Submitted:
17 July 2023
Posted:
18 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
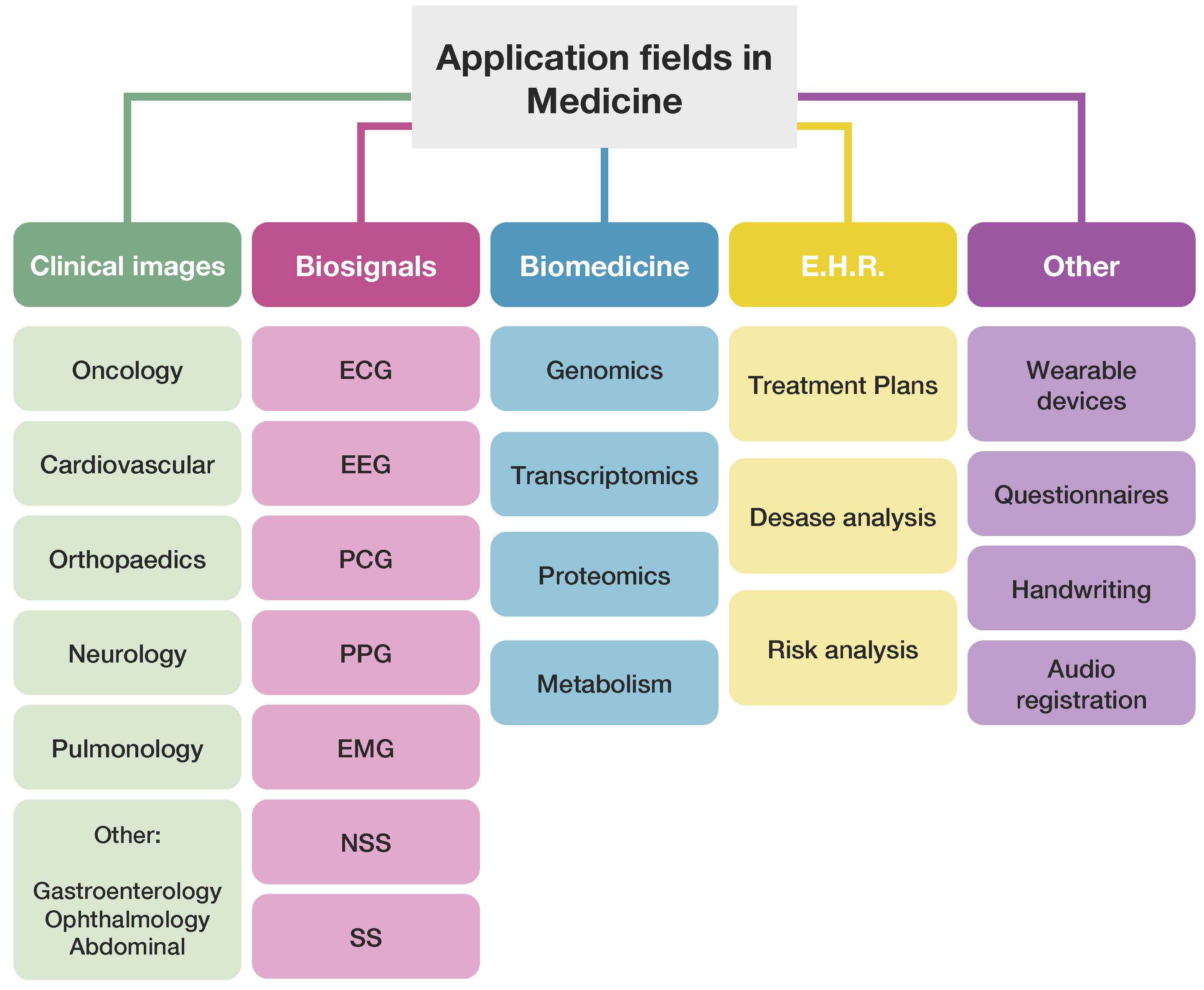
- We will review the state-of-the-art in Italy of recent years (i.e., since 2018), focusing on ML/DL in medicine, including all medical areas.
- We will present a general map of ML/DL research in Italy
- We will propose a categorization of ML/DL approaches in medicine
- We will comprehensively classify the most relevant medicine-related ML/DL applications
2. Methods
2.1. The framework
- type of paper: only research journal paper (i.e., we excluded review/survey paper and conference paper)
- subject area: we have considered only relevant subject areas in SCOPUS, i.e., Medicine, Computer Science, Engineering, Biochemistry, Genetics and Molecular Biology, Neuroscience, Pharmacology, Toxicology and Pharmaceutics, Health Profession, Nursing, Dentistry, Immunology and Microbiology, Multidisciplinary
2.2. Limitations
2.3. Analysis Criteria
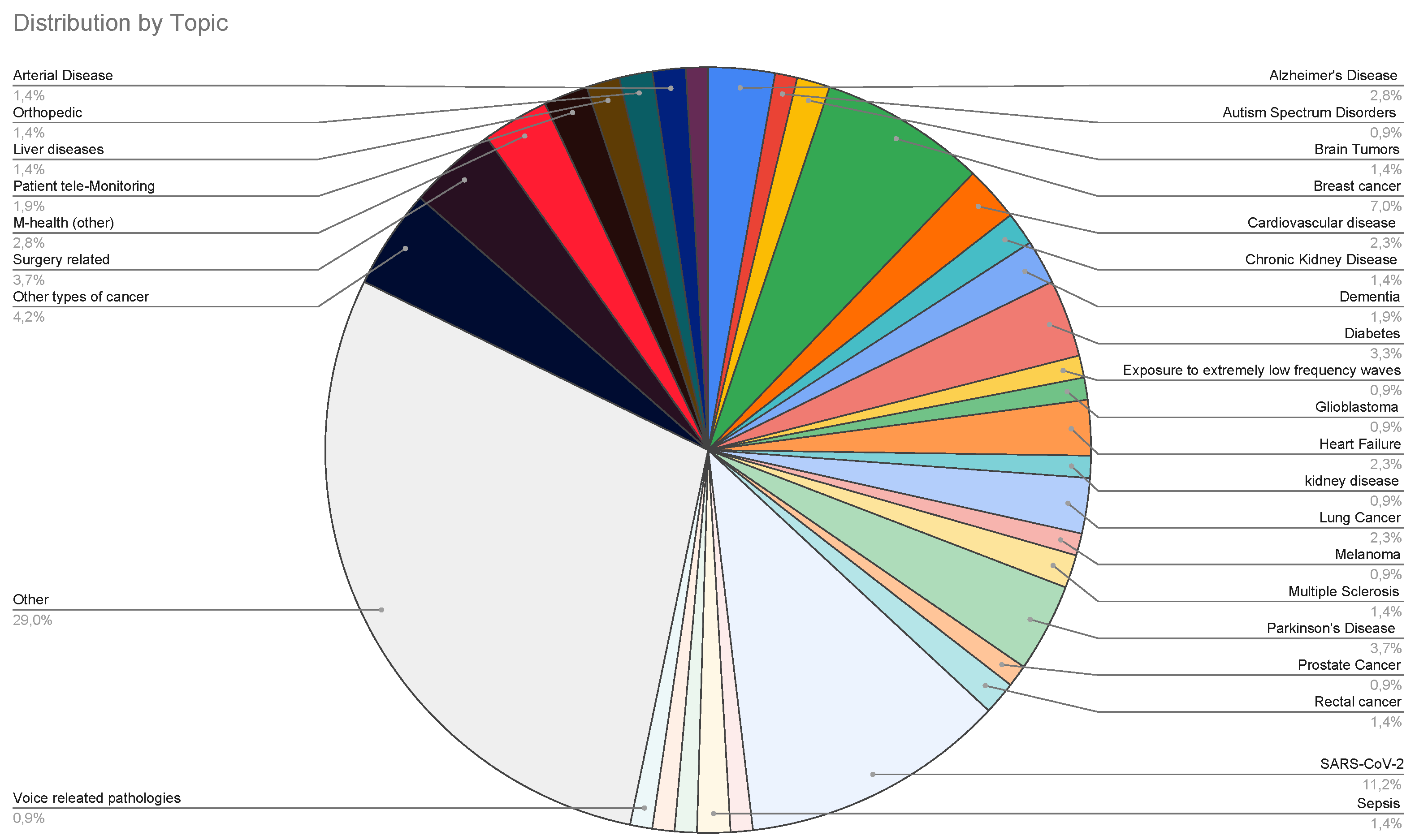
- the medical topics;
- the type of data;
- the type of preprocessing methods;
- the learning methods;
- the evaluation methods.
- feature selection;
- feature extraction;
- feature reduction;
- data filtering;
- data normalization;
- missing data management;
- undersampling;
- oversampling;
- other.
- ML supervised
- ML unsupervised
- ML semisupervised
- ML reinforcement learning
- DL supervised
- DL unsupervised
- DL semisupervised
- DL reinforcement learning
3. Results
- in Section 3.1 we provide a general analysis on all papers to provide a (simple) general snapshot of ML/DL Italian research in the medical area;
- in Section 3.2 we provide a systematic analysis of the selected papers as described in Section 2.
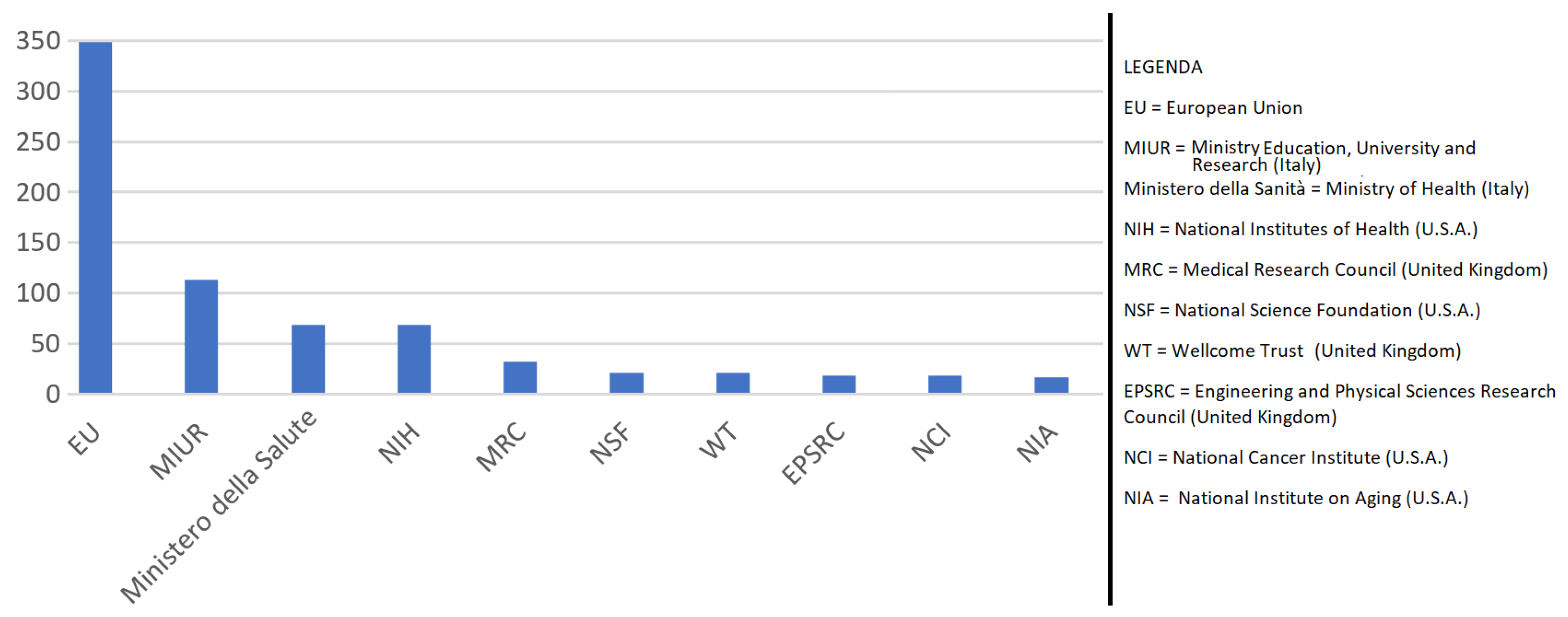
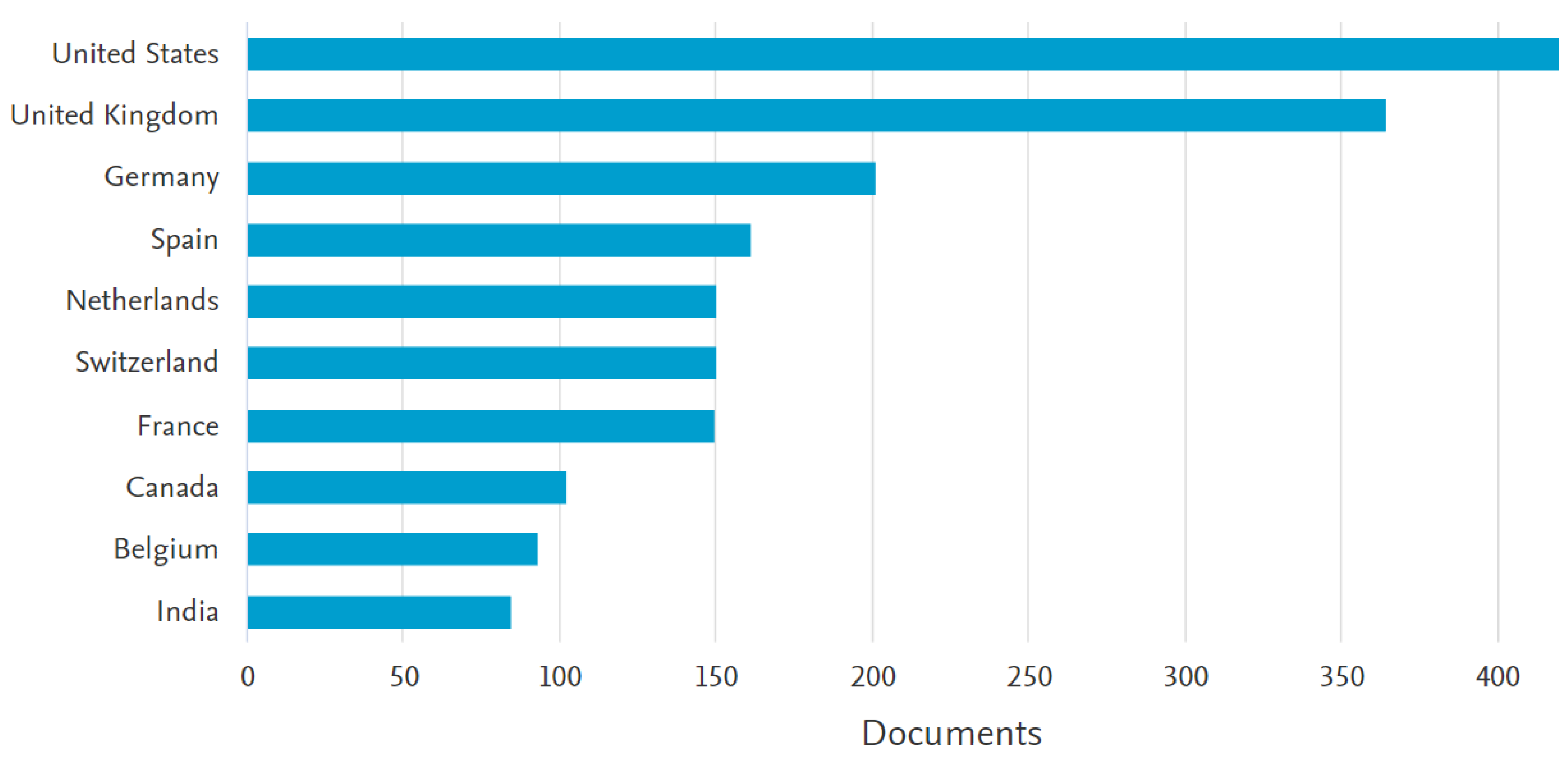
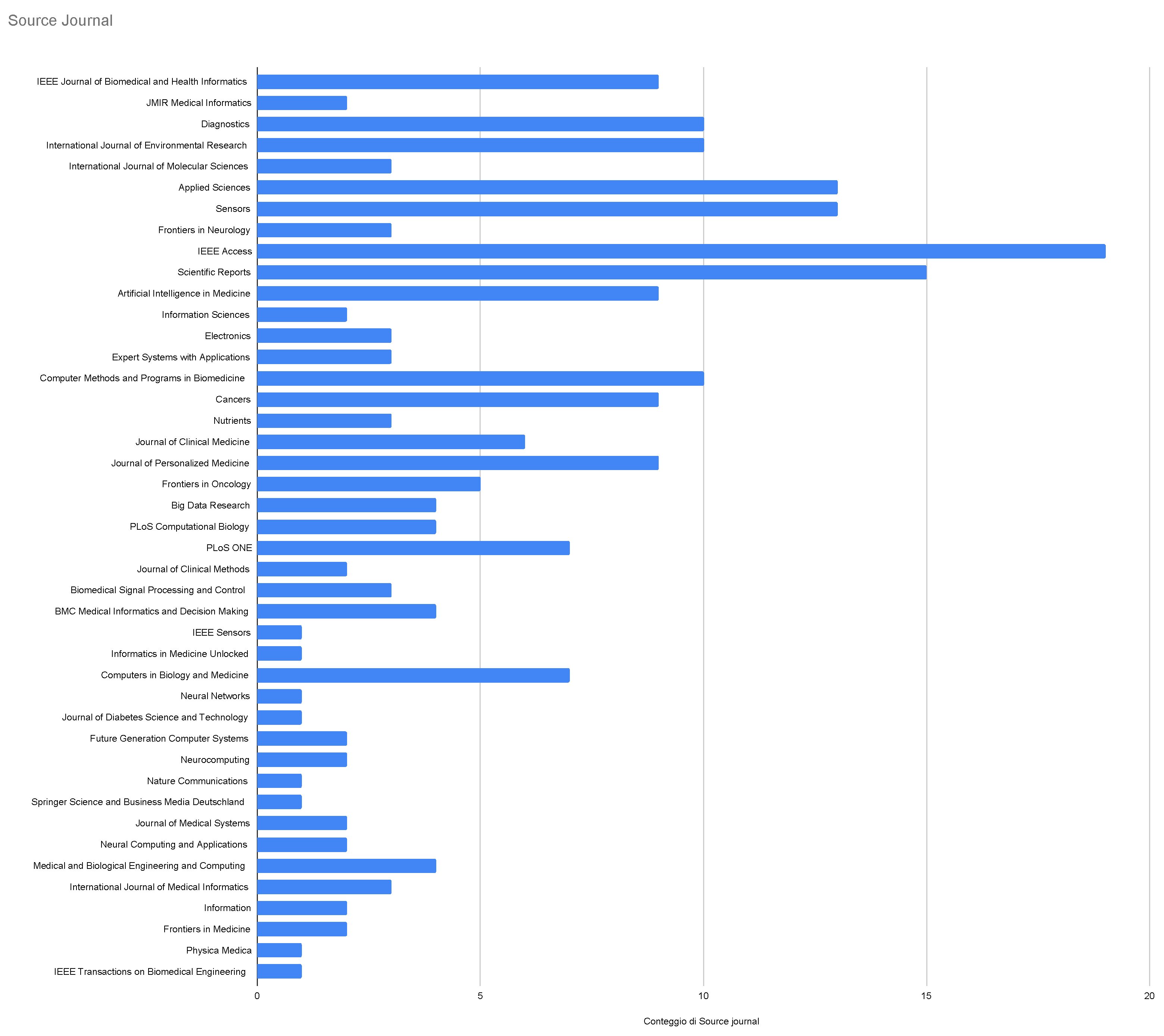
3.1. A description of Italian Machine Learning/Deep Learning research in the medical area at a glance
3.2. Systematic analysis
| Topic | Reference |
|---|---|
| Alzheimer’s disease | [41,42,43,174,191,226] |
| Autism Spectrum Disorders | [47,227] |
| %midrule Brain Tumors | [50,188,195] |
| Breast Cancer | [15,16,17,18,19,52,53,54,55,56,57,175,196,197,198] |
| Cardiovascular disease | [60,61,62,176,201] |
| Chronic Kidney Disease | [67,68,202] |
| Dementia | [77,78,79,179] |
| Diabetes | [80,81,82,83,84,85,204] |
| Exposure to extremely low frequency waves | [20,21] |
| Glioblastoma | [91,206] |
| Heart Failure | [92,93,94,182,207] |
| kidney disease | [100,228] |
| Lung Cancer | [23,103,104,105,106] |
| Melanoma | [109,110] |
| Multiple Sclerosis | [24,113,211] |
| Parkinson’s Disease | [25,123,124,125,126,127,128,129] |
| Prostate Cancer | [130,131] |
| Rectal cancer | [134,135,184] |
| SARS-CoV-2 | [27,34,35,46,143,144,145,146,147,185,186,215,216] |
| [148,149,150,151,152,153,154,155,187,193,217] | |
| Seasonal flu | [28,156] |
| Sepsis | [157,158,159] |
| Stroke | [165,219] |
| Varicella Zoster | [169,224] |
| Voice releated pathologies | [172,173] |
| Other types of cancer | [29,31,58,59,66,69,160,199,214] |
| Surgery related | [63,99,121,132,161,162,192,220] |
| M-health | [48,71,76,107,164,180] |
| Patient tele-Monitoring | [26,33,112,210] |
| Liver diseases | [89,95,102] |
| Orthopedic | [90,122,133] |
| Arterial Disease | [44,45,73] |
| Trauma | [70,168] |
| Other | [14,36,37,38,39,40,49,51,64,65,177,194,200,203] |
| [22,32,72,74,75,86,87,88,178,181,205,208] | |
| [96,97,98,101,108,111,114,115,116,117,189,190,209] | |
| [118,119,120,136,137,138,139,140,141,142,183,212,213,218] | |
| [30,163,166,167,170,171,221,222,223] |
4. Discussion
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| ML | Machine Learning |
| DL | Deep Learning |
| AUC-ROC | Area under the curve Receiver Operating Characteristic |
References
- Samuel, A.L. Some Studies in Machine Learning Using the Game of Checkers. IBM Journal 1959, 3, 535–554.
- Shortliffe, E.H. A rule-based computer program for advising physicians regarding antimicrobial therapy selection. Proceedings of the 1974 Annual ACM Conference, ACM 1974 1974, p. 739. [CrossRef]
- Piccialli, F.; Somma, V.D.; Giampaolo, F.; Cuomo, S.; Fortino, G. A survey on deep learning in medicine: Why, how and when? Information Fusion 2021, 66, 111–137. [CrossRef]
- Ferrari, E.; Retico, A.; Bacciu, D. Measuring the effects of confounders in medical supervised classification problems: the Confounding Index (CI). Artificial Intelligence in Medicine 2020, 103. cited By 5, . [CrossRef]
- Cabitza, F.; Campagner, A.; Albano, D.; Aliprandi, A.; Bruno, A.; Chianca, V.; Corazza, A.; Pietto, F.; Gambino, A.; Gitto, S.; Messina, C.; Orlandi, D.; Pedone, L.; Zappia, M.; Sconfienza, L. The elephant in the machine: Proposing a new metric of data reliability and its application to a medical case to assess classification reliability. Applied Sciences (Switzerland) 2020, 10. cited By 11, . [CrossRef]
- Cabitza, F.; Campagner, A.; Sconfienza, L. As if sand were stone. New concepts and metrics to probe the ground on which to build trustable AI. BMC Medical Informatics and Decision Making 2020, 20. cited By 18, . [CrossRef]
- Tavazzi, E.; Daberdaku, S.; Vasta, R.; Calvo, A.; Chiò, A.; Di Camillo, B. Exploiting mutual information for the imputation of static and dynamic mixed-type clinical data with an adaptive k-nearest neighbours approach. BMC Medical Informatics and Decision Making 2020, 20. cited By 10, . [CrossRef]
- Campagner, A.; Ciucci, D.; Svensson, C.M.; Figge, M.; Cabitza, F. Ground truthing from multi-rater labeling with three-way decision and possibility theory. Information Sciences 2021, 545, 771–790. cited By 25, . [CrossRef]
- Chicco, D.; Warrens, M.; Jurman, G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Computer Science 2021, 7, 1–24. cited By 414, . [CrossRef]
- La Gatta, V.; Moscato, V.; Postiglione, M.; Sperlì, G. CASTLE: Cluster-aided space transformation for local explanations. Expert Systems with Applications 2021, 179. cited By 5, . [CrossRef]
- Campagner, A.; Cabitza, F.; Berjano, P.; Ciucci, D. Three-way decision and conformal prediction: Isomorphisms, differences and theoretical properties of cautious learning approaches. Information Sciences 2021, 579, 347–367. cited By 8, . [CrossRef]
- Campagner, A.; Sternini, F.; Cabitza, F. Decisions are not all equal—Introducing a utility metric based on case-wise raters’ perceptions. Computer Methods and Programs in Biomedicine 2022, 221. cited By 0, . [CrossRef]
- Parimbelli, E.; Buonocore, T.; Nicora, G.; Michalowski, W.; Wilk, S.; Bellazzi, R. Why did AI get this one wrong? — Tree-based explanations of machine learning model predictions. Artificial Intelligence in Medicine 2023, 135. cited By 0, . [CrossRef]
- Falsetti, L.; Rucco, M.; Proietti, M.; Viticchi, G.; Zaccone, V.; Scarponi, M.; Giovenali, L.; Moroncini, G.; Nitti, C.; Salvi, A. Risk prediction of clinical adverse outcomes with machine learning in a cohort of critically ill patients with atrial fibrillation. Scientific Reports 2021, 11. cited By 4, . [CrossRef]
- Amoroso, N.; Pomarico, D.; Fanizzi, A.; Didonna, V.; Giotta, F.; La Forgia, D.; Latorre, A.; Monaco, A.; Pantaleo, E.; Petruzzellis, N.; Tamborra, P.; Zito, A.; Lorusso, V.; Bellotti, R.; Massafra, R. A roadmap towards breast cancer therapies supported by explainable artificial intelligence. Applied Sciences (Switzerland) 2021, 11. cited By 9, . [CrossRef]
- de Sire, A.; Gallelli, L.; Marotta, N.; Lippi, L.; Fusco, N.; Calafiore, D.; Cione, E.; Muraca, L.; Maconi, A.; De Sarro, G.; Ammendolia, A.; Invernizzi, M. Vitamin D Deficiency in Women with Breast Cancer: A Correlation with Osteoporosis? A Machine Learning Approach with Multiple Factor Analysis. Nutrients 2022, 14. cited By 7, . [CrossRef]
- Ferro, S.; Bottigliengo, D.; Gregori, D.; Fabricio, A.; Gion, M.; Baldi, I. Phenomapping of patients with primary breast cancer using machine learning-based unsupervised cluster analysis. Journal of Personalized Medicine 2021, 11. cited By 4, . [CrossRef]
- Militello, C.; Ranieri, A.; Rundo, L.; D’angelo, I.; Marinozzi, F.; Bartolotta, T.; Bini, F.; Russo, G. On unsupervised methods for medical image segmentation: Investigating classic approaches in breast cancer dce-mri. Applied Sciences (Switzerland) 2022, 12. cited By 5, . [CrossRef]
- Pozzoli, S.; Soliman, A.; Bahri, L.; Branca, R.; Girdzijauskas, S.; Brambilla, M. Domain expertise–agnostic feature selection for the analysis of breast cancer data*. Artificial Intelligence in Medicine 2020, 108. cited By 6, . [CrossRef]
