Submitted:
17 July 2023
Posted:
18 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data Set Acquisition

2.2. Methods
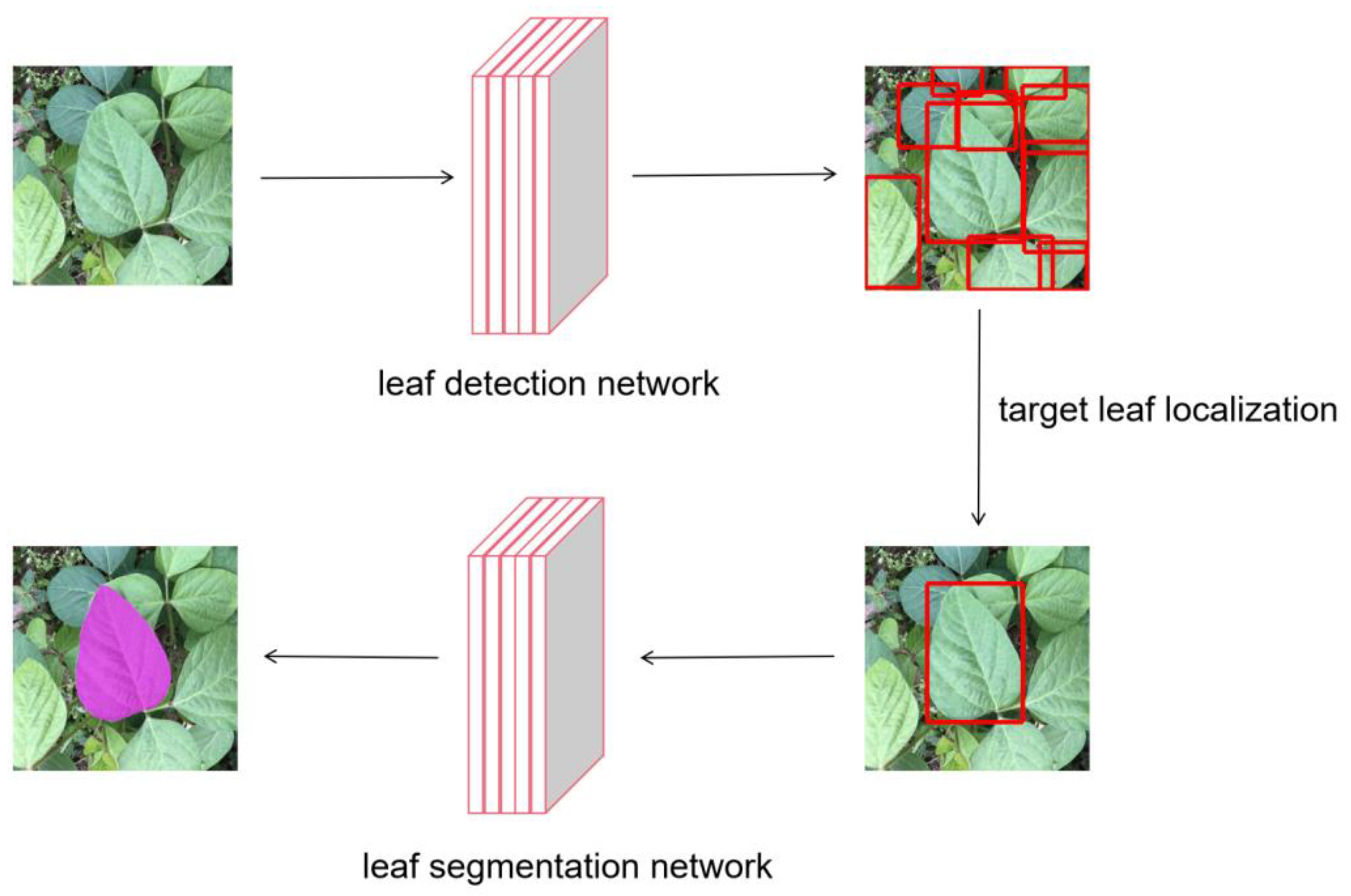
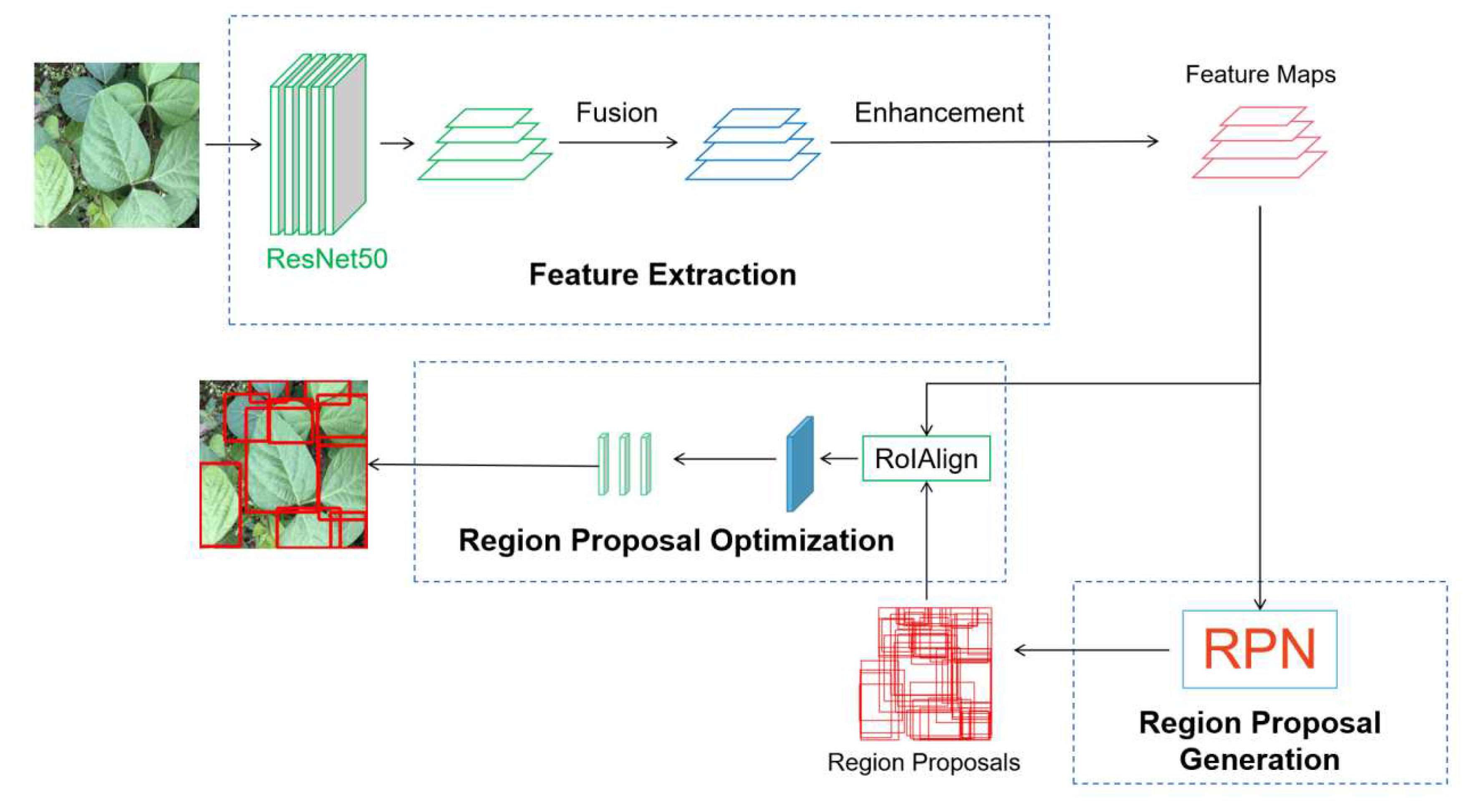
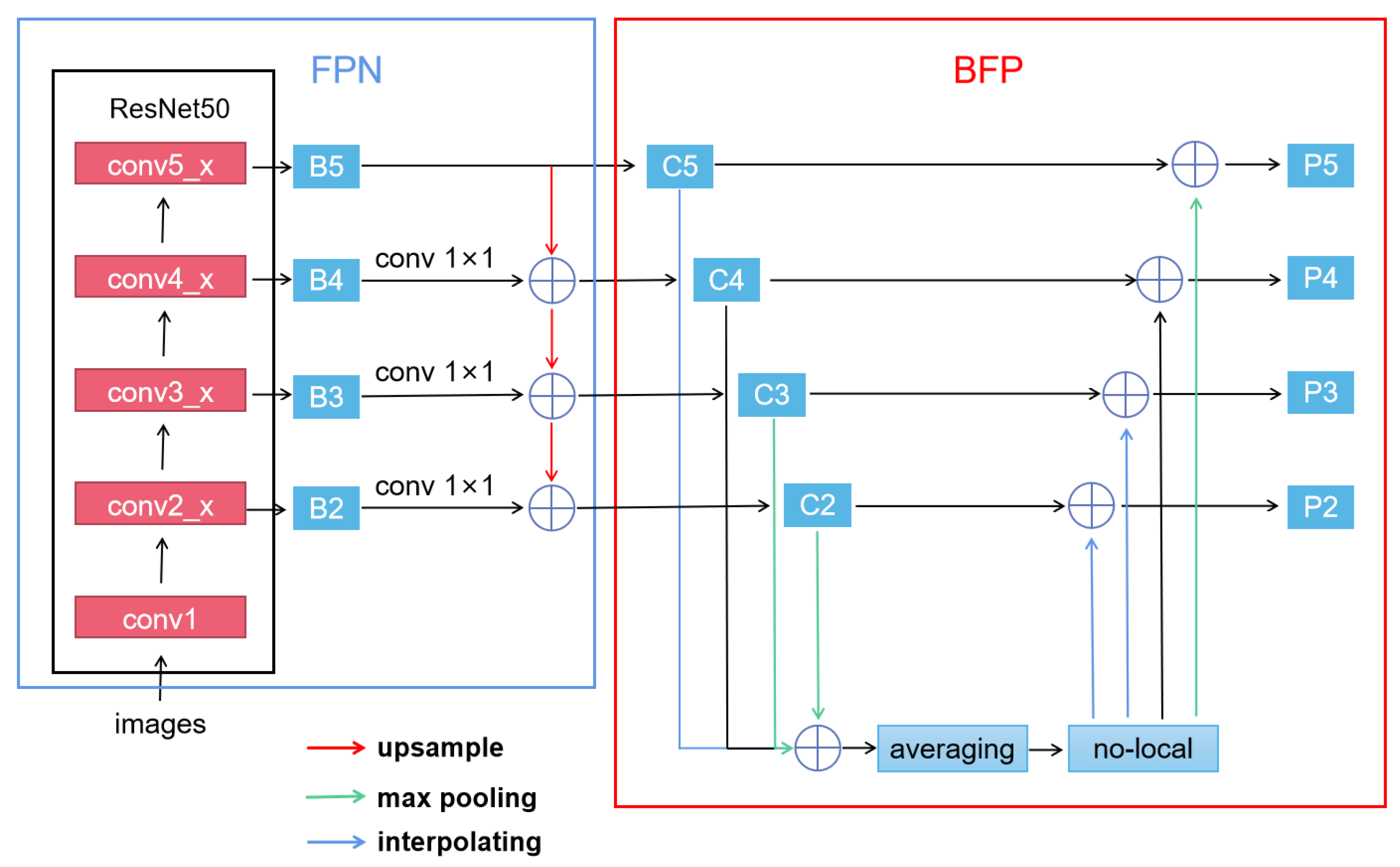
