panchal, R. G.; Ruthel, G.; Kenny, T. A.; Kallstrom, G. H.; Lane, D.; Badie, S. S.; Li, L. M.; Bavari, S.; Aman, M. J. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proceedings of the National Academy of Sciences of the United States of America 2003, 100(26), 15936–15941. [Google Scholar] [CrossRef] [PubMed]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P. D. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nature Medicine 2001, 7(12), 1313–1319. [Google Scholar] [CrossRef]
- Freed, E. O. Viral late domains. Journal of Virology 2002, 76(10), 4679–4687. [Google Scholar] [CrossRef]
- Garrus, J. E.; von Schwedler, U. K.; Pornillos, O. W.; Morham, S. G.; Zavitz, K. H.; Wang, H. E.; Wettstein, D. A.; Stray, K. M.; Cote, M.; Rich, R. L.; Myszka, D. G.; Sundquist, W. I. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107(1), 55–65. [Google Scholar] [CrossRef]
- Licata, J. M.; Simpson-Holley, M.; Wright, N. T.; Han, Z. Y.; Paragas, J.; Harty, R. N. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: Involvement of host proteins TSG101 and VPS-4. Journal of Virology 2003, 77(3), 1812–1819. [Google Scholar] [CrossRef]
- Lee, S.; Joshi, A.; Nagashima, K.; Freed, E. O.; Hurley, J. H. Structural basis for viral late-domain binding to Alix. Nature Structural & Molecular Biology 2007, 14(3), 194–199. [Google Scholar]
- Martin-Serrano, J.; Marsh, M. ALIX catches HIV. Cell Host & Microbe 2007, 1(1), 5–7. [Google Scholar]
- Zhai, Q.; Landesman, M. B.; Chung, H.-Y.; Dierkers, A.; Jeffries, C. M.; Trewhella, J.; Hill, C. P.; Sundquist, W. I. Activation of the Retroviral Budding Factor ALIX. Journal of Virology 2011, 85(17), 9222–9226. [Google Scholar] [CrossRef] [PubMed]
- Irie, T.; Shimazu, Y.; Yoshida, T.; Sakaguchi, T. The YLDL sequence within Sendai virus m protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. Journal of Virology 2007, 81(5), 2263–2273. [Google Scholar] [CrossRef]
- Han, Z.; Madara, J. J.; Liu, Y.; Liu, W.; Ruthel, G.; Freedman, B. D.; Harty, R. N. ALIX Rescues Budding of a Double PTAP/PPEY L-Domain Deletion Mutant of Ebola VP40: A Role for ALIX in Ebola Virus Egress. Journal of Infectious Diseases 2015, 212, S138–S145. [Google Scholar] [CrossRef]
- Stieler, J. T.; Prange, R. Involvement of ESCRT-II in Hepatitis B Virus Morphogenesis. Plos One 2014, 9(3), e91279. [Google Scholar] [CrossRef]
- Ringelhan, M.; O'Connor, T.; Protzer, U.; Heikenwalder, M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. Journal of Pathology 2015, 235(2), 355–367. [Google Scholar] [CrossRef]
- Thomas, E.; Yoneda, M.; Schiff, E. R. Viral Hepatitis: Past and Future of HBV and HDV. Cold Spring Harbor Perspectives in Medicine 2015, 5(2), a021345. [Google Scholar] [CrossRef]
- Ganem, D.; Prince, A. M. Mechanisms of disease: Hepatitis B virus infection - Natural history and clinical consequences. New England Journal of Medicine 2004, 350(11), 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Tuttleman, J. S.; Pourcel, C.; Summers, J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 1986, 47(3), 451–60. [Google Scholar] [CrossRef]
- Chou, S.-F.; Tsai, M.-L.; Huang, J.-Y.; Chang, Y.-S.; Shih, C. The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion. PLoS Pathogens 2015, 11(10), e1005123. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, M.; Li, S.; Bu, Y.; Xu, Z.; Zhu, G.; Wu, C.; Zhao, K.; Li, A.; Chen, Q.; Wang, J.; Hua, R.; Teng, Y.; Zhao, L.; Cheng, X.; Xia, Y. Hepatitis B virus hijacks TSG101 to facilitate egress via multiple vesicle bodies. Plos Pathogens 2023, 19(5), e1011382. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, Z. H. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 2018, 360(6384), eaao7298. [Google Scholar] [CrossRef]
- Bigalke, J. M.; Heldwein, E. E. Have NEC Coat, Will Travel: Structural Basis of Membrane Budding During Nuclear Egress in Herpesviruses. In Advances in Virus Research, Vol 97; Kielian, M., Mettenleiter, T. C., Roossinck, M. J., Eds.; 2017; Vol. 97, pp. 107–141. [Google Scholar]
- Hollinshead, M.; Johns, H. L.; Sayers, C. L.; Gonzalez-Lopez, C.; Smith, G. L.; Elliott, G. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. Embo Journal 2012, 31(21), 4204–4220. [Google Scholar] [CrossRef]
- Crump, C. A.; Yates, C.; Minson, T. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. Journal of Virology 2007, 81(14), 7380–7387. [Google Scholar] [CrossRef]
- Pawliczek, T.; Crump, C. M. Herpes Simplex Virus Type 1 Production Requires a Functional ESCRT-III Complex but Is Independent of TSG101 and ALIX Expression. Journal of Virology 2009, 83(21), 11254–11264. [Google Scholar] [CrossRef]
- Barnes, J.; Wilson, D. W. The ESCRT-II Subunit EAP20/VPS25 and the Bro1 Domain Proteins HD-PTP and BROX Are Individually Dispensable for Herpes Simplex Virus 1 Replication. Journal of Virology 2020, 94(4), e01641–19. [Google Scholar] [CrossRef]
- Crump, C. Virus Assembly and Egress of HSV. In Human Herpesviruses; Kawaguchi, Y., Mori, Y., Kimura, H., Eds.; 2018; Vol. 1045, pp. 23–44. [Google Scholar]
- Barnes, J.; Wilson, D. W. Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus? Journal of Virology 2019, 93(13), e00392–19. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).