- Tognola, G.; Bonato, M.; Chiaramello, E.; Fiocchi, S.; Magne, I.; Souques, M.; Parazzini, M.; Ravazzani, P. Use of machine learning in the analysis of indoor ELF MF exposure in children. International Journal of Environmental Research and Public Health 2019, 16. cited By 15, . [CrossRef]
- Tognola, G.; Chiaramello, E.; Bonato, M.; Magne, I.; Souques, M.; Fiocchi, S.; Parazzini, M.; Ravazzani, P. Cluster analysis of residential personal exposure to ELF magnetic field in children: Effect of environmental variables. International Journal of Environmental Research and Public Health 2019, 16. cited By 6, . [CrossRef]
- Durán, C.; Ciucci, S.; Palladini, A.; Ijaz, U.Z.; Zippo, A.G.; Sterbini, F.P.; Masucci, L.; Cammarota, G.; Ianiro, G.; Spuul, P.; Schroeder, M.; Grill, S.W.; Parsons, B.N.; Pritchard, D.M.; Posteraro, B.; Sanguinetti, M.; Gasbarrini, G.; Gasbarrini, A.; Cannistraci, C.V. Nonlinear machine learning pattern recognition and bacteria-metabolite multilayer network analysis of perturbed gastric microbiome. Nature Communications 2021 12:1 2021, 12, 1–22. [CrossRef]
- Tortora, M.; Cordelli, E.; Sicilia, R.; Miele, M.; Matteucci, P.; Iannello, G.; Ramella, S.; Soda, P. Deep Reinforcement Learning for Fractionated Radiotherapy in Non-Small Cell Lung Carcinoma. Artificial Intelligence in Medicine 2021, 119. cited By 3, . [CrossRef]
- Fiorentino, G.; Visintainer, R.; Domenici, E.; Lauria, M.; Marchetti, L. MOUSSE: Multi-omics using subject-specific signatures. Cancers 2021, 13. cited By 1, . [CrossRef]
- Termine, A.; Fabrizio, C.; Strafella, C.; Caputo, V.; Petrosini, L.; Caltagirone, C.; Cascella, R.; Giardina, E. A Hybrid Machine Learning and Network Analysis Approach Reveals Two Parkinson’s Disease Subtypes from 115 RNA-Seq Post-Mortem Brain Samples. International Journal of Molecular Sciences 2022, 23. cited By 1, . [CrossRef]
- Arpaia, P.; Crauso, F.; De Benedetto, E.; Duraccio, L.; Improta, G.; Serino, F. Soft Transducer for Patient’s Vitals Telemonitoring with Deep Learning-Based Personalized Anomaly Detection. Sensors 2022, 22. cited By 2, . [CrossRef]
- Bertacchini, F.; Bilotta, E.; Pantano, P. On the temporal spreading of the SARS-CoV-2. PLoS ONE 2020, 15. cited By 7, . [CrossRef]
- Kalimeri, K.; Delfino, M.; Cattuto, C.; Perrotta, D.; Colizza, V.; Guerrisi, C.; Turbelin, C.; Duggan, J.; Edmunds, J.; Obi, C.; Pebody, R.; Franco, A.; Moreno, Y.; Meloni, S.; Koppeschaar, C.; Kjelsø, C.; Mexia, R.; Paolotti, D. Unsupervised extraction of epidemic syndromes from participatory influenza surveillance self-reported symptoms. PLoS Computational Biology 2019, 15. cited By 11, . [CrossRef]
- Ferrari, M.; Mattavelli, D.; Schreiber, A.; Gualtieri, T.; Rampinelli, V.; Tomasoni, M.; Taboni, S.; Ardighieri, L.; Battocchio, S.; Bozzola, A.; Ravanelli, M.; Maroldi, R.; Piazza, C.; Bossi, P.; Deganello, A.; Nicolai, P. Does Reorganization of Clinicopathological Information Improve Prognostic Stratification and Prediction of Chemoradiosensitivity in Sinonasal Carcinomas? A Retrospective Study on 145 Patients. Frontiers in Oncology 2022, 12. cited By 0, . [CrossRef]
- Nobile, M.; Capitoli, G.; Sowirono, V.; Clerici, F.; Piga, I.; van Abeelen, K.; Magni, F.; Pagni, F.; Galimberti, S.; Cazzaniga, P.; Besozzi, D. Unsupervised neural networks as a support tool for pathology diagnosis in MALDI-MSI experiments: A case study on thyroid biopsies. Expert Systems with Applications 2023, 215. cited By 0, . [CrossRef]
- Di Santo, R.; Vaccaro, M.; Romanò, S.; Di Giacinto, F.; Papi, M.; Rapaccini, G.; De Spirito, M.; Miele, L.; Basile, U.; Ciasca, G. Machine Learning-Assisted FTIR Analysis of Circulating Extracellular Vesicles for Cancer Liquid Biopsy. Journal of Personalized Medicine 2022, 12. cited By 5, . [CrossRef]
- Comito, C.; Falcone, D.; Forestiero, A. AI-Driven Clinical Decision Support: Enhancing Disease Diagnosis Exploiting Patients Similarity. IEEE Access 2022, 10, 6878–6888. cited By 1, . [CrossRef]
- Monteriù, A.; Prist, M.; Frontoni, E.; Longhi, S.; Pietroni, F.; Casaccia, S.; Scalise, L.; Cenci, A.; Romeo, L.; Berta, R.; Pescosolido, L.; Orlandi, G.; Revel, G. Smart sensing architecture for domestic monitoring: Methodological approach and experimental validation. Sensors (Switzerland) 2018, 18. cited By 36, . [CrossRef]
- Biondi, R.; Curti, N.; Coppola, F.; Giampieri, E.; Vara, G.; Bartoletti, M.; Cattabriga, A.; Cocozza, M.; Ciccarese, F.; De Benedittis, C.; Cercenelli, L.; Bortolani, B.; Marcelli, E.; Pierotti, L.; Strigari, L.; Viale, P.; Golfieri, R.; Castellani, G. Classification performance for covid patient prognosis from automatic ai segmentation—a single-center study. Applied Sciences (Switzerland) 2021, 11. cited By 1, . [CrossRef]
- Bottrighi, A.; Pennisi, M.; Roveta, A.; Massarino, C.; Cassinari, A.; Betti, M.; Bolgeo, T.; Bertolotti, M.; Rava, E.; Maconi, A. A machine learning approach for predicting high risk hospitalized patients with COVID-19 SARS-Cov-2. BMC Medical Informatics and Decision Making 2022, 22. cited By 0, . [CrossRef]
- Qi, W.; Aliverti, A. A multimodal wearable system for continuous and real-time breathing pattern monitoring during daily activity. IEEE Journal of Biomedical and Health Informatics 2020, 24, 2199–2207. cited By 48, . [CrossRef]
- Tarekegn, A.; Ricceri, F.; Costa, G.; Ferracin, E.; Giacobini, M. Predictive modeling for frailty conditions in Elderly People: Machine learning approaches. JMIR Medical Informatics 2020, 8. cited By 16, . [CrossRef]
- Ciliberti, F.; Guerrini, L.; Gunnarsson, A.; Recenti, M.; Jacob, D.; Cangiano, V.; Tesfahunegn, Y.; Islind, A.; Tortorella, F.; Tsirilaki, M.; Jónsson, H., J.; Gargiulo, P.; Aubonnet, R. CT-and MRI-Based 3D Reconstruction of Knee Joint to Assess Cartilage and Bone. Diagnostics 2022, 12. cited By 7, . [CrossRef]
- Crocamo, C.; Viviani, M.; Bartoli, F.; Carrà, G.; Pasi, G. Detecting binge drinking and alcohol-related risky behaviours from twitter’s users: An exploratory content-and topology-based analysis. International Journal of Environmental Research and Public Health 2020, 17. cited By 7, . [CrossRef]
- Spiga, O.; Cicaloni, V.; Visibelli, A.; Davoli, A.; Paparo, M.; Orlandini, M.; Vecchi, B.; Santucci, A. Towards a precision medicine approach based on machine learning for tailoring medical treatment in alkaptonuria. International Journal of Molecular Sciences 2021, 22, 1–10. cited By 3, . [CrossRef]
- Alongi, P.; Laudicella, R.; Panasiti, F.; Stefano, A.; Comelli, A.; Giaccone, P.; Arnone, A.; Minutoli, F.; Quartuccio, N.; Cupidi, C.; Arnone, G.; Piccoli, T.; Grimaldi, L.; Baldari, S.; Russo, G. Radiomics Analysis of Brain [18 F]FDG PET/CT to Predict Alzheimer’s Disease in Patients with Amyloid PET Positivity: A Preliminary Report on the Application of SPM Cortical Segmentation, Pyradiomics and Machine-Learning Analysis. Diagnostics 2022, 12. cited By 4, . [CrossRef]
- Grassi, M.; Rouleaux, N.; Caldirola, D.; Loewenstein, D.; Schruers, K.; Perna, G.; Dumontier, M.; Initiative, A.D.N. A novel ensemble-based machine learning algorithm to predict the conversion from mild cognitive impairment to Alzheimer’s disease using socio-demographic characteristics, clinical information, and neuropsychological measures. Frontiers in Neurology 2019, 10. cited By 34, . [CrossRef]
- Lella, E.; Lombardi, A.; Amoroso, N.; Diacono, D.; Maggipinto, T.; Monaco, A.; Bellotti, R.; Tangaro, S. Machine learning and DWI brain communicability networks for Alzheimer’s disease detection. Applied Sciences (Switzerland) 2020, 10. cited By 14, . [CrossRef]
- Chicco, D.; Jurman, G. Arterial Disease Computational Prediction and Health Record Feature Ranking among Patients Diagnosed with Inflammatory Bowel Disease. IEEE Access 2021, 9, 78648–78657. cited By 1, . [CrossRef]
- Cavallo, A.; Troisi, J.; Muscogiuri, E.; Cavallo, P.; Rajagopalan, S.; Citro, R.; Bossone, E.; McVeigh, N.; Forte, V.; Di Donna, C.; Giannini, F.; Floris, R.; Garaci, F.; Sperandio, M. Cardiac Computed Tomography Radiomics-Based Approach for the Detection of Left Ventricular Remodeling in Patients with Arterial Hypertension. Diagnostics 2022, 12. cited By 1, . [CrossRef]
- Ferrari, D.; Milic, J.; Tonelli, R.; Ghinelli, F.; Meschiari, M.; Volpi, S.; Faltoni, M.; Franceschi, G.; Iadisernia, V.; Yaacoub, D.; Ciusa, G.; Bacca, E.; Rogati, C.; Tutone, M.; Burastero, G.; Raimondi, A.; Menozzi, M.; Franceschini, E.; Cuomo, G.; Corradi, L.; Orlando, G.; Santoro, A.; Digaetano, M.; Puzzolante, C.; Carli, F.; Borghi, V.; Bedini, A.; Fantini, R.; Tabbì, L.; Castaniere, I.; Busani, S.; Clini, E.; Girardis, M.; Sarti, M.; Cossarizza, A.; Mussini, C.; Mandreoli, F.; Missier, P.; Guaraldi, G. Machine learning in predicting respiratory failure in patients with COVID-19 pneumonia —Challenges, strengths, and opportunities in a global health emergency. PLoS ONE 2020, 15. cited By 23, . [CrossRef]