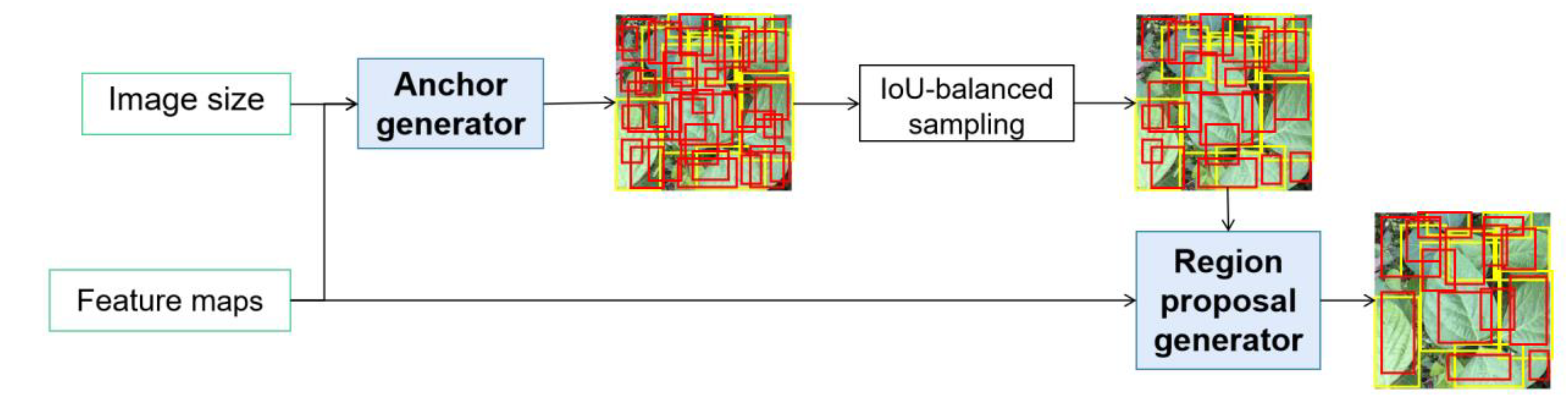
2.2.1. Libra R-CNN Network for Leaf Detection
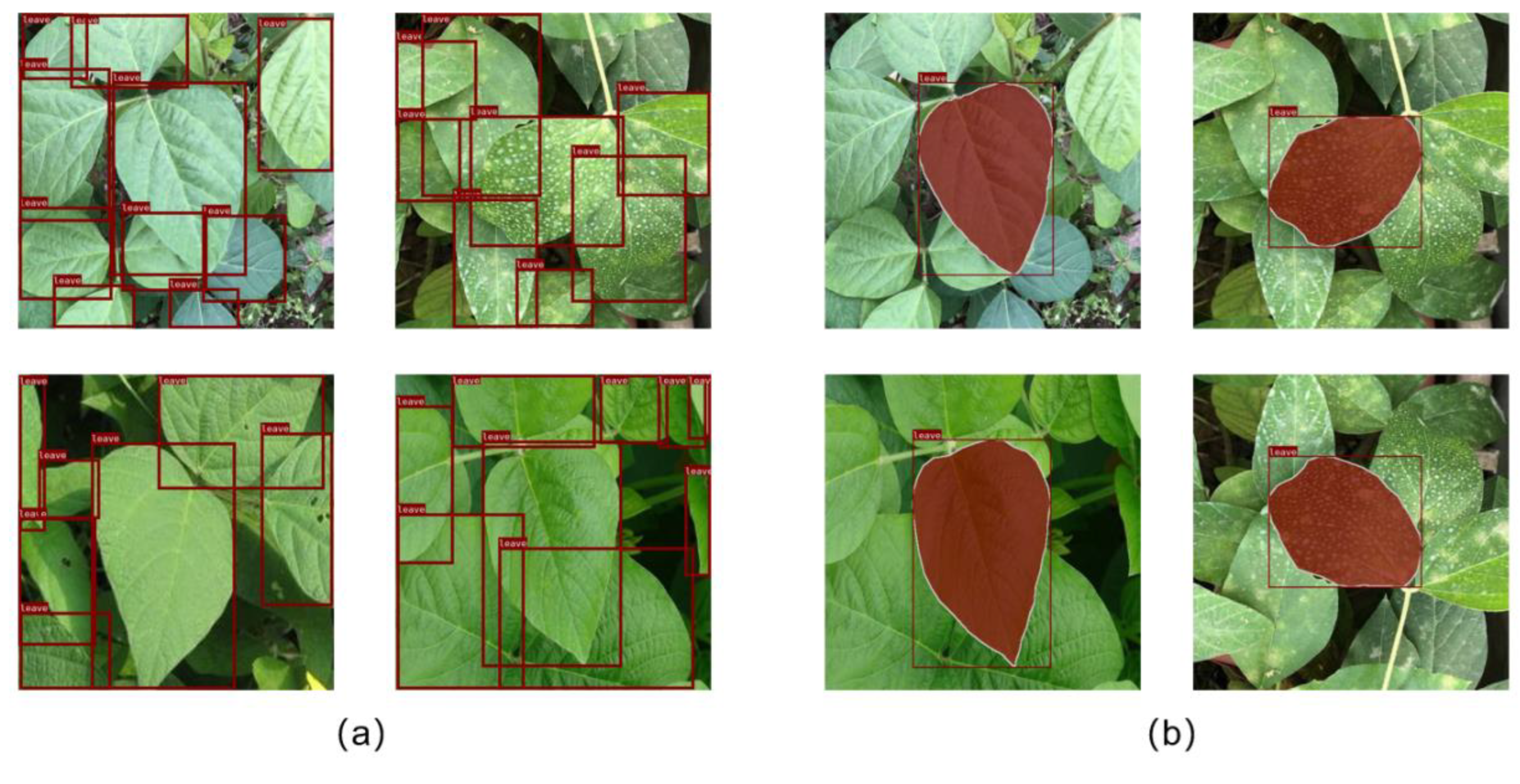
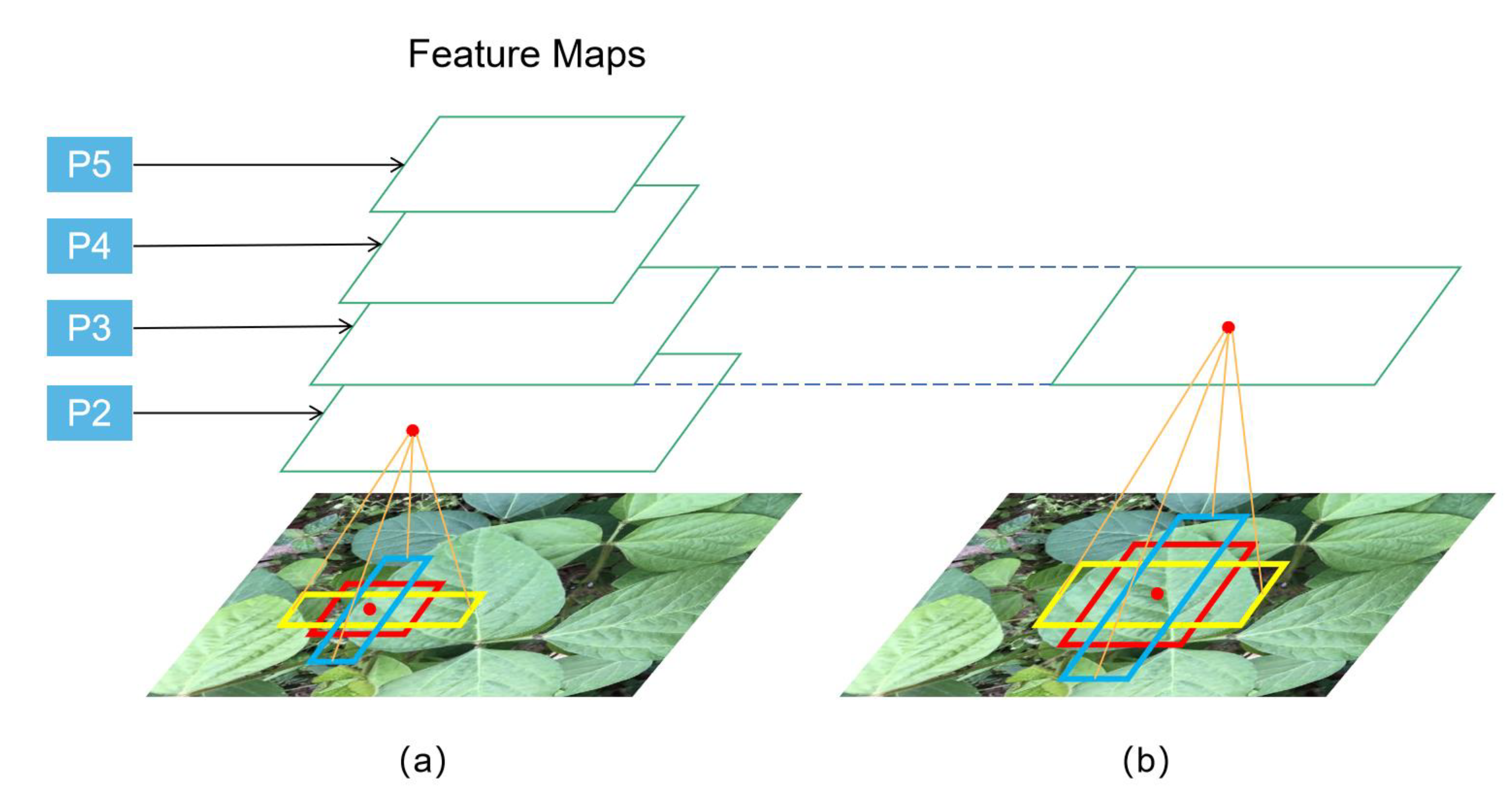
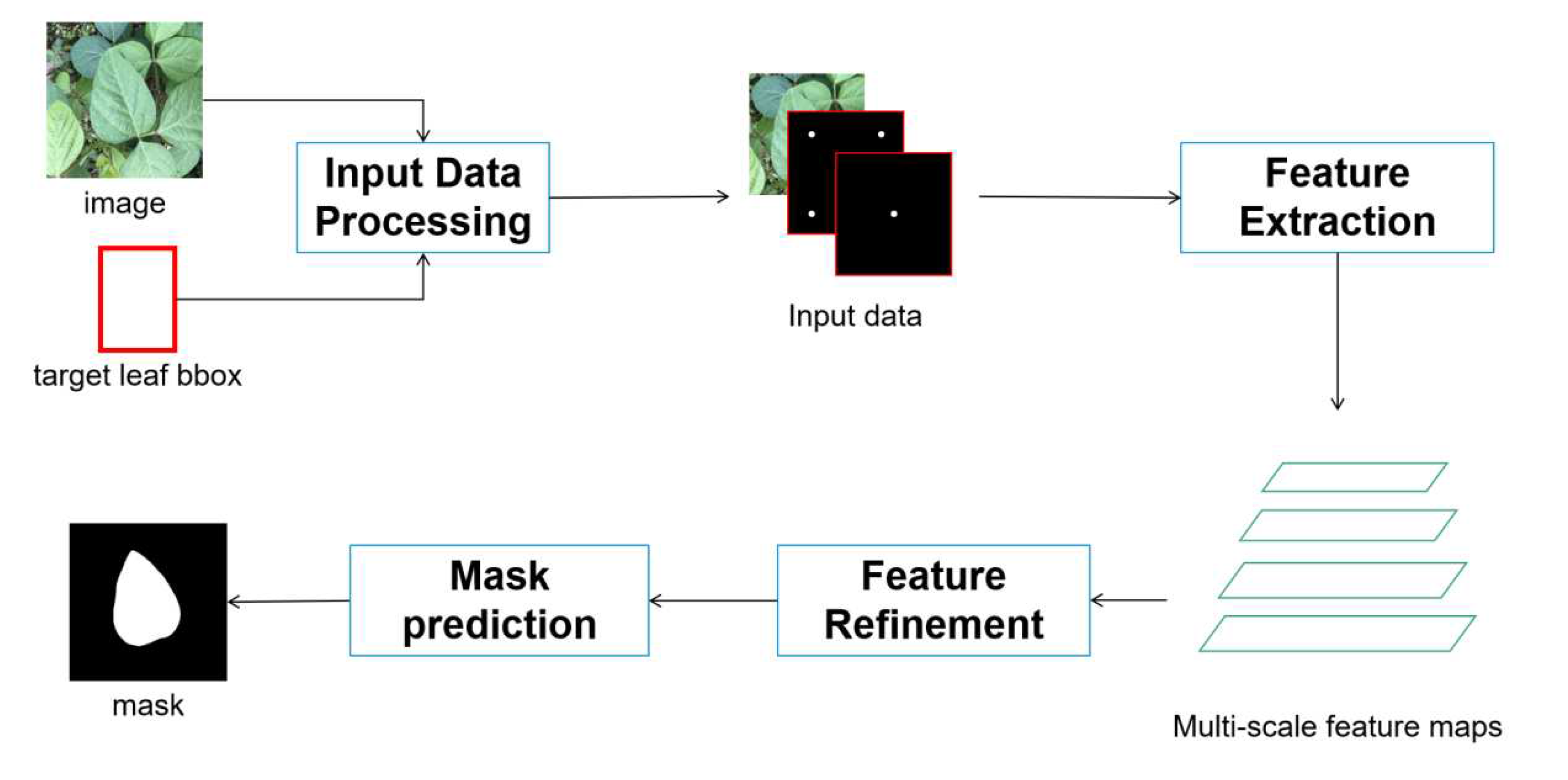