- Tartarisco, G.; Cicceri, G.; Di Pietro, D.; Leonardi, E.; Aiello, S.; Marino, F.; Chiarotti, F.; Gagliano, A.; Arduino, G.; Apicella, F.; Muratori, F.; Bruneo, D.; Allison, C.; Cohen, S.; Vagni, D.; Pioggia, G.; Ruta, L. Use of machine learning to investigate the quantitative checklist for autism in toddlers (Q-CHAT) towards early autism screening. Diagnostics 2021, 11. cited By 4, . [CrossRef]
- Kaczmarek-Majer, K.; Casalino, G.; Castellano, G.; Dominiak, M.; Hryniewicz, O.; Kamińska, O.; Vessio, G.; Díaz-Rodríguez, N. PLENARY: Explaining black-box models in natural language through fuzzy linguistic summaries. Information Sciences 2022, 614, 374–399. cited By 0, . [CrossRef]
- Delnevo, G.; Mancini, G.; Roccetti, M.; Salomoni, P.; Trombini, E.; Andrei, F. The prediction of body mass index from negative affectivity through machine learning: A confirmatory study. Sensors 2021, 21. cited By 5, . [CrossRef]
- Gonella, G.; Binaghi, E.; Nocera, P.; Mordacchini, C. Investigating the behaviour of machine learning techniques to segment brain metastases in radiation therapy planning. Applied Sciences (Switzerland) 2019, 9. cited By 8, . [CrossRef]
- Leoni, J.; Strada, S.; Tanelli, M.; Jiang, K.; Brusa, A.; Proverbio, A. Automatic stimuli classification from ERP data for augmented communication via Brain–Computer Interfaces. Expert Systems with Applications 2021, 184. cited By 4, . [CrossRef]
- Castaldo, R.; Pane, K.; Nicolai, E.; Salvatore, M.; Franzese, M. The impact of normalization approaches to automatically detect radiogenomic phenotypes characterizing breast cancer receptors status. Cancers 2020, 12. cited By 23, . [CrossRef]
- Ferrillo, M.; Migliario, M.; Marotta, N.; Lippi, L.; Antonelli, A.; Calafiore, D.; Ammendolia, V.; Fortunato, L.; Renò, F.; Giudice, A.; Invernizzi, M.; de Sire, A. Oral Health in Breast Cancer Women with Vitamin D Deficiency: A Machine Learning Study. Journal of Clinical Medicine 2022, 11. cited By 2, . [CrossRef]
- Ferroni, P.; Zanzotto, F.; Riondino, S.; Scarpato, N.; Guadagni, F.; Roselli, M. Breast cancer prognosis using a machine learning approach. Cancers 2019, 11. cited By 64, . [CrossRef]
- Gallivanone, F.; Cava, C.; Corsi, F.; Bertoli, G.; Castiglioni, I. In silico approach for the definition of radiomirnomic signatures for breast cancer differential diagnosis. International Journal of Molecular Sciences 2019, 20. cited By 10, . [CrossRef]
- Interlenghi, M.; Salvatore, C.; Magni, V.; Caldara, G.; Schiavon, E.; Cozzi, A.; Schiaffino, S.; Carbonaro, L.; Castiglioni, I.; Sardanelli, F. A Machine Learning Ensemble Based on Radiomics to Predict BI-RADS Category and Reduce the Biopsy Rate of Ultrasound-Detected Suspicious Breast Masses. Diagnostics 2022, 12. cited By 2, . [CrossRef]
- Montemezzi, S.; Benetti, G.; Bisighin, M.; Camera, L.; Zerbato, C.; Caumo, F.; Fiorio, E.; Zanelli, S.; Zuffante, M.; Cavedon, C. 3T DCE-MRI Radiomics Improves Predictive Models of Complete Response to Neoadjuvant Chemotherapy in Breast Cancer. Frontiers in Oncology 2021, 11. cited By 7, . [CrossRef]
- Zonta, G.; Malagù, C.; Gherardi, S.; Giberti, A.; Pezzoli, A.; De Togni, A.; Palmonari, C. Clinical validation results of an innovative non-invasive device for colorectal cancer preventive screening through fecal exhalation analysis. Cancers 2020, 12, 1–14. cited By 6, . [CrossRef]
- Celli, F.; Cumbo, F.; Weitschek, E. Classification of Large DNA Methylation Datasets for Identifying Cancer Drivers. Big Data Research 2018, 13, 21–28. cited By 33, . [CrossRef]
- Dutta, R.; Boudjeltia, K.; Kotsalos, C.; Rousseau, A.; de Sousa, D.; Desmet, J.M.; Van Meerhaeghe, A.; Mira, A.; Chopard, B. Personalized pathology test for Cardiovascular disease: Approximate Bayesian computation with discriminative summary statistics learning. PLoS Computational Biology 2022, 18. cited By 1, . [CrossRef]
- Mezzatesta, S.; Torino, C.; De Meo, P.; Fiumara, G.; Vilasi, A. A machine learning-based approach for predicting the outbreak of cardiovascular diseases in patients on dialysis. Computer Methods and Programs in Biomedicine 2019, 177, 9–15. cited By 60, . [CrossRef]
- Recenti, M.; Ricciardi, C.; Edmunds, K.; Gislason, M.; Gargiulo, P.; Carraro, U. Healthy Aging within an Image: Using Muscle Radiodensitometry and Lifestyle Factors to Predict Diabetes and Hypertension. IEEE Journal of Biomedical and Health Informatics 2021, 25, 2103–2112. cited By 10, . [CrossRef]
- Lanza, M.; Koprowski, R.; Boccia, R.; Ruggiero, A.; De Rosa, L.; Tortori, A.; Wilczyński, S.; Melillo, P.; Sbordone, S.; Simonelli, F. Classification tree to analyze factors connected with post operative complications of cataract surgery in a teaching hospital. Journal of Clinical Medicine 2021, 10. cited By 0, . [CrossRef]
- Danilov, V.; Skirnevskiy, I.; Manakov, R.; Gerget, O.; Melgani, F. Feature selection algorithm based on PDF/PMF area difference. Biomedical Signal Processing and Control 2020, 57. cited By 1, . [CrossRef]
- Piccialli, F.; Calabrò, F.; Crisci, D.; Cuomo, S.; Prezioso, E.; Mandile, R.; Troncone, R.; Greco, L.; Auricchio, R. Precision medicine and machine learning towards the prediction of the outcome of potential celiac disease. Scientific Reports 2021, 11. cited By 10, . [CrossRef]
- Carlini, G.; Curti, N.; Strolin, S.; Giampieri, E.; Sala, C.; Dall’olio, D.; Merlotti, A.; Fanti, S.; Remondini, D.; Nanni, C.; Strigari, L.; Castellani, G. Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features. Applied Sciences (Switzerland) 2022, 12. cited By 0, . [CrossRef]
- Chicco, D.; Lovejoy, C.; Oneto, L. A Machine Learning Analysis of Health Records of Patients with Chronic Kidney Disease at Risk of Cardiovascular Disease. IEEE Access 2021, 9, 165132–165144. cited By 0, . [CrossRef]
- Ventrella, P.; Delgrossi, G.; Ferrario, G.; Righetti, M.; Masseroli, M. Supervised machine learning for the assessment of Chronic Kidney Disease advancement. Computer Methods and Programs in Biomedicine 2021, 209. cited By 4, . [CrossRef]
- Alongi, P.; Stefano, A.; Comelli, A.; Spataro, A.; Formica, G.; Laudicella, R.; Lanzafame, H.; Panasiti, F.; Longo, C.; Midiri, F.; Benfante, V.; La Grutta, L.; Burger, I.; Bartolotta, T.; Baldari, S.; Lagalla, R.; Midiri, M.; Russo, G. Artificial Intelligence Applications on Restaging [18F]FDG PET/CT in Metastatic Colorectal Cancer: A Preliminary Report of Morpho-Functional Radiomics Classification for Prediction of Disease Outcome. Applied Sciences (Switzerland) 2022, 12. cited By 2, . [CrossRef]
- Jacob, D.; Unnsteinsdóttir Kristensen, I.; Aubonnet, R.; Recenti, M.; Donisi, L.; Ricciardi, C.; Svansson, H.; Agnarsdóttir, S.; Colacino, A.; Jónsdóttir, M.; Kristjánsdóttir, H.; Sigurjónsdóttir, H.; Cesarelli, M.; Eggertsdóttir Claessen, L.; Hassan, M.; Petersen, H.; Gargiulo, P. Towards defining biomarkers to evaluate concussions using virtual reality and a moving platform (BioVRSea). Scientific Reports 2022, 12. cited By 6, . [CrossRef]
- Liuzzi, P.; Magliacano, A.; De Bellis, F.; Mannini, A.; Estraneo, A. Predicting outcome of patients with prolonged disorders of consciousness using machine learning models based on medical complexity. Scientific Reports 2022, 12. cited By 3, . [CrossRef]
- Ocagli, H.; Lanera, C.; Lorenzoni, G.; Prosepe, I.; Azzolina, D.; Bortolotto, S.; Stivanello, L.; Degan, M.; Gregori, D. Profiling patients by intensity of nursing care: An operative approach using machine learning. Journal of Personalized Medicine 2020, 10, 1–13. cited By 1, . [CrossRef]
- Ricciardi, C.; Cantoni, V.; Improta, G.; Iuppariello, L.; Latessa, I.; Cesarelli, M.; Triassi, M.; Cuocolo, A. Application of data mining in a cohort of Italian subjects undergoing myocardial perfusion imaging at an academic medical center. Computer Methods and Programs in Biomedicine 2020, 189. cited By 38, . [CrossRef]
- Bottigliengo, D.; Berchialla, P.; Lanera, C.; Azzolina, D.; Lorenzoni, G.; Martinato, M.; Giachino, D.; Baldi, I.; Gregori, D. The role of genetic factors in characterizing extra-intestinal manifestations in Crohn’s disease patients: Are bayesian machine learning methods improving outcome predictions? Journal of Clinical Medicine 2019, 8. cited By 7, . [CrossRef]
- Delmastro, F.; Martino, F.; Dolciotti, C. Cognitive Training and Stress Detection in MCI Frail Older People through Wearable Sensors and Machine Learning. IEEE Access 2020, 8, 65573–65590. cited By 15, . [CrossRef]
- Ocagli, H.; Bottigliengo, D.; Lorenzoni, G.; Azzolina, D.; Acar, A.; Sorgato, S.; Stivanello, L.; Degan, M.; Gregori, D. A machine learning approach for investigating delirium as a multifactorial syndrome. International Journal of Environmental Research and Public Health 2021, 18. cited By 3, . [CrossRef]
- Angelillo, M.; Balducci, F.; Impedovo, D.; Pirlo, G.; Vessio, G. Attentional Pattern Classification for Automatic Dementia Detection. IEEE Access 2019, 7, 57706–57716. cited By 18, . [CrossRef]
- Battineni, G.; Chintalapudi, N.; Amenta, F. Machine learning in medicine: Performance calculation of dementia prediction by support vector machines (SVM). Informatics in Medicine Unlocked 2019, 16. cited By 106, . [CrossRef]
- Ieracitano, C.; Mammone, N.; Hussain, A.; Morabito, F. A novel multi-modal machine learning based approach for automatic classification of EEG recordings in dementia. Neural Networks 2020, 123, 176–190. cited By 131, . [CrossRef]
- Berchialla, P.; Lanera, C.; Sciannameo, V.; Gregori, D.; Baldi, I. Prediction of treatment outcome in clinical trials under a personalized medicine perspective. Scientific Reports 2022, 12. cited By 1, . [CrossRef]
- Bernardini, M.; Romeo, L.; Frontoni, E.; Amini, M.R. A Semi-Supervised Multi-Task Learning Approach for Predicting Short-Term Kidney Disease Evolution. IEEE Journal of Biomedical and Health Informatics 2021, 25, 3983–3994. cited By 3, . [CrossRef]
- Bernardini, M.; Romeo, L.; Misericordia, P.; Frontoni, E. Discovering the Type 2 Diabetes in Electronic Health Records Using the Sparse Balanced Support Vector Machine. IEEE Journal of Biomedical and Health Informatics 2020, 24, 235–246. cited By 43, . [CrossRef]
- Dagliati, A.; Marini, S.; Sacchi, L.; Cogni, G.; Teliti, M.; Tibollo, V.; De Cata, P.; Chiovato, L.; Bellazzi, R. Machine Learning Methods to Predict Diabetes Complications. Journal of Diabetes Science and Technology 2018, 12, 295–302. cited By 144, . [CrossRef]
- Fiorini, S.; Hajati, F.; Barla, A.; Girosi, F. Predicting diabetes second-line therapy initiation in the Australian population via time span-guided neural attention network. PLoS ONE 2019, 14. cited By 6, . [CrossRef]
- D’Angelo, G.; Della-Morte, D.; Pastore, D.; Donadel, G.; De Stefano, A.; Palmieri, F. Identifying patterns in multiple biomarkers to diagnose diabetic foot using an explainable genetic programming-based approach. Future Generation Computer Systems 2023, 140, 138–150. cited By 0, . [CrossRef]
- Bernardini, M.; Romeo, L.; Mancini, A.; Frontoni, E. A Clinical Decision Support System to Stratify the Temporal Risk of Diabetic Retinopathy. IEEE Access 2021, 9, 151864–151872. cited By 0, . [CrossRef]
- Peralta, R.; Garbelli, M.; Bellocchio, F.; Ponce, P.; Stuard, S.; Lodigiani, M.; Matos, J.; Ribeiro, R.; Nikam, M.; Botler, M.; Schumacher, E.; Brancaccio, D.; Neri, L. Development and validation of a machine learning model predicting arteriovenous fistula failure in a large network of dialysis clinics. International Journal of Environmental Research and Public Health 2021, 18. cited By 3, . [CrossRef]
- Amato, F.; Coppolino, L.; Cozzolino, G.; Mazzeo, G.; Moscato, F.; Nardone, R. Enhancing random forest classification with NLP in DAMEH: A system for DAta Management in eHealth Domain. Neurocomputing 2021, 444, 79–91. cited By 6, . [CrossRef]
- Sorino, P.; Caruso, M.; Misciagna, G.; Bonfiglio, C.; Campanella, A.; Mirizzi, A.; Franco, I.; Bianco, A.; Buongiorno, C.; Liuzzi, R.; Cisternino, A.; Notarnicola, M.; Chiloiro, M.; Pascoschi, G.; Osella, A.; Group, M. Selecting the best machine learning algorithm to support the diagnosis of Non-Alcoholic Fatty Liver Disease: A meta learner study. PLoS ONE 2020, 15. cited By 11, . [CrossRef]
- Scala, A.; Borrelli, A.; Improta, G. Predictive analysis of lower limb fractures in the orthopedic complex operative unit using artificial intelligence: the case study of AOU Ruggi. Scientific Reports 2022, 12. cited By 0, . [CrossRef]
- Della Pepa, G.; Caccavella, V.; Menna, G.; Ius, T.; Auricchio, A.; Chiesa, S.; Gaudino, S.; Marchese, E.; Olivi, A. Machine Learning–Based Prediction of 6-Month Postoperative Karnofsky Performance Status in Patients with Glioblastoma: Capturing the Real-Life Interaction of Multiple Clinical and Oncologic Factors. World Neurosurgery 2021, 149, e866–e876. cited By 2, . [CrossRef]
- Chicco, D.; Jurman, G. Survival prediction of patients with sepsis from age, sex, and septic episode number alone. Scientific Reports 2020, 10. cited By 13, . [CrossRef]
- Ishaq, A.; Sadiq, S.; Umer, M.; Ullah, S.; Mirjalili, S.; Rupapara, V.; Nappi, M. Improving the Prediction of Heart Failure Patients’ Survival Using SMOTE and Effective Data Mining Techniques. IEEE Access 2021, 9, 39707–39716. cited By 80, . [CrossRef]
- Lorenzoni, G.; Sabato, S.; Lanera, C.; Bottigliengo, D.; Minto, C.; Ocagli, H.; De Paolis, P.; Gregori, D.; Iliceto, S.; Pisanò, F. Comparison of machine learning techniques for prediction of hospitalization in heart failure patients. Journal of Clinical Medicine 2019, 8. cited By 19, . [CrossRef]
- Chicco, D.; Jurman, G. An ensemble learning approach for enhanced classification of patients with hepatitis and cirrhosis. IEEE Access 2021, 9, 24485–24498. cited By 10, . [CrossRef]
- Montagna, S.; Pengo, M.; Ferretti, S.; Borghi, C.; Ferri, C.; Grassi, G.; Muiesan, M.; Parati, G. Machine Learning in Hypertension Detection: A Study on World Hypertension Day Data. Journal of Medical Systems 2023, 47. cited By 0, . [CrossRef]
- Iadanza, E.; Goretti, F.; Sorelli, M.; Melillo, P.; Pecchia, L.; Simonelli, F.; Gherardelli, M. Automatic Detection of Genetic Diseases in Pediatric Age Using Pupillometry. IEEE Access 2020, 8, 34949–34961. cited By 9, . [CrossRef]
- Bernardini, M.; Morettini, M.; Romeo, L.; Frontoni, E.; Burattini, L. TyG-er: An ensemble Regression Forest approach for identification of clinical factors related to insulin resistance condition using Electronic Health Records. Computers in biology and medicine 2019, 112. [CrossRef]
- Ulivi, M.; Meroni, V.; Orlandini, L.; Prandoni, L.; Rossi, N.; Peretti, G.; Dui, L.; Mangiavini, L.; Ferrante, S. Opportunities to improve feasibility, effectiveness and costs associated with a total joint replacements high-volume hospital registry. Computers in Biology and Medicine 2020, 121. cited By 4, . [CrossRef]
- Bellocchio, F.; Carioni, P.; Lonati, C.; Garbelli, M.; Martínez-Martínez, F.; Stuard, S.; Neri, L. Enhanced sentinel surveillance system for covid-19 outbreak prediction in a large european dialysis clinics network. International Journal of Environmental Research and Public Health 2021, 18. cited By 1, . [CrossRef]
- Patrini, I.; Ruperti, M.; Moccia, S.; Mattos, L.; Frontoni, E.; De Momi, E. Transfer learning for informative-frame selection in laryngoscopic videos through learned features. Medical and Biological Engineering and Computing 2020, 58, 1225–1238. cited By 14, . [CrossRef]
- Hassoun, S.; Bruckmann, C.; Ciardullo, S.; Perseghin, G.; Di Gaudio, F.; Broccolo, F. Setting up of a machine learning algorithm for the identification of severe liver fibrosis profile in the general US population cohort. International Journal of Medical Informatics 2023, 170. cited By 0, . [CrossRef]
- D’Arnese, E.; Donato, G.W.D.; Sozzo, E.D.; Sollini, M.; Sciuto, D.; Santambrogio, M.D. On the Automation of Radiomics-Based Identification and Characterization of NSCLC. IEEE Journal of Biomedical and Health Informatics 2022, 26, 2670–2679. [CrossRef]
- Prelaj, A.; Boeri, M.; Robuschi, A.; Ferrara, R.; Proto, C.; Lo Russo, G.; Galli, G.; De Toma, A.; Brambilla, M.; Occhipinti, M.; Manglaviti, S.; Beninato, T.; Bottiglieri, A.; Massa, G.; Zattarin, E.; Gallucci, R.; Galli, E.; Ganzinelli, M.; Sozzi, G.; de Braud, F.; Garassino, M.; Restelli, M.; Pedrocchi, A.; Trovo’, F. Machine Learning Using Real-World and Translational Data to Improve Treatment Selection for NSCLC Patients Treated with Immunotherapy. Cancers 2022, 14. cited By 5, . [CrossRef]
- Rossi, D.; Dannhauser, D.; Nastri, B.; Ballini, A.; Fiorelli, A.; Santini, M.; Netti, P.; Scacco, S.; Marino, M.; Causa, F.; Boccellino, M.; Di Domenico, M. New trends in precision medicine: A pilot study of pure light scattering analysis as a useful tool for non-small cell lung cancer (nsclc) diagnosis. Journal of Personalized Medicine 2021, 11. cited By 0, . [CrossRef]
- Rundo, L.; Ledda, R.; Noia, C.; Sala, E.; Mauri, G.; Milanese, G.; Sverzellati, N.; Apolone, G.; Gilardi, M.; Messa, M.; Castiglioni, I.; Pastorino, U. A low-dose CT-based radiomic model to improve characterization and screening recall intervals of indeterminate prevalent pulmonary nodules. Diagnostics 2021, 11. cited By 4, . [CrossRef]