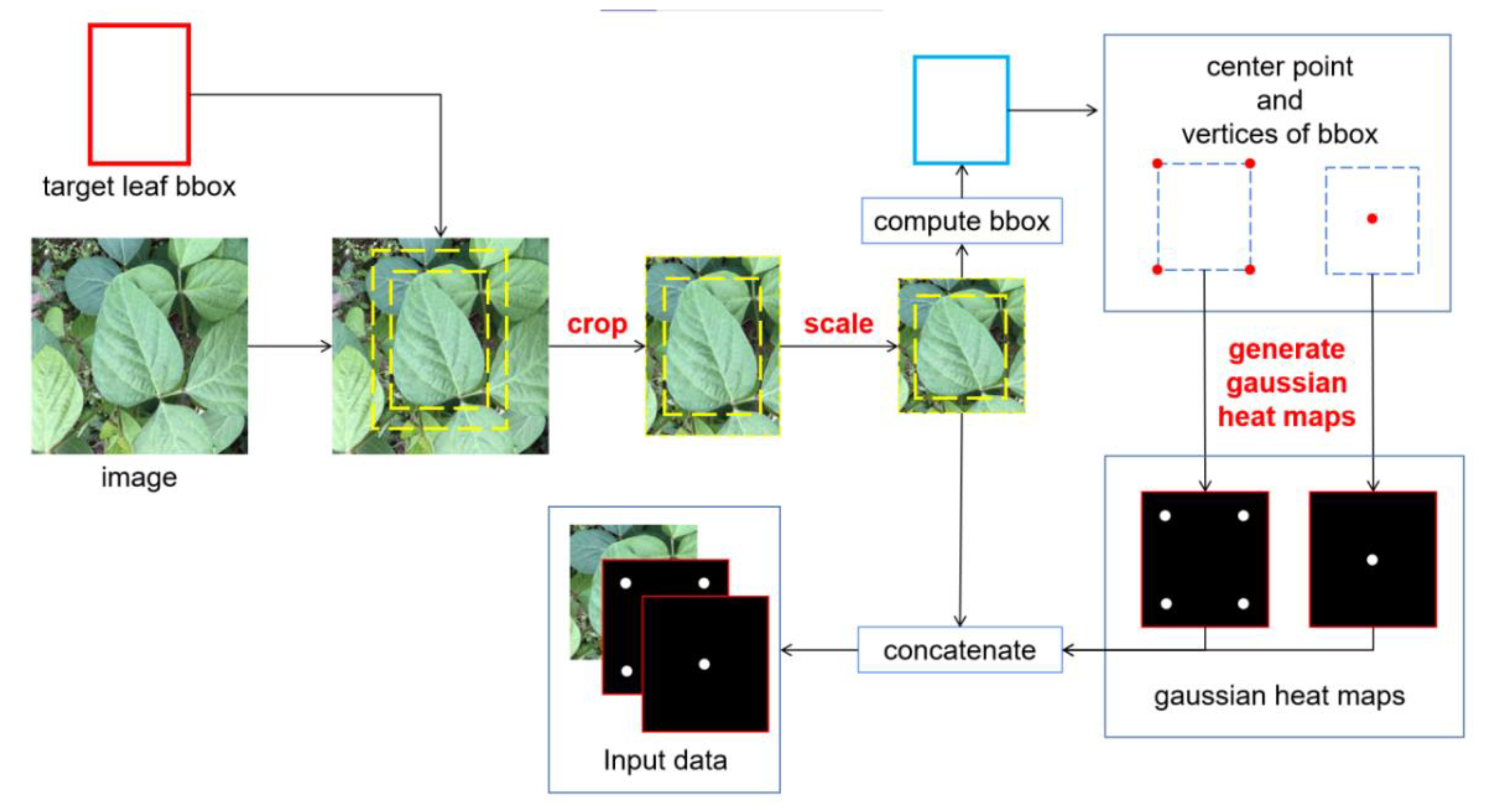
2.2.2. Target Leaf Localization
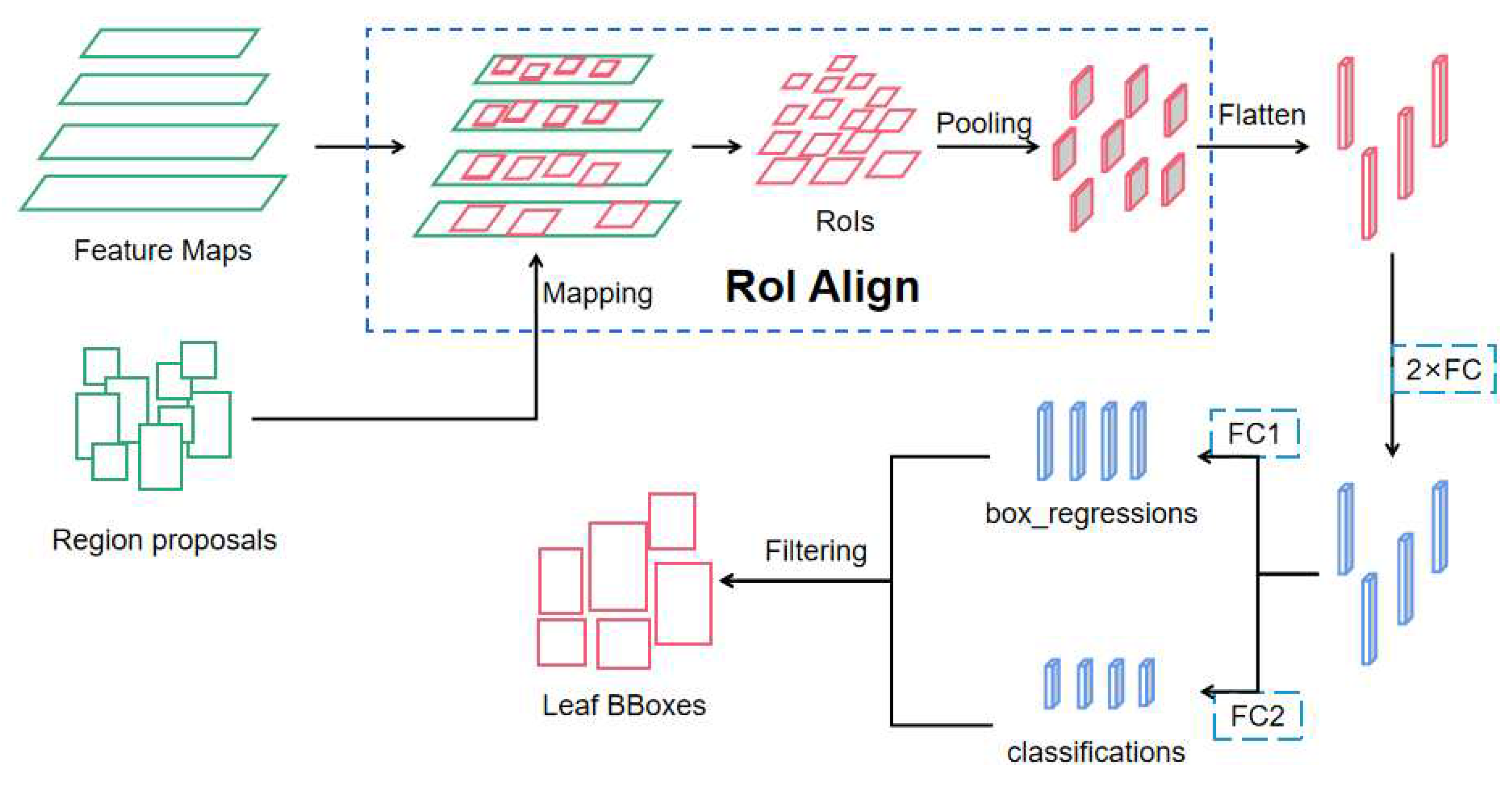
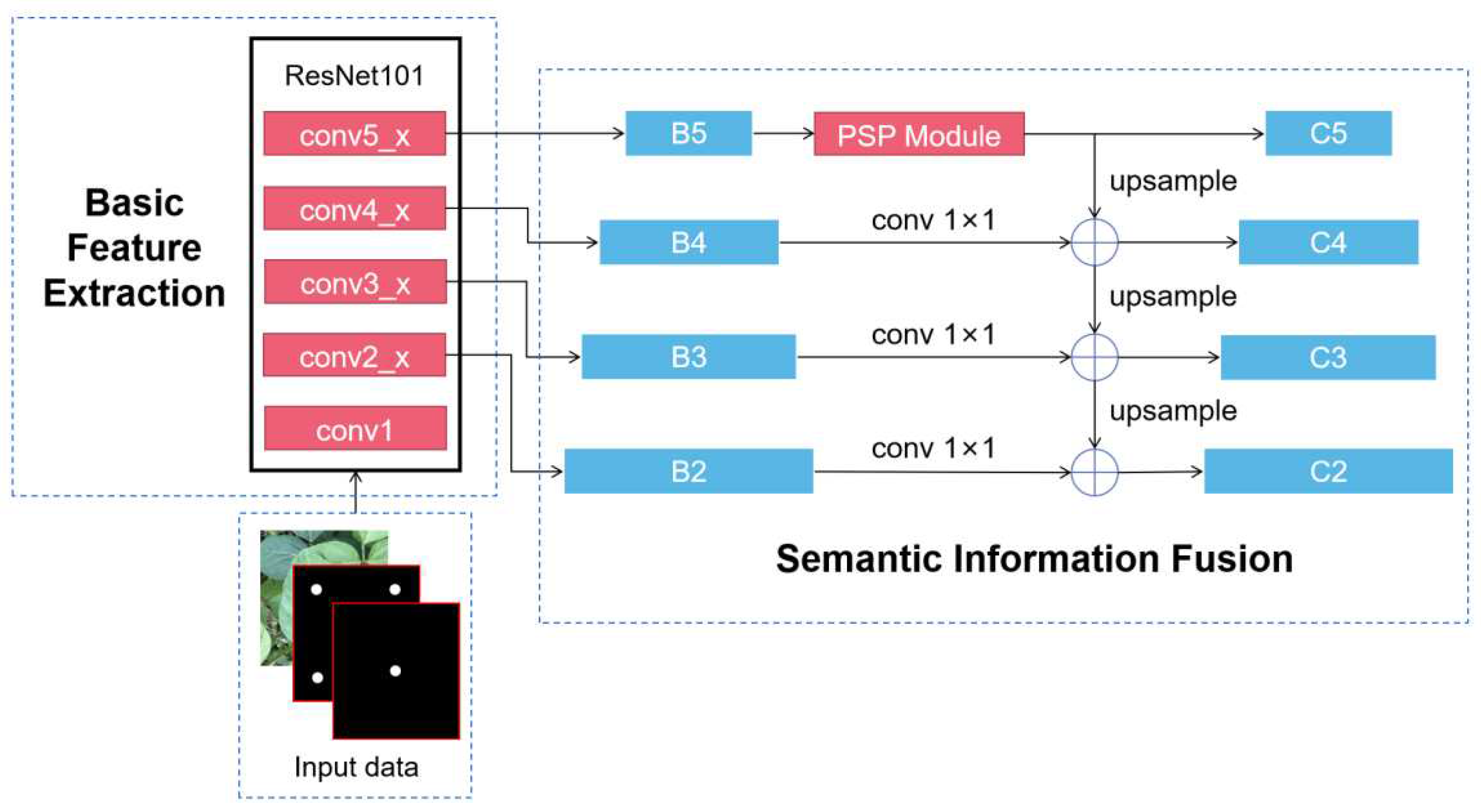
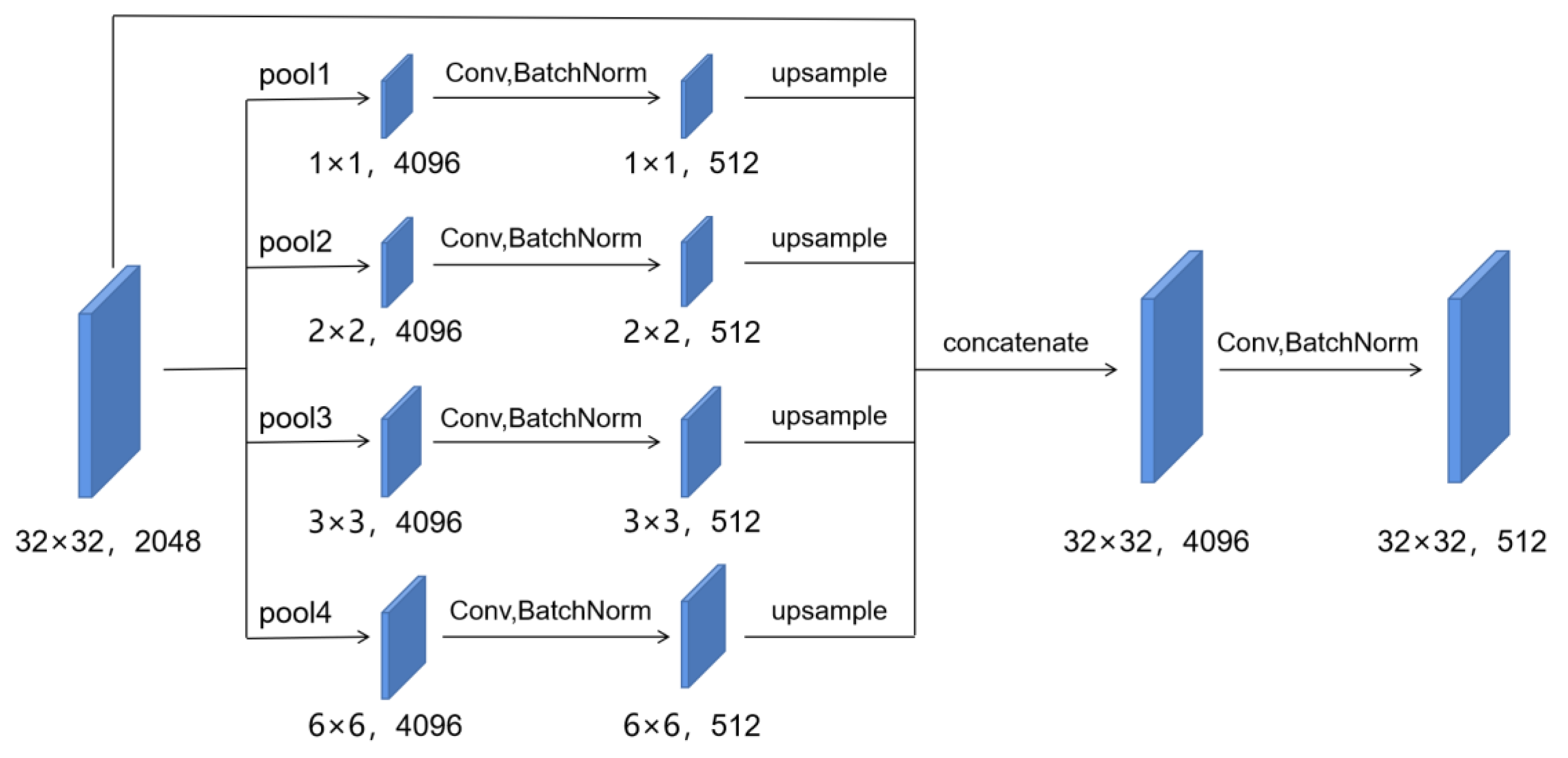
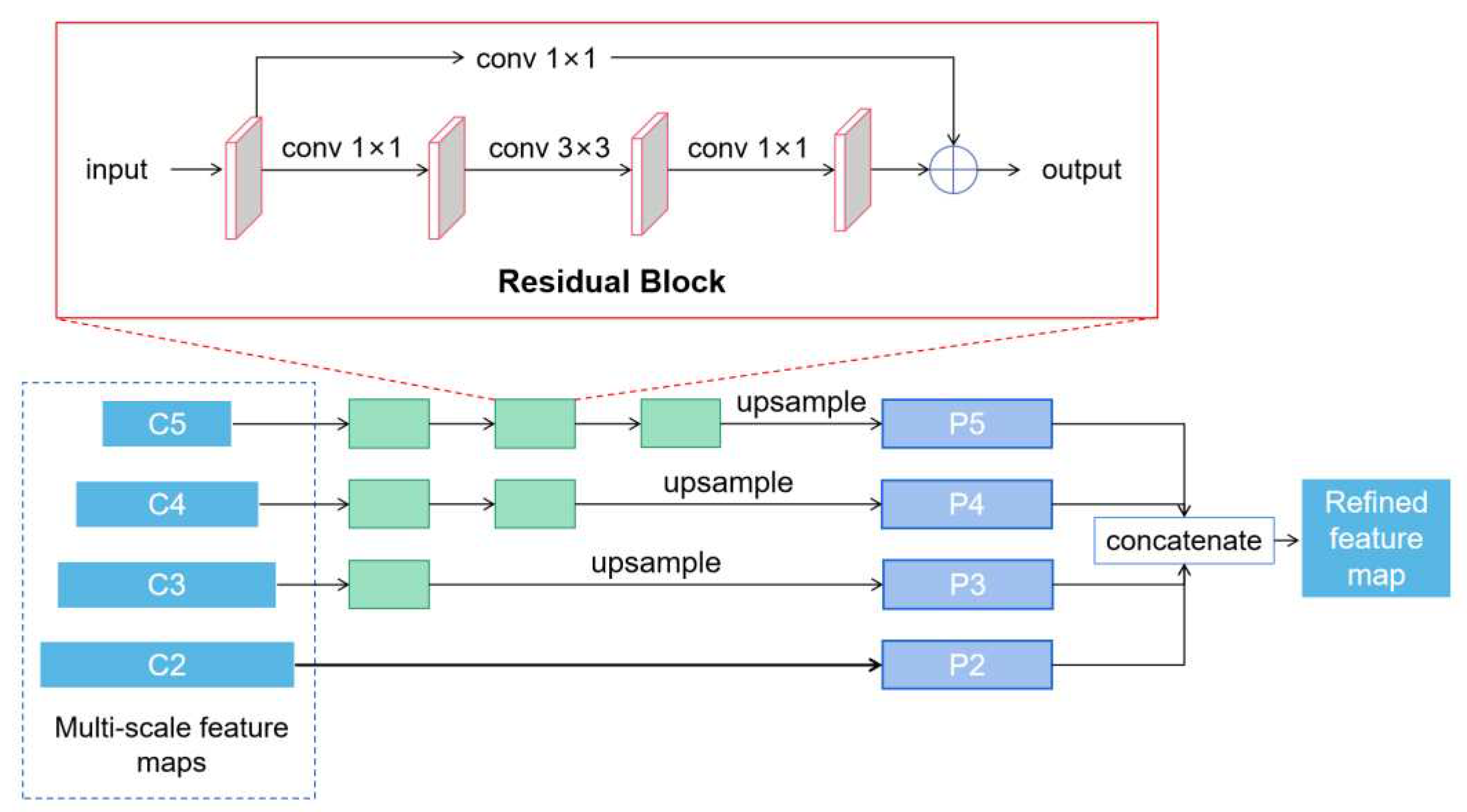
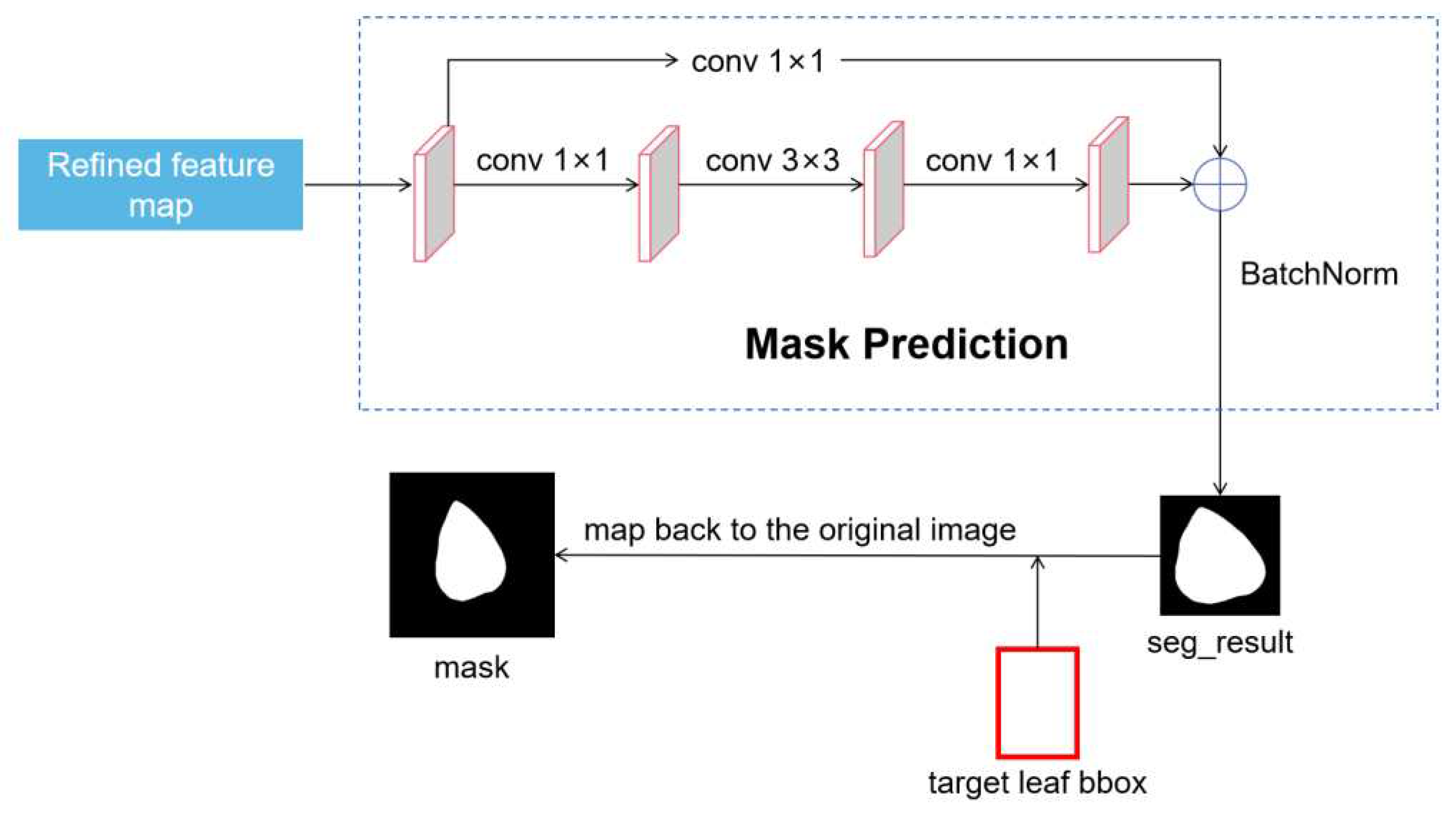