- Naseer Qureshi, K.; Din, S.; Jeon, G.; Piccialli, F. An accurate and dynamic predictive model for a smart M-Health system using machine learning. Information Sciences 2020, 538, 486–502. cited By 28, . [CrossRef]
- Amato, F.; Marrone, S.; Moscato, V.; Piantadosi, G.; Picariello, A.; Sansone, C. HOLMeS: eHealth in the big data and deep learning era. Information (Switzerland) 2019, 10. cited By 16, . [CrossRef]
- Cazzato, G.; Massaro, A.; Colagrande, A.; Lettini, T.; Cicco, S.; Parente, P.; Nacchiero, E.; Lospalluti, L.; Cascardi, E.; Giudice, G.; Ingravallo, G.; Resta, L.; Maiorano, E.; Vacca, A. Dermatopathology of Malignant Melanoma in the Era of Artificial Intelligence: A Single Institutional Experience. Diagnostics 2022, 12. cited By 1, . [CrossRef]
- Madonna, G.; Masucci, G.; Capone, M.; Mallardo, D.; Grimaldi, A.; Simeone, E.; Vanella, V.; Festino, L.; Palla, M.; Scarpato, L.; Tuffanelli, M.; D’angelo, G.; Villabona, L.; Krakowski, I.; Eriksson, H.; Simao, F.; Lewensohn, R.; Ascierto, P. Clinical categorization algorithm (Clical) and machine learning approach (srf-clical) to predict clinical benefit to immunotherapy in metastatic melanoma patients: Real-world evidence from the istituto nazionale tumori irccs fondazione pascale, napoli, italy. Cancers 2021, 13. cited By 2, . [CrossRef]
- Chatterjee, R.; Maitra, T.; Hafizul Islam, S.; Hassan, M.; Alamri, A.; Fortino, G. A novel machine learning based feature selection for motor imagery EEG signal classification in Internet of medical things environment. Future Generation Computer Systems 2019, 98, 419–434. cited By 57, . [CrossRef]
- Ragni, F.; Archetti, L.; Roby-Brami, A.; Amici, C.; Saint-Bauzel, L. Intention prediction and human health condition detection in reaching tasks with machine learning techniques†. Sensors 2021, 21. cited By 1, . [CrossRef]
- Seccia, R.; Gammelli, D.; Dominici, F.; Romano, S.; Landi, A.; Salvetti, M.; Tacchella, A.; Zaccaria, A.; Crisanti, A.; Grassi, F.; Palagi, L. Considering patient clinical history impacts performance of machine learning models in predicting course of multiple sclerosis. PLoS ONE 2020, 15. cited By 21, . [CrossRef]
- Raglio, A.; Imbriani, M.; Imbriani, C.; Baiardi, P.; Manzoni, S.; Gianotti, M.; Castelli, M.; Vanneschi, L.; Vico, F.; Manzoni, L. Machine learning techniques to predict the effectiveness of music therapy: A randomized controlled trial. Computer Methods and Programs in Biomedicine 2020, 185. cited By 6, . [CrossRef]
- Cicirelli, G.; D’Orazio, T. A Low-Cost Video-Based System for Neurodegenerative Disease Detection by Mobility Test Analysis. Applied Sciences (Switzerland) 2023, 13. cited By 0, . [CrossRef]
- Moro, M.; Pastore, V.; Tacchino, C.; Durand, P.; Blanchi, I.; Moretti, P.; Odone, F.; Casadio, M. A markerless pipeline to analyze spontaneous movements of preterm infants. Computer Methods and Programs in Biomedicine 2022, 226. cited By 0, . [CrossRef]
- Amina, M.; Yazdani, J.; Rovetta, S.; Masulli, F. Toward development of PreVoid alerting system for nocturnal enuresis patients: A fuzzy-based approach for determining the level of liquid encased in urinary bladder. Artificial Intelligence in Medicine 2020, 106. cited By 3, . [CrossRef]
- Veneroni, C.; Acciarito, A.; Lombardi, E.; Imeri, G.; Kaminsky, D.; Gobbi, A.; Pompilio, P.; Dellaca’, R. Artificial intelligence for quality control of oscillometry measures. Computers in Biology and Medicine 2021, 138. cited By 1, . [CrossRef]
- Masi, D.; Risi, R.; Biagi, F.; Vasquez Barahona, D.; Watanabe, M.; Zilich, R.; Gabrielli, G.; Santin, P.; Mariani, S.; Lubrano, C.; Gnessi, L. Application of a Machine Learning Technology in the Definition of Metabolically Healthy and Unhealthy Status: A Retrospective Study of 2567 Subjects Suffering from Obesity with or without Metabolic Syndrome. Nutrients 2022, 14. cited By 1, . [CrossRef]
- De Nunzio, G.; Conte, L.; Lupo, R.; Vitale, E.; Calabrò, A.; Ercolani, M.; Carvello, M.; Arigliani, M.; Toraldo, D.; De Benedetto, L. A New Berlin Questionnaire Simplified by Machine Learning Techniques in a Population of Italian Healthcare Workers to Highlight the Suspicion of Obstructive Sleep Apnea. Frontiers in Medicine 2022, 9. cited By 1, . [CrossRef]
- Muzio, F.; Rozzi, G.; Rossi, S.; Luciani, G.; Foresti, R.; Cabassi, A.; Fassina, L.; Miragoli, M. Artificial intelligence supports decision making during open-chest surgery of rare congenital heart defects. Journal of Clinical Medicine 2021, 10. cited By 4, . [CrossRef]
- Milella, F.; Famiglini, L.; Banfi, G.; Cabitza, F. Application of Machine Learning to Improve Appropriateness of Treatment in an Orthopaedic Setting of Personalized Medicine. Journal of Personalized Medicine 2022, 12. cited By 0, . [CrossRef]
- Amato, F.; Borzi, L.; Olmo, G.; Artusi, C.; Imbalzano, G.; Lopiano, L. Speech Impairment in Parkinson’s Disease: Acoustic Analysis of Unvoiced Consonants in Italian Native Speakers. IEEE Access 2021, 9, 166370–166381. cited By 2, . [CrossRef]
- Buongiorno, D.; Bortone, I.; Cascarano, G.; Trotta, G.; Brunetti, A.; Bevilacqua, V. A low-cost vision system based on the analysis of motor features for recognition and severity rating of Parkinson’s Disease. BMC Medical Informatics and Decision Making 2019, 19. cited By 31, . [CrossRef]
- Caramia, C.; Torricelli, D.; Schmid, M.; Munoz-Gonzalez, A.; Gonzalez-Vargas, J.; Grandas, F.; Pons, J. IMU-Based Classification of Parkinson’s Disease from Gait: A Sensitivity Analysis on Sensor Location and Feature Selection. IEEE Journal of Biomedical and Health Informatics 2018, 22, 1765–1774. cited By 103, . [CrossRef]
- Borzì, L.; Mazzetta, I.; Zampogna, A.; Suppa, A.; Irrera, F.; Olmo, G. Predicting Axial Impairment in Parkinson’s Disease through a Single Inertial Sensor. Sensors 2022, Vol. 22, Page 412 2022, 22, 412. [CrossRef]
- Impedovo, D.; Pirlo, G.; Vessio, G. Dynamic handwriting analysis for supporting earlier Parkinson’s disease diagnosis. Information (Switzerland) 2018, 9. cited By 38, . [CrossRef]
- Mileti, I.; Germanotta, M.; Di Sipio, E.; Imbimbo, I.; Pacilli, A.; Erra, C.; Petracca, M.; Rossi, S.; Del Prete, Z.; Bentivoglio, A.; Padua, L.; Palermo, E. Measuring gait quality in Parkinson’s disease through real-time gait phase recognition. Sensors (Switzerland) 2018, 18. cited By 32, . [CrossRef]
- Ricci, M.; Lazzaro, G.; Errico, V.; Pisani, A.; Giannini, F.; Saggio, G. The Impact of Wearable Electronics in Assessing the Effectiveness of Levodopa Treatment in Parkinson’s Disease. IEEE Journal of Biomedical and Health Informatics 2022, 26, 2920–2928. cited By 2, . [CrossRef]
- Cagni, E.; Botti, A.; Wang, Y.; Iori, M.; Petit, S.; Heijmen, B. Pareto-optimal plans as ground truth for validation of a commercial system for knowledge-based DVH-prediction. Physica Medica 2018, 55, 98–106. cited By 17, . [CrossRef]
- Gravina, M.; Spirito, L.; Celentano, G.; Capece, M.; Creta, M.; Califano, G.; Ruvolo, C.; Morra, S.; Imbriaco, M.; Di Bello, F.; Sciuto, A.; Cuocolo, R.; Napolitano, L.; La Rocca, R.; Mirone, V.; Sansone, C.; Longo, N. Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions. Diagnostics 2022, 12. cited By 2, . [CrossRef]
- Sargos, P.; Leduc, N.; Giraud, N.; Gandaglia, G.; Roumiguié, M.; Ploussard, G.; Rozet, F.; Soulié, M.; Mathieu, R.; Artus, P.; Niazi, T.; Vinh-Hung, V.; Beauval, J.B. Deep Neural Networks Outperform the CAPRA Score in Predicting Biochemical Recurrence After Prostatectomy. Frontiers in Oncology 2021, 10. cited By 5, . [CrossRef]
- Trunfio, T.; Borrelli, A.; Improta, G. Is It Possible to Predict the Length of Stay of Patients Undergoing Hip-Replacement Surgery? International Journal of Environmental Research and Public Health 2022, 19. cited By 2, . [CrossRef]
- Delli Pizzi, A.; Chiarelli, A.; Chiacchiaretta, P.; d’Annibale, M.; Croce, P.; Rosa, C.; Mastrodicasa, D.; Trebeschi, S.; Lambregts, D.; Caposiena, D.; Serafini, F.; Basilico, R.; Cocco, G.; Di Sebastiano, P.; Cinalli, S.; Ferretti, A.; Wise, R.; Genovesi, D.; Beets-Tan, R.; Caulo, M. MRI-based clinical-radiomics model predicts tumor response before treatment in locally advanced rectal cancer. Scientific Reports 2021, 11. cited By 500, . [CrossRef]
- Gatta, R.; Vallati, M.; Dinapoli, N.; Masciocchi, C.; Lenkowicz, J.; Cusumano, D.; Casá, C.; Farchione, A.; Damiani, A.; van Soest, J.; Dekker, A.; Valentini, V. Towards a modular decision support system for radiomics: A case study on rectal cancer. Artificial Intelligence in Medicine 2019, 96, 145–153. cited By 26, . [CrossRef]
- Altilio, R.; Rossetti, A.; Fang, Q.; Gu, X.; Panella, M. A comparison of machine learning classifiers for smartphone-based gait analysis. Medical and Biological Engineering and Computing 2021, 59, 535–546. cited By 5, . [CrossRef]
- Galassi, A.; Fasulo, E.; Ciceri, P.; Casazza, R.; Bonelli, F.; Zierold, C.; Calleri, M.; Blocki, F.; Palmieri, M.; Mastronardo, C.; Cozzolino, M. 1,25-dihydroxyvitamin D as Predictor of Renal Worsening Function in Chronic Kidney Disease. Results From the PASCaL-1,25D Study. Frontiers in Medicine 2022, 9. cited By 1, . [CrossRef]
- Yang, Y.; Li, Y.; Chen, R.; Zheng, J.; Cai, Y.; Fortino, G. Risk Prediction of Renal Failure for Chronic Disease Population Based on Electronic Health Record Big Data. Big Data Research 2021, 25. cited By 2, . [CrossRef]
- Mollalo, A.; Vahedi, B.; Bhattarai, S.; Hopkins, L.; Banik, S.; Vahedi, B. Predicting the hotspots of age-adjusted mortality rates of lower respiratory infection across the continental United States: Integration of GIS, spatial statistics and machine learning algorithms. International Journal of Medical Informatics 2020, 142. cited By 12, . [CrossRef]
- Monaco, A.; Amoroso, N.; Bellantuono, L.; Pantaleo, E.; Tangaro, S.; Bellotti, R. Multi-time-scale features for accurate respiratory sound classification. Applied Sciences (Switzerland) 2020, 10, 1–17. cited By 11, . [CrossRef]
- Brancati, N.; Frucci, M.; Gragnaniello, D.; Riccio, D.; Di Iorio, V.; Di Perna, L.; Simonelli, F. Learning-based approach to segment pigment signs in fundus images for Retinitis Pigmentosa analysis. Neurocomputing 2018, 308, 159–171. cited By 6, . [CrossRef]
- Leone, A.; Rescio, G.; Manni, A.; Siciliano, P.; Caroppo, A. Comparative Analysis of Supervised Classifiers for the Evaluation of Sarcopenia Using a sEMG-Based Platform. Sensors 2022, 22. cited By 4, . [CrossRef]
- Adamo, S.; Ambrosino, P.; Ricciardi, C.; Accardo, M.; Mosella, M.; Cesarelli, M.; D’addio, G.; Maniscalco, M. A Machine Learning Approach to Predict the Rehabilitation Outcome in Convalescent COVID-19 Patients. Journal of Personalized Medicine 2022, 12. cited By 1, . [CrossRef]
- Ahmed, I.; Ahmad, M.; Jeon, G.; Piccialli, F. A Framework for Pandemic Prediction Using Big Data Analytics. Big Data Research 2021, 25. cited By 34, . [CrossRef]
- Bartolucci, M.; Benelli, M.; Betti, M.; Bicchi, S.; Fedeli, L.; Giannelli, F.; Aquilini, D.; Baldini, A.; Consales, G.; Di Natale, M.; Lotti, P.; Vannucchi, L.; Trezzi, M.; Mazzoni, L.; Santini, S.; Carpi, R.; Matarrese, D.; Bernardi, L.; Mascalchi, M.; Cavigli, E.; Bindi, A.; Cozzi, D.; Miele, V.; Busoni, S.; Taddeucci, A.; Allescia, G.; Zini, C.; Dedola, G.; Mazzocchi, S.; Pozzessere, C.; Viviani, A.; the COVID Working Group. The incremental value of computed tomography of COVID-19 pneumonia in predicting ICU admission. Scientific Reports 2021, 11. cited By 4, . [CrossRef]
- Bellocchio, F.; Lonati, C.; Titapiccolo, J.; Nadal, J.; Meiselbach, H.; Schmid, M.; Baerthlein, B.; Tschulena, U.; Schneider, M.; Schultheiss, U.; Barbieri, C.; Moore, C.; Steppan, S.; Eckardt, K.U.; Stuard, S.; Neri, L. Validation of a novel predictive algorithm for kidney failure in patients suffering from chronic kidney disease: The prognostic reasoning system for chronic kidney disease (PROGRES-CKD). International Journal of Environmental Research and Public Health 2021, 18. cited By 3, . [CrossRef]
- De Falco, I.; De Pietro, G.; Sannino, G. Classification of Covid-19 chest X-ray images by means of an interpretable evolutionary rule-based approach. Neural Computing and Applications 2022. cited By 6, . [CrossRef]
- Flesia, L.; Monaro, M.; Mazza, C.; Fietta, V.; Colicino, E.; Segatto, B.; Roma, P. Predicting perceived stress related to the covid-19 outbreak through stable psychological traits and machine learning models. Journal of Clinical Medicine 2020, 9, 1–17. cited By 116, . [CrossRef]
- Giotta, M.; Trerotoli, P.; Palmieri, V.; Passerini, F.; Portincasa, P.; Dargenio, I.; Mokhtari, J.; Montagna, M.; De Vito, D. Application of a Decision Tree Model to Predict the Outcome of Non-Intensive Inpatients Hospitalized for COVID-19. International Journal of Environmental Research and Public Health 2022, 19. cited By 0, . [CrossRef]
- Gumaei, A.; Ismail, W.; Rafiul Hassan, M.; Hassan, M.; Mohamed, E.; Alelaiwi, A.; Fortino, G. A Decision-Level Fusion Method for COVID-19 Patient Health Prediction. Big Data Research 2022, 27. cited By 8, . [CrossRef]
- Iori, M.; Di Castelnuovo, C.; Verzellesi, L.; Meglioli, G.; Lippolis, D.; Nitrosi, A.; Monelli, F.; Besutti, G.; Trojani, V.; Bertolini, M.; Botti, A.; Castellani, G.; Remondini, D.; Sghedoni, R.; Croci, S.; Salvarani, C. Mortality Prediction of COVID-19 Patients Using Radiomic and Neural Network Features Extracted from a Wide Chest X-ray Sample Size: A Robust Approach for Different Medical Imbalanced Scenarios. Applied Sciences (Switzerland) 2022, 12. cited By 3, . [CrossRef]
- Loddo, A.; Meloni, G.; Pes, B. Using Artificial Intelligence for COVID-19 Detection in Blood Exams: A Comparative Analysis. IEEE Access 2022, 10, 119593–119606. cited By 0, . [CrossRef]
- Monaco, A.; Pantaleo, E.; Amoroso, N.; Bellantuono, L.; Stella, A.; Bellotti, R. Country-level factors dynamics and ABO/Rh blood groups contribution to COVID-19 mortality. Scientific Reports 2021, 11. cited By 1, . [CrossRef]
- Schiaffino, S.; Codari, M.; Cozzi, A.; Albano, D.; Alì, M.; Arioli, R.; Avola, E.; Bnà, C.; Cariati, M.; Carriero, S.; Cressoni, M.; Danna, P.; Della Pepa, G.; Di Leo, G.; Dolci, F.; Falaschi, Z.; Flor, N.; Foà, R.; Gitto, S.; Leati, G.; Magni, V.; Malavazos, A.; Mauri, G.; Messina, C.; Monfardini, L.; Paschè, A.; Pesapane, F.; Sconfienza, L.; Secchi, F.; Segalini, E.; Spinazzola, A.; Tombini, V.; Tresoldi, S.; Vanzulli, A.; Vicentin, I.; Zagaria, D.; Fleischmann, D.; Sardanelli, F. Machine learning to predict in-hospital mortality in covid-19 patients using computed tomography-derived pulmonary and vascular features. Journal of Personalized Medicine 2021, 11. cited By 9, . [CrossRef]
- Verde, L.; De Pietro, G.; Ghoneim, A.; Alrashoud, M.; Al-Mutib, K.; Sannino, G. Exploring the Use of Artificial Intelligence Techniques to Detect the Presence of Coronavirus Covid-19 through Speech and Voice Analysis. IEEE Access 2021, 9, 65750–65757. cited By 23, . [CrossRef]
- Gozzi, N.; Perrotta, D.; Paolotti, D.; Perra, N. Towards a data-driven characterization of behavioral changes induced by the seasonal flu. PLoS Computational Biology 2020, 16. cited By 11, . [CrossRef]
- Camacho-Cogollo, J.; Bonet, I.; Gil, B.; Iadanza, E. Machine Learning Models for Early Prediction of Sepsis on Large Healthcare Datasets. Electronics (Switzerland) 2022, 11. cited By 0, . [CrossRef]
- Chicco, D.; Jurman, G. Machine learning can predict survival of patients with heart failure from serum creatinine and ejection fraction alone. BMC Medical Informatics and Decision Making 2020, 20. cited By 149, . [CrossRef]
- Wernly, B.; Mamandipoor, B.; Baldia, P.; Jung, C.; Osmani, V. Machine learning predicts mortality in septic patients using only routinely available ABG variables: a multi-centre evaluation. International Journal of Medical Informatics 2021, 145. cited By 18, . [CrossRef]
- Buizza, G.; Paganelli, C.; D’ippolito, E.; Fontana, G.; Molinelli, S.; Preda, L.; Riva, G.; Iannalfi, A.; Valvo, F.; Orlandi, E.; Baroni, G. Radiomics and dosiomics for predicting local control after carbon-ion radiotherapy in skull-base chordoma. Cancers 2021, 13, 1–15. cited By 9, . [CrossRef]
- Berjano, P.; Langella, F.; Ventriglia, L.; Compagnone, D.; Barletta, P.; Huber, D.; Mangili, F.; Licandro, G.; Galbusera, F.; Cina, A.; Bassani, T.; Lamartina, C.; Scaramuzzo, L.; Bassani, R.; Brayda-Bruno, M.; Villafañe, J.; Monti, L.; Azzimonti, L. The Influence of Baseline Clinical Status and Surgical Strategy on Early Good to Excellent Result in Spinal Lumbar Arthrodesis: A Machine Learning Approach. Journal of Personalized Medicine 2021, 11. cited By 3, . [CrossRef]
- Campagner, A.; Berjano, P.; Lamartina, C.; Langella, F.; Lombardi, G.; Cabitza, F. Assessment and prediction of spine surgery invasiveness with machine learning techniques. Computers in Biology and Medicine 2020, 121. cited By 10, . [CrossRef]
- Rossi, A.; Pappalardo, L.; Cintia, P.; Iaia, F.; Fernàndez, J.; Medina, D. Effective injury forecasting in soccer with GPS training data and machine learning. PLoS ONE 2018, 13. cited By 92, . [CrossRef]
- Betti, S.; Lova, R.; Rovini, E.; Acerbi, G.; Santarelli, L.; Cabiati, M.; Ry, S.; Cavallo, F. Evaluation of an integrated system of wearable physiological sensors for stress monitoring in working environments by using biological markers. IEEE Transactions on Biomedical Engineering 2018, 65, 1748–1758. cited By 97, . [CrossRef]