2.2.3. Target Leaf Segmentation Network
3. Experimental Results and Analysis
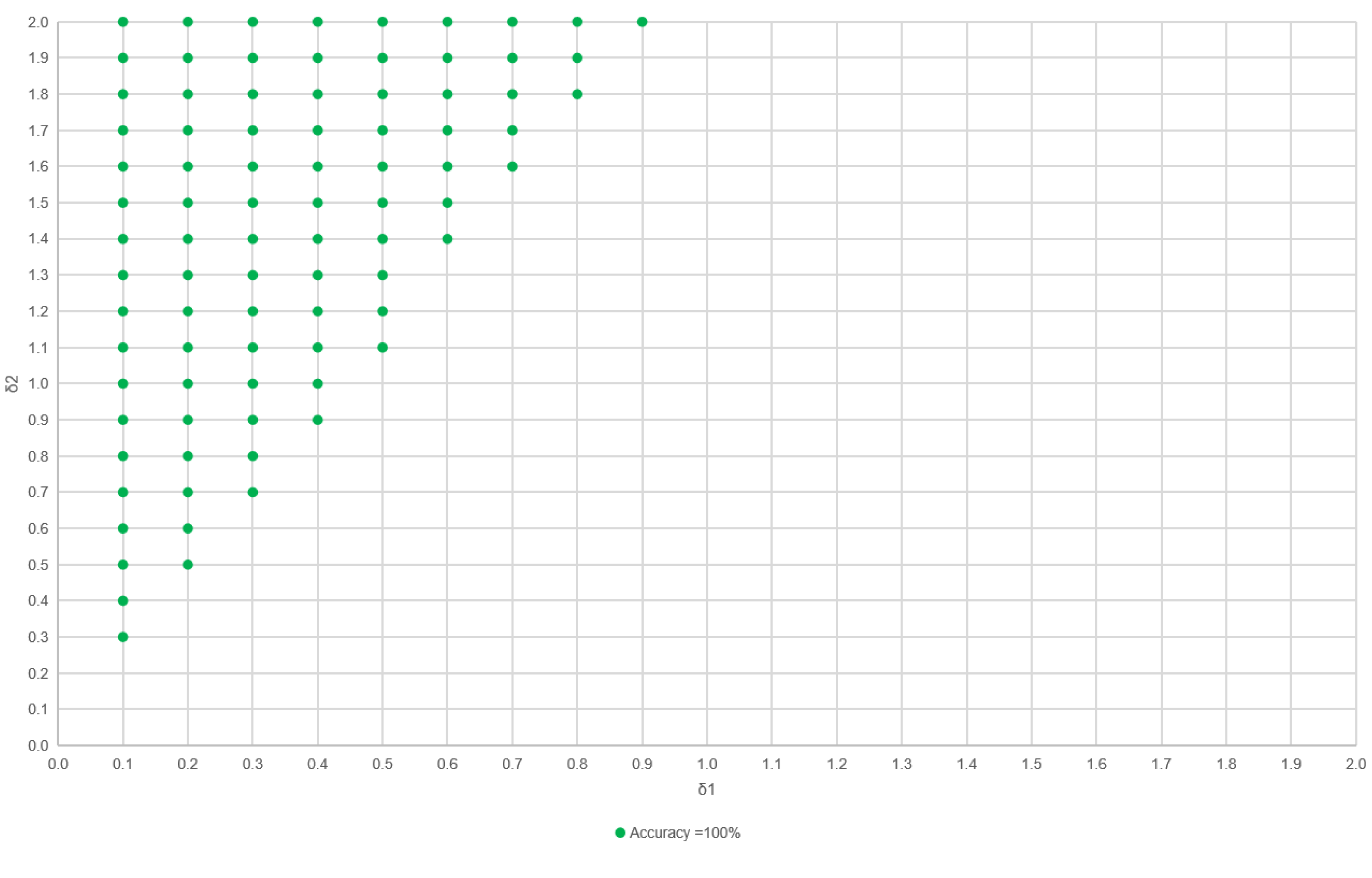
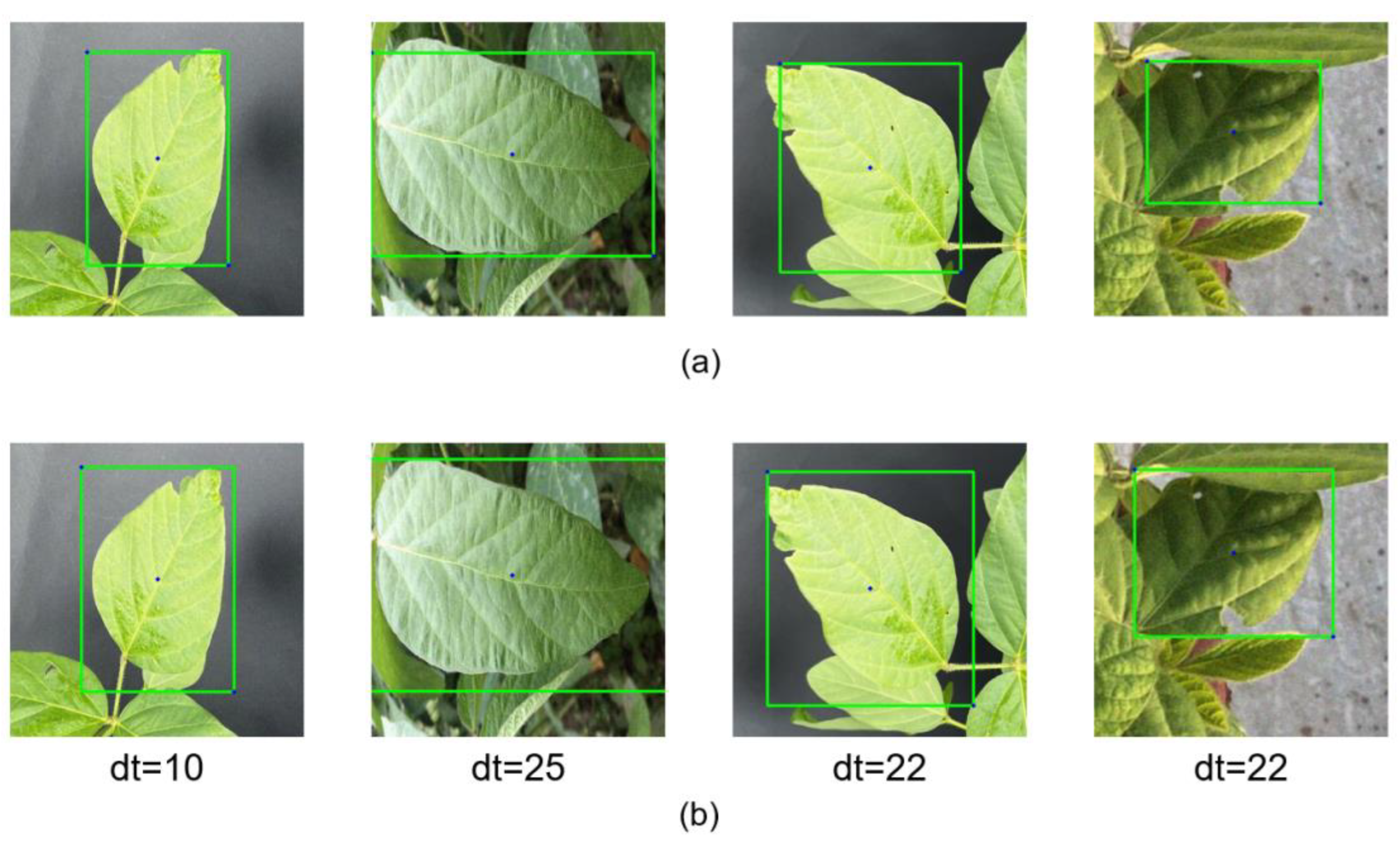
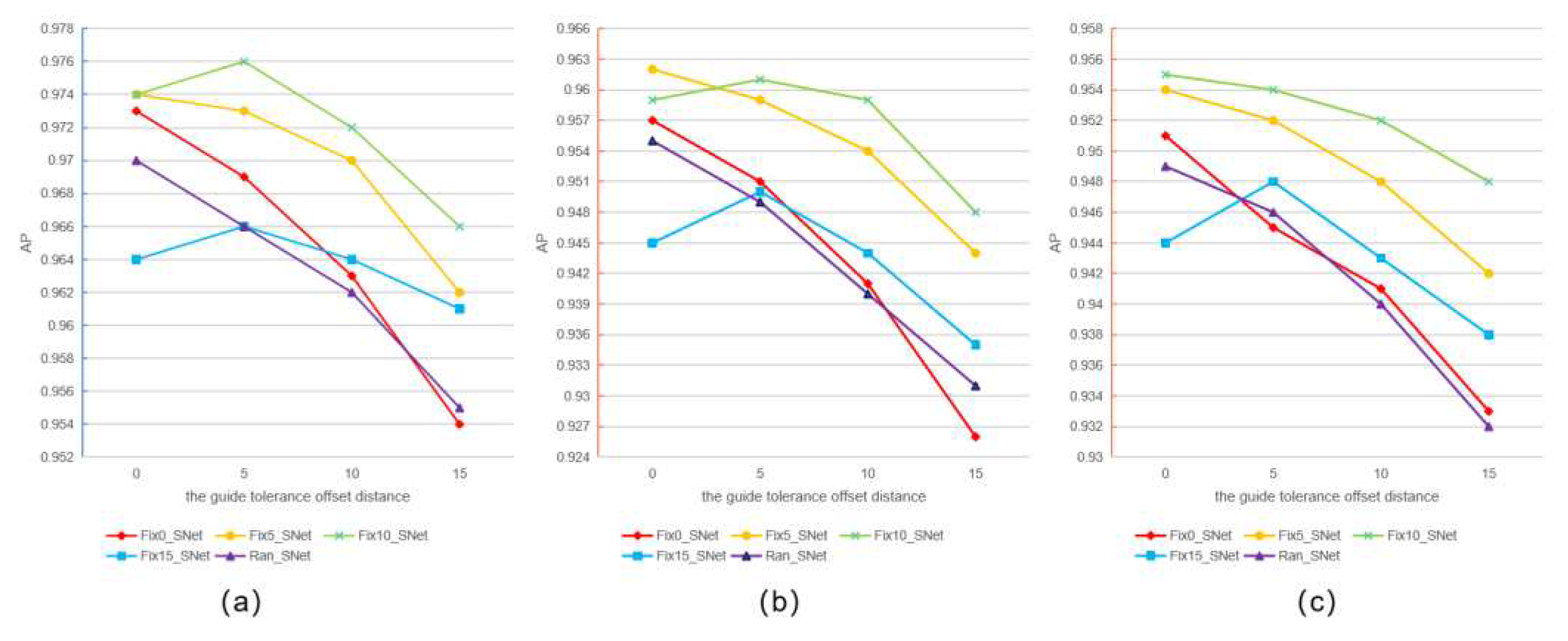