- Scrutinio, D.; Ricciardi, C.; Donisi, L.; Losavio, E.; Battista, P.; Guida, P.; Cesarelli, M.; Pagano, G.; D’Addio, G. Machine learning to predict mortality after rehabilitation among patients with severe stroke. Scientific Reports 2020, 10. cited By 37, . [CrossRef]
- Martorell-Marugán, J.; Chierici, M.; Jurman, G.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. Differential diagnosis of systemic lupus erythematosus and Sjögren’s syndrome using machine learning and multi-omics data. Computers in Biology and Medicine 2023, 152, 106373. [CrossRef]
- Murdaca, G.; Caprioli, S.; Tonacci, A.; Billeci, L.; Greco, M.; Negrini, S.; Cittadini, G.; Zentilin, P.; Spagnolo, E.; Gangemi, S. A machine learning application to predict early lung involvement in scleroderma: A feasibility evaluation. Diagnostics 2021, 11. cited By 9, . [CrossRef]
- Montagna, S.; Mariani, S.; Gamberini, E. Augmenting BDI Agency with a Cognitive Service: Architecture and Validation in Healthcare Domain. Journal of Medical Systems 2021, 45. cited By 0, . [CrossRef]
- Lanera, C.; Berchialla, P.; Baldi, I.; Lorenzoni, G.; Tramontan, L.; Scamarcia, A.; Cantarutti, L.; Giaquinto, C.; Gregori, D. Use of machine learning techniques for case-detection of varicella zoster using routinely collected textual ambulatory records: Pilot observational study. JMIR Medical Informatics 2020, 8. cited By 4, . [CrossRef]
- Polignano, M.; Narducci, F.; Iovine, A.; Musto, C.; De Gemmis, M.; Semeraro, G. HealthAssistantBot: A Personal Health Assistant for the Italian Language. IEEE Access 2020, 8, 107479–107497. cited By 18, . [CrossRef]
- Ferrante, G.; Fasola, S.; Piazza, M.; Tenero, L.; Zaffanello, M.; La Grutta, S.; Piacentini, G. Vitamin D and Healthcare Service Utilization in Children: Insights from a Machine Learning Approach. Journal of Clinical Medicine 2022, 11. cited By 0, . [CrossRef]
- Verde, L.; De Pietro, G.; Sannino, G. Voice Disorder Identification by Using Machine Learning Techniques. IEEE Access 2018, 6, 16246–16255. cited By 61, . [CrossRef]
- Verde, L.; De Pietro, G.; Alrashoud, M.; Ghoneim, A.; Al-Mutib, K.; Sannino, G. Leveraging artificial intelligence to improve voice disorder identification through the use of a reliable mobile app. IEEE Access 2019, 7, 124048–124054. cited By 15, . [CrossRef]
- Redolfi, A.; De Francesco, S.; Palesi, F.; Galluzzi, S.; Muscio, C.; Castellazzi, G.; Tiraboschi, P.; Savini, G.; Nigri, A.; Bottini, G.; Bruzzone, M.; Ramusino, M.; Ferraro, S.; Gandini Wheeler-Kingshott, C.; Tagliavini, F.; Frisoni, G.; Ryvlin, P.; Demonet, J.F.; Kherif, F.; Cappa, S.; D’Angelo, E.; of Neuroscience Neurorehabilitation (RIN) Initiatives, A.H.I.N. Medical Informatics Platform (MIP): A Pilot Study Across Clinical Italian Cohorts. Frontiers in Neurology 2020, 11. cited By 5, . [CrossRef]
- Comes, M.; Forgia, D.; Didonna, V.; Fanizzi, A.; Giotta, F.; Latorre, A.; Martinelli, E.; Mencattini, A.; Paradiso, A.; Tamborra, P.; Terenzio, A.; Zito, A.; Lorusso, V.; Massafra, R. Early prediction of breast cancer recurrence for patients treated with neoadjuvant chemotherapy: A transfer learning approach on dce-mris. Cancers 2021, 13. cited By 13, . [CrossRef]
- De Marco, F.; Ferrucci, F.; Risi, M.; Tortora, G. Classification of QRS complexes to detect Premature Ventricular Contraction using machine learning techniques. PLoS ONE 2022, 17. cited By 0, . [CrossRef]
- Awais, M.; Chiari, L.; Ihlen, E.; Helbostad, J.; Palmerini, L. Classical machine learning versus deep learning for the older adults free-living activity classification. Sensors 2021, 21. cited By 4, . [CrossRef]
- Vaccari, I.; Orani, V.; Paglialonga, A.; Cambiaso, E.; Mongelli, M. A generative adversarial network (GAN) technique for internet of medical things data. Sensors 2021, 21. cited By 15, . [CrossRef]
- Di Benedetto, M.; Carrara, F.; Tafuri, B.; Nigro, S.; De Blasi, R.; Falchi, F.; Gennaro, C.; Gigli, G.; Logroscino, G.; Amato, G.; Initiative, F.L.D.N. Deep networks for behavioral variant frontotemporal dementia identification from multiple acquisition sources. Computers in Biology and Medicine 2022, 148. cited By 0, . [CrossRef]
- Wang, R.; Hao, Y.; Yu, Q.; Chen, M.; Humar, I.; Fortino, G. Depression Analysis and Recognition Based on Functional Near-Infrared Spectroscopy. IEEE Journal of Biomedical and Health Informatics 2021, 25, 4289–4299. cited By 6, . [CrossRef]
- Piccialli, F.; Cuomo, S.; Crisci, D.; Prezioso, E.; Mei, G. A deep learning approach for facility patient attendance prediction based on medical booking data. Scientific Reports 2020, 10. cited By 4, . [CrossRef]
- Umer, M.; Sadiq, S.; Karamti, H.; Karamti, W.; Majeed, R.; Nappi, M. IoT Based Smart Monitoring of Patients’ with Acute Heart Failure. Sensors 2022, 22. cited By 5, . [CrossRef]
- Gerussi, A.; Verda, D.; Cappadona, C.; Cristoferi, L.; Bernasconi, D.; Bottaro, S.; Carbone, M.; Muselli, M.; Invernizzi, P.; Asselta, R.; on behalf of The Italian PBC Genetics Study Group. LLM-PBC: Logic Learning Machine-Based Explainable Rules Accurately Stratify the Genetic Risk of Primary Biliary Cholangitis. Journal of Personalized Medicine 2022, 12. cited By 0, . [CrossRef]
- Filitto, G.; Coppola, F.; Curti, N.; Giampieri, E.; Dall’olio, D.; Merlotti, A.; Cattabriga, A.; Cocozza, M.; Taninokuchi Tomassoni, M.; Remondini, D.; Pierotti, L.; Strigari, L.; Cuicchi, D.; Guido, A.; Rihawi, K.; D’errico, A.; Di Fabio, F.; Poggioli, G.; Morganti, A.; Ricciardiello, L.; Golfieri, R.; Castellani, G. Automated Prediction of the Response to Neoadjuvant Chemoradiotherapy in Patients Affected by Rectal Cancer. Cancers 2022, 14. cited By 1, . [CrossRef]
- Chieregato, M.; Frangiamore, F.; Morassi, M.; Baresi, C.; Nici, S.; Bassetti, C.; Bnà, C.; Galelli, M. A hybrid machine learning/deep learning COVID-19 severity predictive model from CT images and clinical data. Scientific Reports 2022, 12. cited By 13, . [CrossRef]
- Fadja, A.; Fraccaroli, M.; Bizzarri, A.; Mazzuchelli, G.; Lamma, E. Neural-Symbolic Ensemble Learning for early-stage prediction of critical state of Covid-19 patients. Medical and Biological Engineering and Computing 2022, 60, 3461–3474. cited By 0, . [CrossRef]
- Sîrbu, A.; Barbieri, G.; Faita, F.; Ferragina, P.; Gargani, L.; Ghiadoni, L.; Priami, C. Early outcome detection for COVID-19 patients. Scientific Reports 2021 11:1 2021, 11, 1–14. [CrossRef]
- Donelli, M.; Espa, G.; Feraco, P. A Semi-Unsupervised Segmentation Methodology Based on Texture Recognition for Radiomics: A Preliminary Study on Brain Tumours. Electronics (Switzerland) 2022, 11. cited By 0, . [CrossRef]
- Kumar, V.; Recupero, D.; Riboni, D.; Helaoui, R. Ensembling Classical Machine Learning and Deep Learning Approaches for Morbidity Identification from Clinical Notes. IEEE Access 2021, 9, 7107–7126. cited By 32, . [CrossRef]
- Stoianov, I.; Pennartz, C.; Lansink, C.; Pezzulo, G. Model-based spatial navigation in the hippocampus-ventral striatum circuit: A computational analysis. PLoS Computational Biology 2018, 14. cited By 14, . [CrossRef]
- De Falco, I.; De Pietro, G.; Sannino, G. Article A Two-Step Approach for Classification in Alzheimer’s Disease. Sensors 2022, 22. cited By 2, . [CrossRef]
- Marzullo, A.; Moccia, S.; Catellani, M.; Calimeri, F.; Momi, E. Towards realistic image generation using image-domain translation. Computer Methods and Programs in Biomedicine 2021, 200. cited By 12, . [CrossRef]
- Sherwani, M.; Marzullo, A.; De Momi, E.; Calimeri, F. Lesion segmentation in lung CT scans using unsupervised adversarial learning. Medical and Biological Engineering and Computing 2022, 60, 3203–3215. cited By 0, . [CrossRef]
- Silvestri, S.; Gargiulo, F.; Ciampi, M. Iterative Annotation of Biomedical NER Corpora with Deep Neural Networks and Knowledge Bases. Applied Sciences (Switzerland) 2022, 12. cited By 1, . [CrossRef]
- Munir, K.; Frezza, F.; Rizzi, A. Deep Learning Hybrid Techniques for Brain Tumor Segmentation. Sensors 2022, 22. cited By 0, . [CrossRef]
- Aljuaid, H.; Alturki, N.; Alsubaie, N.; Cavallaro, L.; Liotta, A. Computer-aided diagnosis for breast cancer classification using deep neural networks and transfer learning. Computer Methods and Programs in Biomedicine 2022, 223. cited By 5, . [CrossRef]
- Costanzo, S.; Flores, A.; Buonanno, G. Machine Learning Approach to Quadratic Programming-Based Microwave Imaging for Breast Cancer Detection. Sensors 2022, 22. cited By 2, . [CrossRef]
- Famouri, S.; Morra, L.; Mangia, L.; Lamberti, F. Breast Mass Detection with Faster R-CNN: On the Feasibility of Learning from Noisy Annotations. IEEE Access 2021, 9, 66163–66175. cited By 1, . [CrossRef]