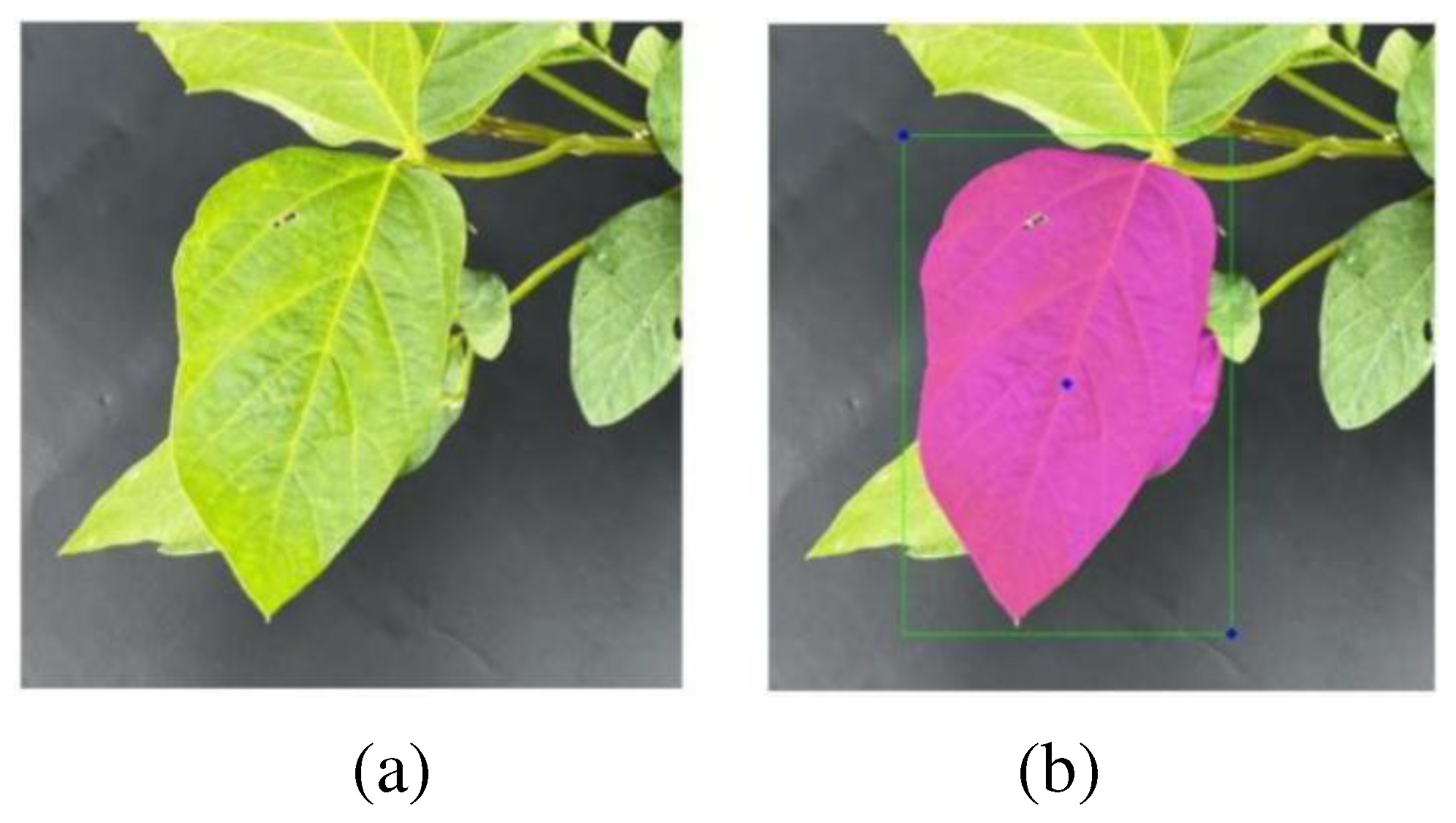
3.1. Vertex Offset Strategy for Bounding Box
3.2. The Affection of Leaf Detectors on the Segmentation
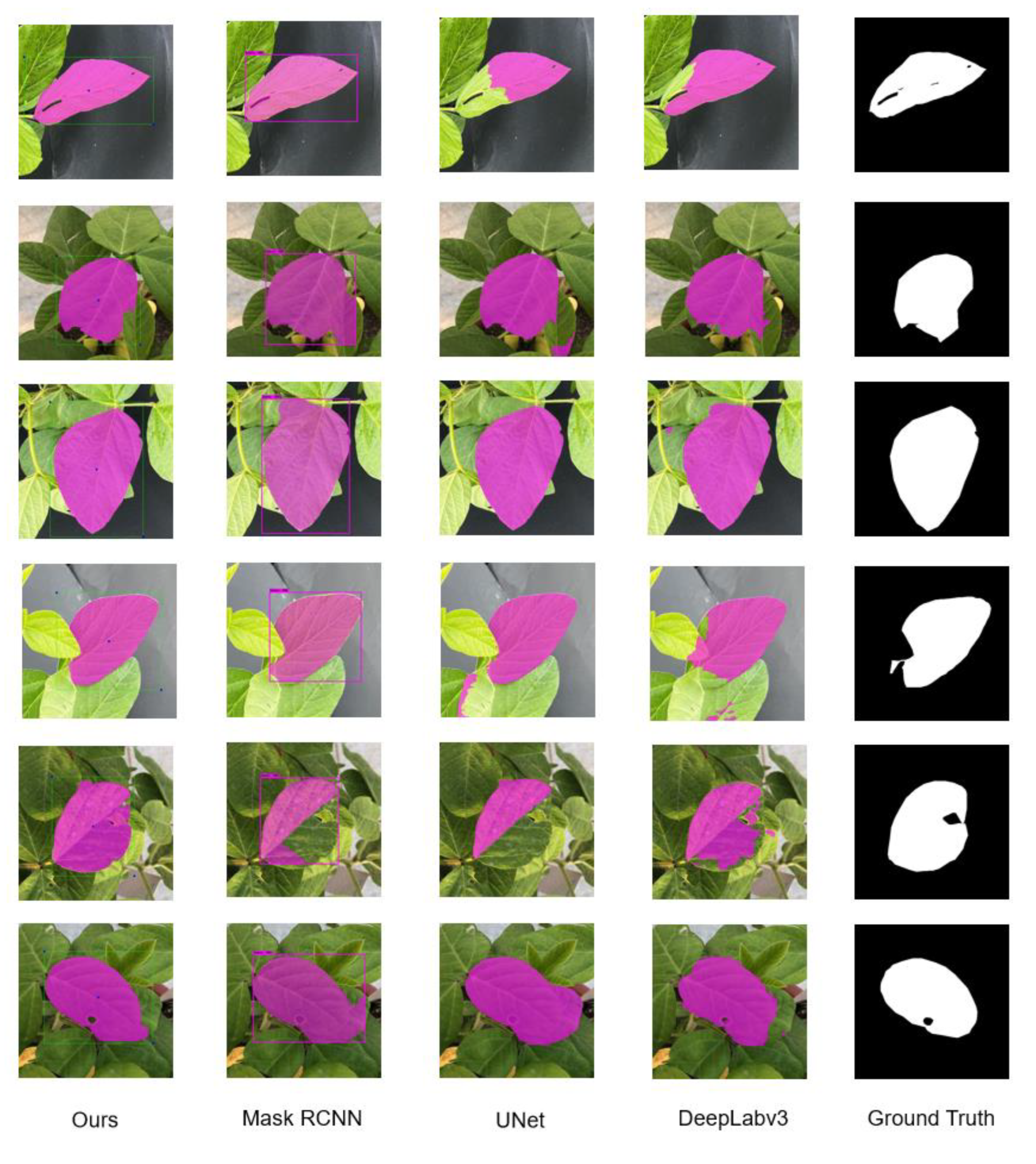
3.3. Comparative Experiment
3.3.1. Quantitative Analysis
3.3.2. Quantitative Comparison
4. Conclusions
References
- Bai, X., Li, X., Fu, Z., Lv, X., & Zhang, L. (2017). A fuzzy clustering segmentation method based on neighborhood grayscale information for defining cucumber leaf spot disease images. Computers and Electronics in Agriculture, 136, 157-165. [CrossRef]
- Benenson, R., Popov, S., & Ferrari, V. (2019). Large-scale interactive object segmentation with human annotators. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 11700-11709.