- Bruno, P.; Calimeri, F.; Kitanidis, A.; De Momi, E. Data reduction and data visualization for automatic diagnosis using gene expression and clinical data. Artificial Intelligence in Medicine 2020, 107. cited By 2, . [CrossRef]
- Piantadosi, G.; Sansone, M.; Fusco, R.; Sansone, C. Multi-planar 3D breast segmentation in MRI via deep convolutional neural networks. Artificial Intelligence in Medicine 2020, 103. cited By 33, . [CrossRef]
- Esposito, S.; Gialluisi, A.; Costanzo, S.; Di Castelnuovo, A.; Ruggiero, E.; De Curtis, A.; Persichillo, M.; Cerletti, C.; Donati, M.; de Gaetano, G.; Iacoviello, L.; Bonaccio, M. Dietary polyphenol intake is associated with biological aging, a novel predictor of cardiovascular disease: Cross-sectional findings from the moli-sani study. Nutrients 2021, 13. cited By 6, . [CrossRef]
- Cascarano, G.; Debitonto, F.; Lemma, R.; Brunetti, A.; Buongiorno, D.; De Feudis, I.; Guerriero, A.; Venere, U.; Matino, S.; Rocchetti, M.; Rossini, M.; Pesce, F.; Gesualdo, L.; Bevilacqua, V. A neural network for glomerulus classification based on histological images of kidney biopsy. BMC Medical Informatics and Decision Making 2021, 21. cited By 1, . [CrossRef]
- Ponzio, F.; Urgese, G.; Ficarra, E.; Di Cataldo, S. Dealing with lack of training data for convolutional neural networks: The case of digital pathology. Electronics (Switzerland) 2019, 8. cited By 10, . [CrossRef]
- Aliberti, A.; Pupillo, I.; Terna, S.; MacIi, E.; Di Cataldo, S.; Patti, E.; Acquaviva, A. A Multi-Patient Data-Driven Approach to Blood Glucose Prediction. IEEE Access 2019, 7, 69311–69325. cited By 52, . [CrossRef]
- Mazzoleni, M.; Previdi, F.; Bonfiglio, N. Classification algorithms analysis for brain–computer interface in drug craving therapy. Biomedical Signal Processing and Control 2019, 52, 463–472. cited By 6, . [CrossRef]
- Tariciotti, L.; Caccavella, V.; Fiore, G.; Schisano, L.; Carrabba, G.; Borsa, S.; Giordano, M.; Palmisciano, P.; Remoli, G.; Remore, L.; Pluderi, M.; Caroli, M.; Conte, G.; Triulzi, F.; Locatelli, M.; Bertani, G. A Deep Learning Model for Preoperative Differentiation of Glioblastoma, Brain Metastasis and Primary Central Nervous System Lymphoma: A Pilot Study. Frontiers in Oncology 2022, 12. cited By 5, . [CrossRef]
- Sbrollini, A.; Barocci, M.; Mancinelli, M.; Paris, M.; Raffaelli, S.; Marcantoni, I.; Morettini, M.; Swenne, C.; Burattini, L. Automatic diagnosis of newly emerged heart failure from serial electrocardiography by repeated structuring & learning procedure. Biomedical Signal Processing and Control 2023, 79. cited By 0, . [CrossRef]
- Merone, M.; Sansone, C.; Soda, P. A computer-aided diagnosis system for HEp-2 fluorescence intensity classification. Artificial Intelligence in Medicine 2019, 97, 71–78. cited By 12, . [CrossRef]
- Son, L.; Ciaramella, A.; Thu Huyen, D.; Staiano, A.; Tuan, T.; Van Hai, P. Predictive reliability and validity of hospital cost analysis with dynamic neural network and genetic algorithm. Neural Computing and Applications 2020, 32, 15237–15248. cited By 5, . [CrossRef]
- Coro, G.; Bardelli, S.; Cuttano, A.; Scaramuzzo, R.; Ciantelli, M. A self-training automatic infant-cry detector. Neural Computing and Applications 2022. cited By 0, . [CrossRef]
- Barile, B.; Marzullo, A.; Stamile, C.; Durand-Dubief, F.; Sappey-Marinier, D. Data augmentation using generative adversarial neural networks on brain structural connectivity in multiple sclerosis. Computer Methods and Programs in Biomedicine 2021, 206. cited By 17, . [CrossRef]
- de Farias, E.; di Noia, C.; Han, C.; Sala, E.; Castelli, M.; Rundo, L. Impact of GAN-based lesion-focused medical image super-resolution on the robustness of radiomic features. Scientific Reports 2021, 11. cited By 9, . [CrossRef]
- Ismail, W.; Hassan, M.; Alsalamah, H.; Fortino, G. CNN-based health model for regular health factors analysis in internet-of-medical things environment. IEEE Access 2020, 8, 52541–52549. cited By 49, . [CrossRef]
- Prezioso, E.; Izzo, S.; Giampaolo, F.; Piccialli, F.; Dell’aversana Orabona, G.; Cuocolo, R.; Abbate, V.; Ugga, L.; Califano, L. Predictive Medicine for Salivary Gland Tumours Identification Through Deep Learning. IEEE Journal of Biomedical and Health Informatics 2022, 26, 4869–4879. cited By 3, . [CrossRef]
- Ahmad, M.; Sadiq, S.; Eshmawi, A.; Alluhaidan, A.; Umer, M.; Ullah, S.; Nappi, M. Industry 4.0 technologies and their applications in fighting COVID-19 pandemic using deep learning techniques. Computers in Biology and Medicine 2022, 145. cited By 3, . [CrossRef]
- Bougourzi, F.; Contino, R.; Distante, C.; Taleb-Ahmed, A. Recognition of COVID-19 from CT scans using two-stage deep-learning-based approach: CNR-IEMN. Sensors 2021, 21. cited By 6, . [CrossRef]
- Kumar, S.; Chaube, M.; Alsamhi, S.; Gupta, S.; Guizani, M.; Gravina, R.; Fortino, G. A novel multimodal fusion framework for early diagnosis and accurate classification of COVID-19 patients using X-ray images and speech signal processing techniques. Computer Methods and Programs in Biomedicine 2022, 226. cited By 2, . [CrossRef]
- Chirikhina, E.; Chirikhin, A.; Dewsbury-Ennis, S.; Bianconi, F.; Xiao, P. Skin characterizations by using contact capacitive imaging and high-resolution ultrasound imaging with machine learning algorithms. Applied Sciences (Switzerland) 2021, 11. cited By 3, . [CrossRef]
- Iosa, M.; Capodaglio, E.; Pelà, S.; Persechino, B.; Morone, G.; Antonucci, G.; Paolucci, S.; Panigazzi, M. Artificial Neural Network Analyzing Wearable Device Gait Data for Identifying Patients With Stroke Unable to Return to Work. Frontiers in Neurology 2021, 12. cited By 11, . [CrossRef]
- Bombieri, M.; Rospocher, M.; Ponzetto, S.; Fiorini, P. Machine understanding surgical actions from intervention procedure textbooks. Computers in Biology and Medicine 2023, 152. cited By 0, . [CrossRef]
- Dipaola, F.; Gatti, M.; Pacetti, V.; Bottaccioli, A.; Shiffer, D.; Minonzio, M.; Menè, R.; Levra, A.; Solbiati, M.; Costantino, G.; Anastasio, M.; Sini, E.; Barbic, F.; Brunetta, E.; Furlan, R. Artificial intelligence algorithms and natural language processing for the recognition of syncope patients on emergency department medical records. Journal of Clinical Medicine 2019, 8. cited By 9, . [CrossRef]
- Rizzo, S.; Savastano, A.; Lenkowicz, J.; Savastano, M.; Boldrini, L.; Bacherini, D.; Falsini, B.; Valentini, V. Artificial intelligence and oct angiography in full thickness macular hole. New developments for personalized medicine. Diagnostics 2021, 11. cited By 2, . [CrossRef]
- Duong, L.; Le, N.; Tran, T.; Ngo, V.; Nguyen, P. Detection of tuberculosis from chest X-ray images: Boosting the performance with vision transformer and transfer learning. Expert Systems with Applications 2021, 184. cited By 27, . [CrossRef]
- Lanera, C.; Baldi, I.; Francavilla, A.; Barbieri, E.; Tramontan, L.; Scamarcia, A.; Cantarutti, L.; Giaquinto, C.; Gregori, D. A Deep Learning Approach to Estimate the Incidence of Infectious Disease Cases for Routinely Collected Ambulatory Records: The Example of Varicella-Zoster. International Journal of Environmental Research and Public Health 2022, 19. cited By 0, . [CrossRef]
- Europe—canceratlas.cancer.org. Available online: https://canceratlas.cancer.org/the-burden/europe/ (accessed on 19 June 2023).
- Dachena, C.; Casu, S.; Fanti, A.; Lodi, M.; Mazzarella, G. Combined use of MRI, fMRIand cognitive data for Alzheimer’s Disease: Preliminary results. Applied Sciences (Switzerland) 2019, 9. cited By 16, . [CrossRef]
- Ferrari, E.; Bosco, P.; Calderoni, S.; Oliva, P.; Palumbo, L.; Spera, G.; Fantacci, M.; Retico, A. Dealing with confounders and outliers in classification medical studies: The Autism Spectrum Disorders case study. Artificial Intelligence in Medicine 2020, 108. cited By 10, . [CrossRef]
- Bernardini, M.; Romeo, L.; Frontoni, E.; Amini, M.R. A Semi-Supervised Multi-Task Learning Approach for Predicting Short-Term Kidney Disease Evolution. IEEE journal of biomedical and health informatics 2021, 25, 3983–3994. [CrossRef]
| 1 | All the queries have been performed on the 13th January 2023 |
| 2 | |
| 3 | see also the discussion in Section 3.1 about international funding and international co-authoring. |
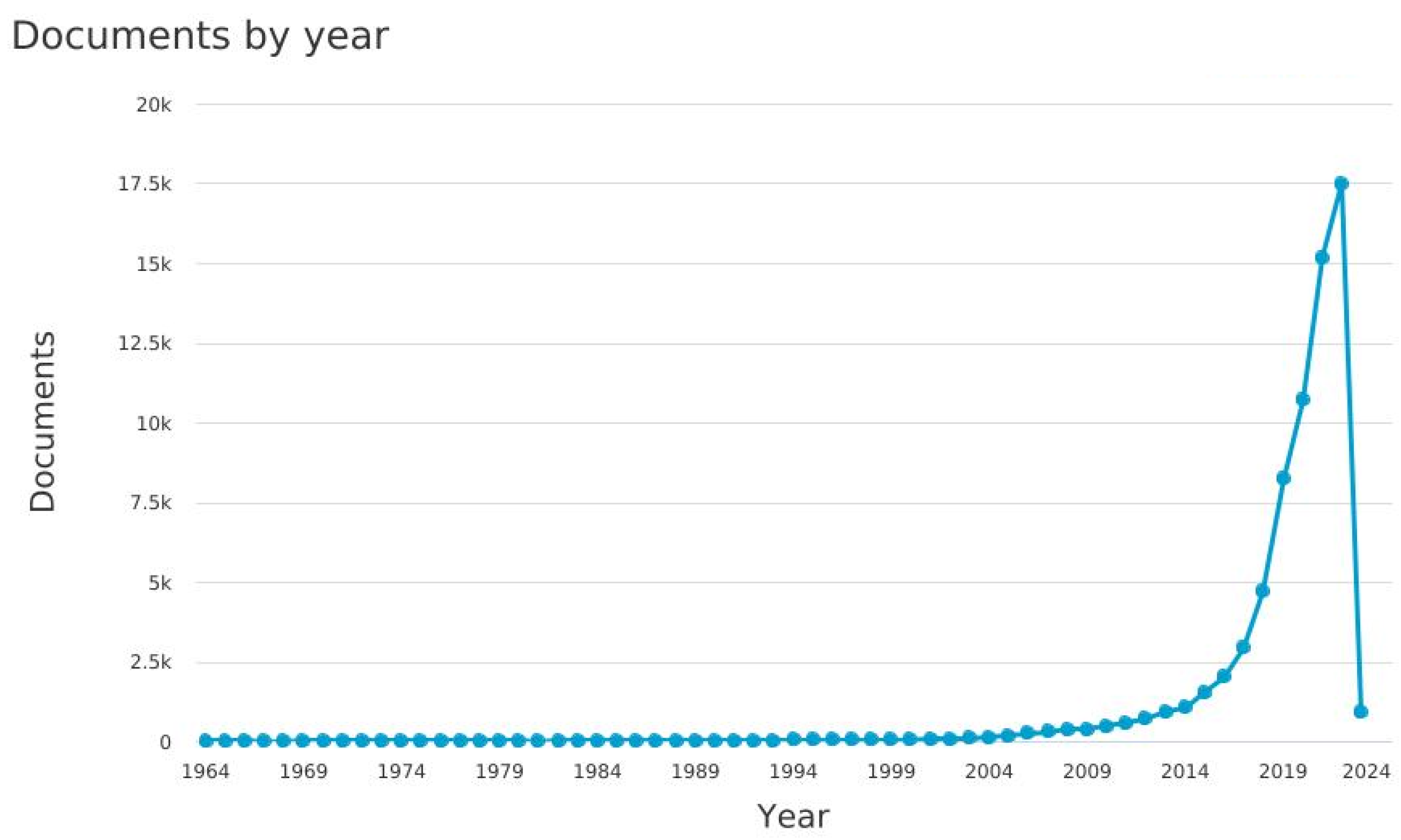
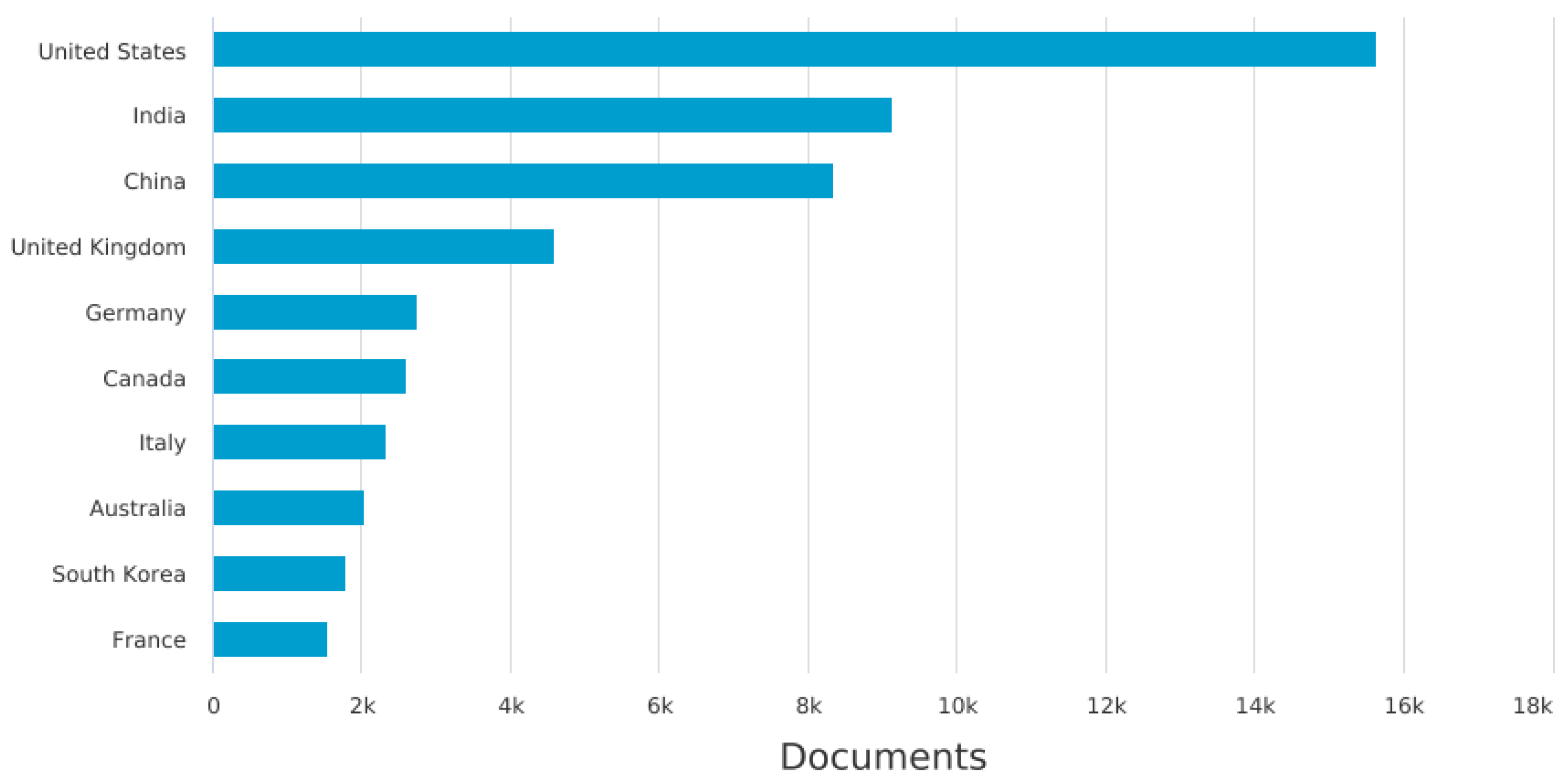

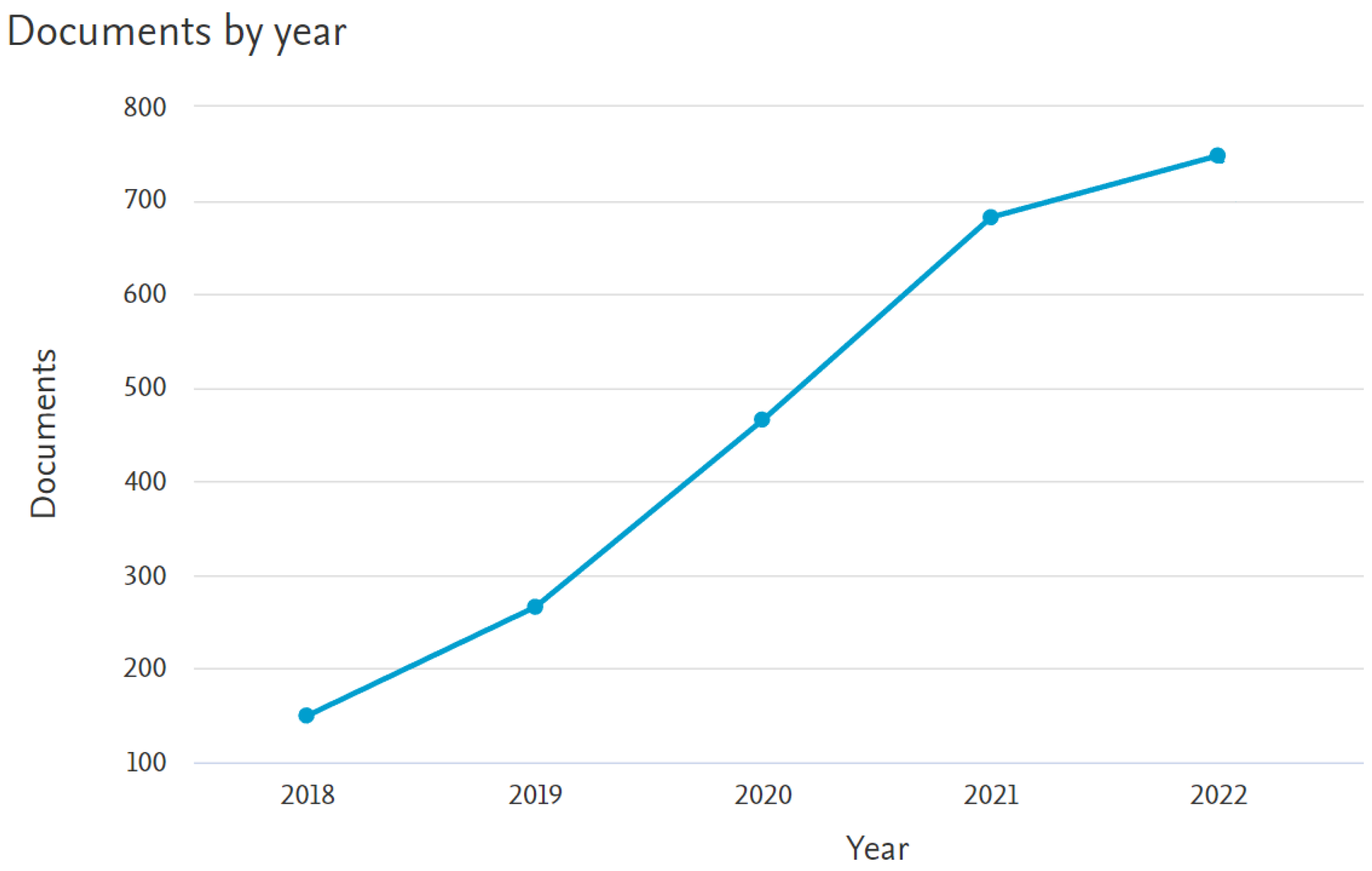
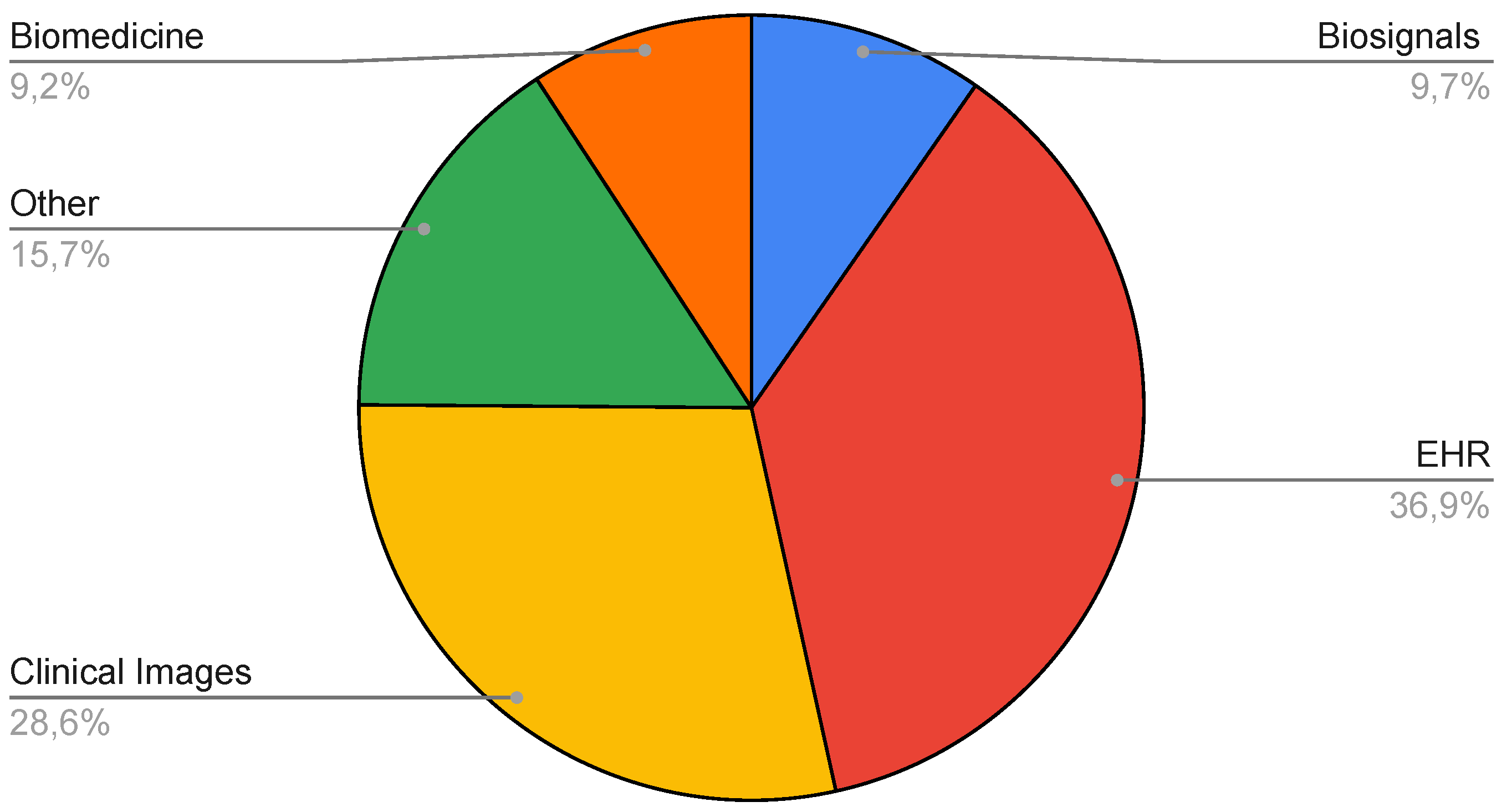
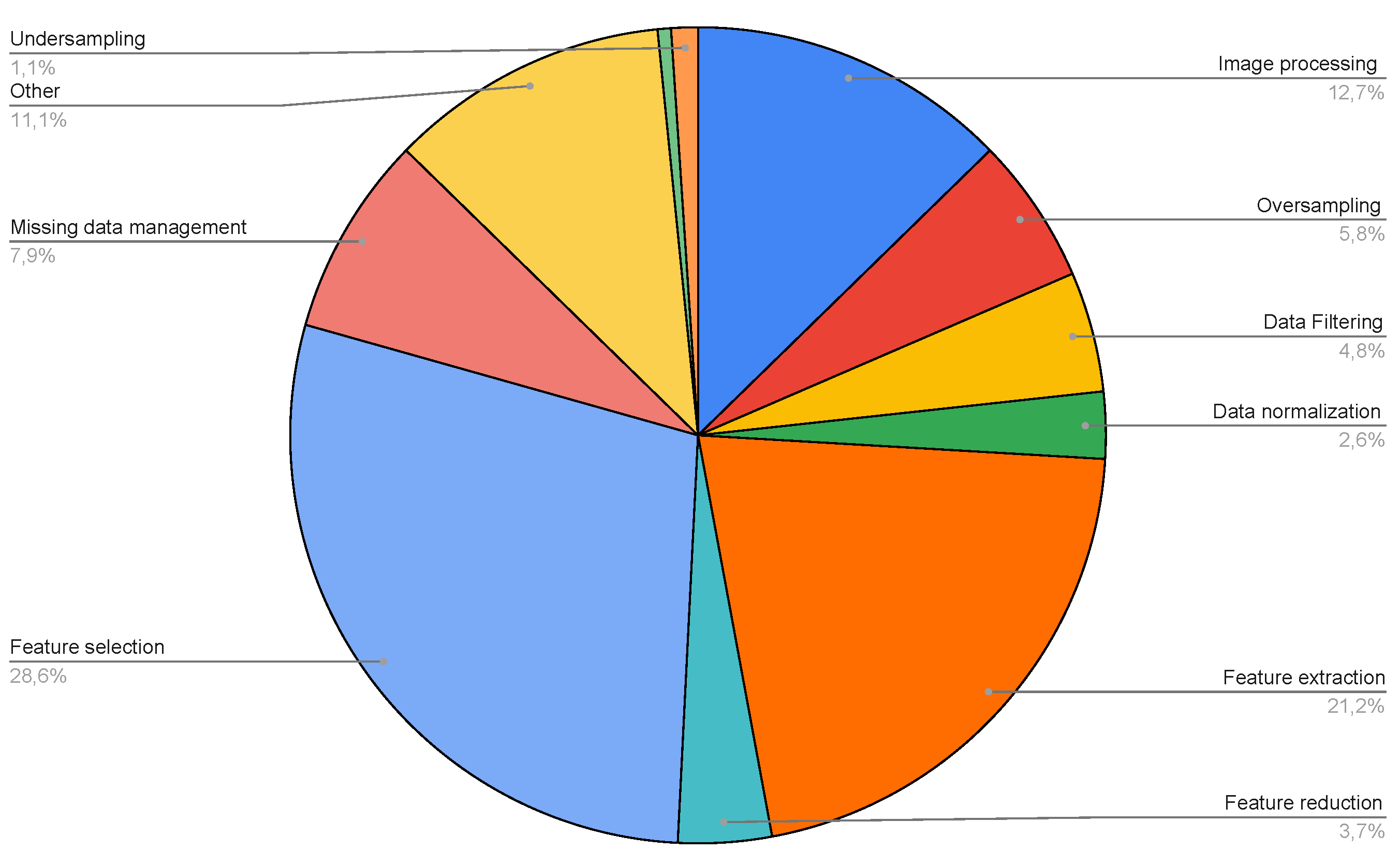
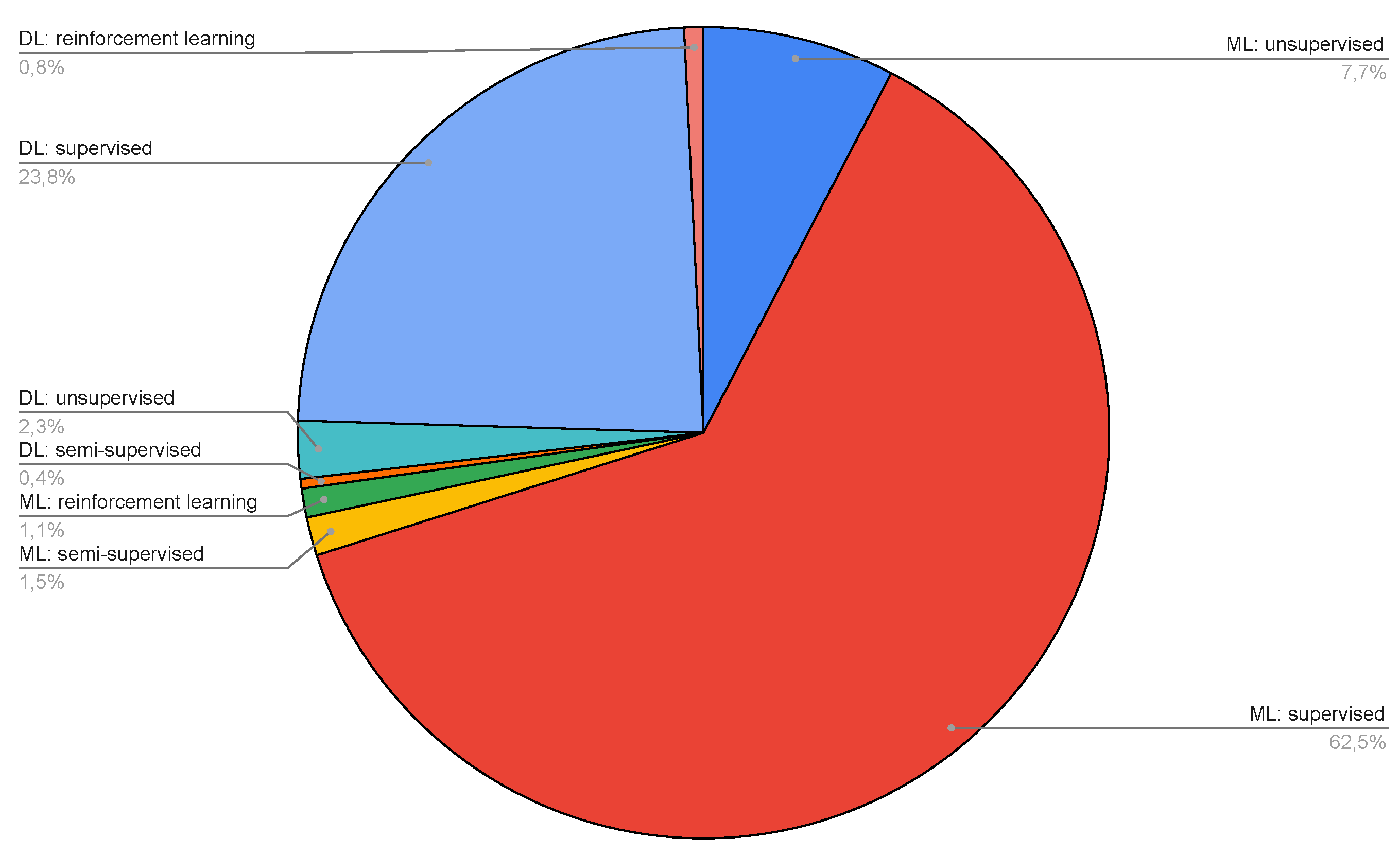
| Number of data types | Number of Papers |
|---|---|
| 1 | 176 |
| 2 | 32 |
| 3 | 6 |
| Number of pre-processing methods | Number of Papers |
|---|---|
| 0 | 86 |
| 1 | 87 |
| 2 | 29 |
| 3 | 10 |
| 4 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