- Bhagat, S. , Kokare, M., Haswani, V., Hambarde, P., & Kamble, R. (2022). Eff-UNet++: A novel architecture for plant leaf segmentation and counting. Ecological Informatics, 68, 101583. [CrossRef]
- Cai, Z., & Vasconcelos, N. (2018). Cascade r-cnn: Delving into high quality object detection. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 6154-6162.
- Chandio, A. , Gui, G., Kumar, T., Ullah, I., Ranjbarzadeh, R., Roy, A. M., Shen, Y. (2022). Precise single-stage detector. arXiv preprint, arXiv:2210.04252.
- Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F., & Adam, H. (2018). Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV), pp. 801-818.
- Gao, L., & Lin, X. (2018). A method for accurately segmenting images of medicinal plant leaves with complex backgrounds. Computers and Electronics in Agriculture, 155, 426-445. [CrossRef]
- Girshick, R. (2015). Fast r-cnn. In Proceedings of the IEEE international conference on computer vision, pp. 1440-1448.
- Girshick, R., Donahue, J., Darrell, T., & Malik, J. (2014). Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 580-587. [CrossRef]
- He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
- He, K., Gkioxari, G., Dollár, P., & Girshick, R. (2017). Mask r-cnn. In Proceedings of the IEEE international conference on computer vision, pp. 2961-2969.
- Kumar, J. P., & Domnic, S. (2019). Image based leaf segmentation and counting in rosette plants. Information processing in agriculture, 6(2), 233-246. 2). [CrossRef]
- Li, Z. , Chen, Q. , & Koltun, V. (2018). Interactive image segmentation with latent diversity. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; pp. 577–585. [Google Scholar]
- Lin, T. Y. , Dollár, P. , Girshick, R., He, K., Hariharan, B., & Belongie, S. (2017). Feature pyramid networks for object detection. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2117-2125). [Google Scholar]
- Lin, Z. , Zhang, Z. , Chen, L. Z., Cheng, M. M., & Lu, S. P. (2020). Interactive image segmentation with first click attention. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; pp. 13339–13348. [Google Scholar]
- Liu, W. , Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C. Y., & Berg, A. C. (2016). Ssd: Single shot multibox detector. In Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, October 11–14, 2016, Proceedings, Part I 14 (pp. 21-37). Springer International Publishing.
- Liu, X. , Hu, C., & Li, P. (2020). Automatic segmentation of overlapped poplar seedling leaves combining Mask R-CNN and DBSCAN. Computers and Electronics in Agriculture, 178, 105753. [CrossRef]
- Long, J. , Shelhamer, E. , & Darrell, T. (2015). Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 3431–3440. [Google Scholar]
- Maninis, K. K. , Caelles, S. , Pont-Tuset, J., & Van Gool, L. (2018). Deep extreme cut: From extreme points to object segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 616–625. [Google Scholar]
- Pang, J. , Chen, K. , Shi, J., Feng, H., Ouyang, W., & Lin, D. (2019). Libra r-cnn: Towards balanced learning for object detection. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; pp. 821–830. [Google Scholar]
- Redmon, J. , & Farhadi, A. (2017). YOLO9000: better, faster, stronger. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 7263–7271.
- Redmon, J., Divvala, S., Girshick, R., & Farhadi, A. (2016). You only look once: Unified, real-time object detection. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 779-788.
- Ren, S. , He, K., Girshick, R., & Sun, J. (2015). Faster r-cnn: Towards real-time object detection with region proposal networks. Advances in neural information processing systems, 28.
- Reynolds, M. , Chapman, S., Crespo-Herrera, L., Molero, G., Mondal, S., Pequeno, D. N., Pinto, F., Pinera-Chavez, F. J., Poland, J., Rivera-Amado, C., et al. Breeder friendly phenotyping. Plant Science, pp. 110396, 2020. [CrossRef]
- Ronneberger, O. , Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18 (pp. 234-241). Springer International Publishing.
- Rother, C. , Kolmogorov, V., & Blake, A. (2004). “ GrabCut” interactive foreground extraction using iterated graph cuts. ACM transactions on graphics (TOG), 23(3), 309-314. [CrossRef]
- Song, G. , Myeong, H. , & Lee, K. M. (2018). Seednet: Automatic seed generation with deep reinforcement learning for robust interactive segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 1760–1768. [Google Scholar] [CrossRef]
- Tassis, L. M. , de Souza, J. E. T., & Krohling, R. A. (2021). A deep learning approach combining instance and semantic segmentation to identify diseases and pests of coffee leaves from in-field images. Computers and Electronics in Agriculture, 186, 106191. [CrossRef]
- Tian, K. , Li, J., Zeng, J., Evans, A., & Zhang, L. (2019). Segmentation of tomato leaf images based on adaptive clustering number of K-means algorithm. Computers and Electronics in Agriculture, 165, 104962. [CrossRef]
- Tian, Y. , Yang, G., Wang, Z., Li, E., & Liang, Z. (2020). Instance segmentation of apple flowers using the improved mask R–CNN model. Biosystems engineering, 193, 264-278. [CrossRef]
- Wang, C. , Du, P., Wu, H., Li, J., Zhao, C., & Zhu, H. (2021). A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+ and U-Net. Computers and Electronics in Agriculture, 189, 106373. [CrossRef]
- Wang, P., Zhang, Y., Jiang, B., & Hou, J. (2020). An maize leaf segmentation algorithm based on image repairing technology. Computers and electronics in agriculture, 172, 105349. [CrossRef]
- Wang, X. , Girshick, R. , Gupta, A., & He, K. (2018). Non-local neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 7794–7803. [Google Scholar]
- Ward, B. , Brien, C., Oakey, H., Pearson, A., Negrão, S., Schilling, R. K.,... & van den Hengel, A. (2019). High-throughput 3D modelling to dissect the genetic control of leaf elongation in barley (Hordeum vulgare). The Plant Journal, 98(3), 555-570. [CrossRef]
- Xie, X. , Cheng, G. , Wang, J., Yao, X., & Han, J. (2021). Oriented R-CNN for object detection. In Proceedings of the IEEE/CVF international conference on computer vision; pp. 3520–3529. [Google Scholar]
- Xu, N. , Price, B., Cohen, S., Yang, J., & Huang, T. (2017). Deep grabcut for object selection. arXiv preprint, arXiv:1707.00243. [CrossRef]
- Xu, N. , Price, B. , Cohen, S., Yang, J., & Huang, T. S. (2016). Deep interactive object selection. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 373–381. [Google Scholar]
- Yang, W. , Feng, H., Zhang, X., Zhang, J., Doonan, J. H., Batchelor, W. D.,... & Yan, J. (2020). Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Molecular Plant, 13(2), 187-214. [CrossRef]
- Zhang, S. , Liew, J. H., Wei, Y., Wei, S., & Zhao, Y. (2020). Interactive object segmentation with inside-outside guidance. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; pp. 12234–12244. [Google Scholar]
- Zhao, H. , Shi, J. , Qi, X., Wang, X., & Jia, J. (2017). Pyramid scene parsing network. In Proceedings of the IEEE conference on computer vision and pattern recognition; pp. 2881–2890. [Google Scholar]
| Configuration | Parameter |
|---|---|
| CPU | Intel(R) Core(TM) i7 - 6700 CPU |
| GPU | GeForce GTX 1080 Ti |
| Operating system | Ubuntu 22.04 LTS |
| Base environment | CUDA : 11.6 |
| Development environment | Pycharm2022 |
| Parameter | Leaf detector | Leaf segmentation network |
|---|---|---|
| Epoch | 60 | 100 |
| Learning rate | 0.001 | 1×10-8 |
| Batch | 4 | 5 |
| Weight decay | 0.0005 | 0.005 |
| Momentum | 0.9 | 0.9 |
| Model | AP | AR | Accuracy | Precision | Recall | F1 |
|---|---|---|---|---|---|---|
| Ours | 0.976 | 0.981 | 0.993 | 0.9899 | 0.9901 | 0.99 |
| Mask R-CNN | 0.921 | 0.936 | 0.9838 | 0.9759 | 0.9778 | 0.9769 |
| DeepLabv3 | 0.767 | 0.815 | 0.9645 | 0.9422 | 0.9584 | 0.9769 |
| UNet | 0.794 | 0.834 | 0.9675 | 0.9544 | 0.9521 | 0.9532 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




