1. Introduction
Tardigrade, also called water bears, is a phylum consisting of ca. 1,500 species 1–4 that inhabit terrestrial and aquatic environments throughout the world5. They are mostly found in mosses, lichens, soil, leaf litter, sediments and on aquatic plants5–7. The phylum consists of two classes, i.e., Heterotardigrada and Eutardigrada5. Eutardigrada is further divided into two limnoterrestrial orders, i.e., Apochela and Parachela. Moreover, order Parachela consists of various superfamilies and families, one of them being Macrobiotidae (Thulin, 1928)8 with genus Paramacrobiotus Guidetti, Schill, Bertolani, Dandekar and Wolf, 20099. The genus was erected in 2009 from the genus Macrobiotus and till date 45 species have been described: Paramacrobiotus alekseevi (Tumanov, 2005)10, Pam. arduus Guidetti, Cesari, Bertolani, Altiero & Rebecchi, 201911, Pam. areolatus (Murray, 1907)12, Pam. beotiae (Durante Pasa & Maucci, 1979)13, Pam. celsus Guidetti, Cesari, Bertolani, Altiero & Rebecchi, 201911, Pam. centesimus (Pilato, 2000)14, Pam. chieregoi (Maucci & Durante Pasa, 1980)15, Pam. corgatensis (Pilato, Binda & Lisi, 2004)16, Pam. csotiensis (Iharos, 1966)17, Pam. danielae (Pilato, Binda, Napolitano & Moncada, 2001)18, Pam. danielisae (Pilato, Binda & Lisi, 2006)19, Pam. depressus Guidetti, Cesari, Bertolani, Altiero & Rebecchi, 201911, Pam. derkai (Degma, Michalczyk & Kaczmarek, 2008)20, Pam. experimentalis Kaczmarek, Mioduchowska, Poprawa & Roszkowska, 202021, Pam. fairbanksi Schill, Förster, Dandekar & Wolf, 201022, Pam. filipi Dudziak, Stec & Michalczyk 202023, Pam. gadabouti Kayastha, Stec, Mioduchowska and Kaczmarek 202324, Pam. garynahi (Kaczmarek, Michalczyk & Diduszko, 2005)25, Pam. gerlachae (Pilato, Binda & Lisi, 2004)16, Pam. halei (Bartels, Pilato, Lisi & Nelson, 2009)26, Pam. hapukuensis (Pilato, Binda & Lisi, 2006)19, Pam. huziori (Michalczyk & Kaczmarek, 2006)27, Pam. intii Kaczmarek, Cytan, Zawierucha, Diduszko & Michalczyk, 201428, Pam. kenianus Schill, Förster, Dandekar & Wolf, 201022, Pam. klymenki Pilato, Kiosya, Lisi & Sabella, 201229, Pam. lachowskae Stec, Roszkowska, Kaczmarek & Michalczyk, 201830, Pam. lorenae (Biserov, 1996)31, Pam. magdalenae (Michalczyk & Kaczmarek, 2006)27, Pam. metropolitanus Sugiura, Matsumoto & Kunieda, 202232 Pam. palaui Schill, Förster, Dandekar & Wolf, 201022, Pam. peteri (Pilato, Claxton & Binda, 1989)33, Pam. pius Lisi, Binda & Pilato, 201634, Pam. priviterae (Binda, Pilato, Moncada & Napolitano, 2001)35, Pam. richtersi (Murray, 1911)36, Pam. rioplatensis (Claps & Rossi, 1997)37, Pam. sagani Daza, Caicedo, Lisi & Quiroga, 201738, Pam. savai (Binda & Pilato, 2001)39, Pam. sklodowskae (Michalczyk, Kaczmarek & Węglarska, 2006)40, Pam. spatialis Guidetti, Cesari, Bertolani, Altiero & Rebecchi, 201911, Pam. spinosus Kaczmarek, Gawlak, Bartels, Nelson & Roszkowska, 201741, Pam. submorulatus (Iharos, 1966)17, Pam. tonollii (Ramazzotti, 1956)42, Pam. vanescens (Pilato, Binda & Catanzaro, 1991)43, Pam. walteri (Biserov, 1997/98)44 and Pam. wauensis (Iharos, 1973)45. Furthermore, the genus is divided into two species groups, i.e., richtersi group with presence of microplacoid within the pharynx, and areolatus group without microplacoid within the pharynx. In turn, Kaczmarek et al.41 proposed to separate subgenera for which specific names were clarified by Marley et al.46. However, the two subgenera are not valid according to Guidetti et al.11 and Stec et al.47.
In this paper we summarise the data on taxonomy, distribution, mode of reproduction, microbiome study, feeding behaviour, life history, morphological taxonomy, phylogeny and cryptobiotic studies along with new key for species identification in genus Paramacrobiotus.
2. Cryptobiosis
A stage of an organism's life known as cryptobiosis is one in which no activity is apparent48. Many organisms go through cryptobiosis to survive the harsh environmental conditions they encounter 49–51. These conditions can include anhydrobiosis (lack of water), anoxybiosis (lack of oxygen), cryobiosis (low temperature), or osmobiosis (change in osmotic conditions). Tardigrades have a remarkable capacity for undergoing and surviving several types of cryptobiosis 48,52. The majority of anhydrobiosis, or absence of water, has been studied in the species of the genus Paramacrobiotus, although there has also been research on famine, freezing, and bet-hedging53–57. Reuner et al.53 studied how the influence of starvation and anhydrobiosis affects the size and number of storage cells in Paramacrobiotus tonollii to understand the energetic side of anhydrobiosis. Starving Pam. tonollii for seven days led to reduction in storage cell size by 46.41% but no significant reduction in storage cell number was observed. Furthermore, when storage cells size and number were investigated after inducing anhydrobiosis for seven days where no significant changes in storage cell size and number of Pam. tonollii was observed. Also, the mortality was checked using prolonged starvation and Pam. tonollii reached 50% mortality after 30 days. Likewise, Rizzo et al.54 investigated antioxidant defenses (capable of counteracting reactive oxygen species (ROS)) in Pam. richtersi in both active and dehydrated states. Activity of several antioxidant enzymes, the fatty acid composition and heat shock protein (Hsp) expression were compared in these two states. The increase in both antioxidant enzyme (superoxide dismutase due to induction of both glutathione peroxidase and glutathione during desiccation) and the fatty acid composition (polyunsaturated fatty acids and the amount of thiobarbituric acid reactive substances) were observed in desiccated Pam. richtersi specimens but no significant differences in the relative level of heat shock proteins were observed (Hsp70 and Hsp90). In addition, Tsujimoto et al.55 performed a study where the production of reactive oxygen species and involvement of bioprotectants during anhydrobiosis in Pam. spatialis was investigated. The study provides evidence of increase in ROS production relative to time spent in anhydrobiosis which is due to oxidative stress in the animals. Using RNA interference, involvement of bioprotectants, including those combating ROS was assessed. As Rizzo et al.54 concluded the role of glutathione peroxidase in desiccation in Pam. richtersi, this gene was targeted and what was observed is that glutathione peroxidase gene compromised survival during drying and rehydration of Pam. spatialis. This furthermore strengthened the evidence that glutathione reductase and catalase play important roles during desiccation and rehydration. Also, involvement of aquaporins 3 and 10 during rehydration of Pam. spatialis was observed. And recently Roszkowska et al.57 study the length that different tardigrades survive in the anhydrobiotic state including Pam. experimentalis. The study concludes that anhydrobiotic competence is dependent on habitat instead of nutritional behavior and the time taken to return to activity after anhydrobiosis is dependent upon the length of the anhydrobiosis.
3. Distribution
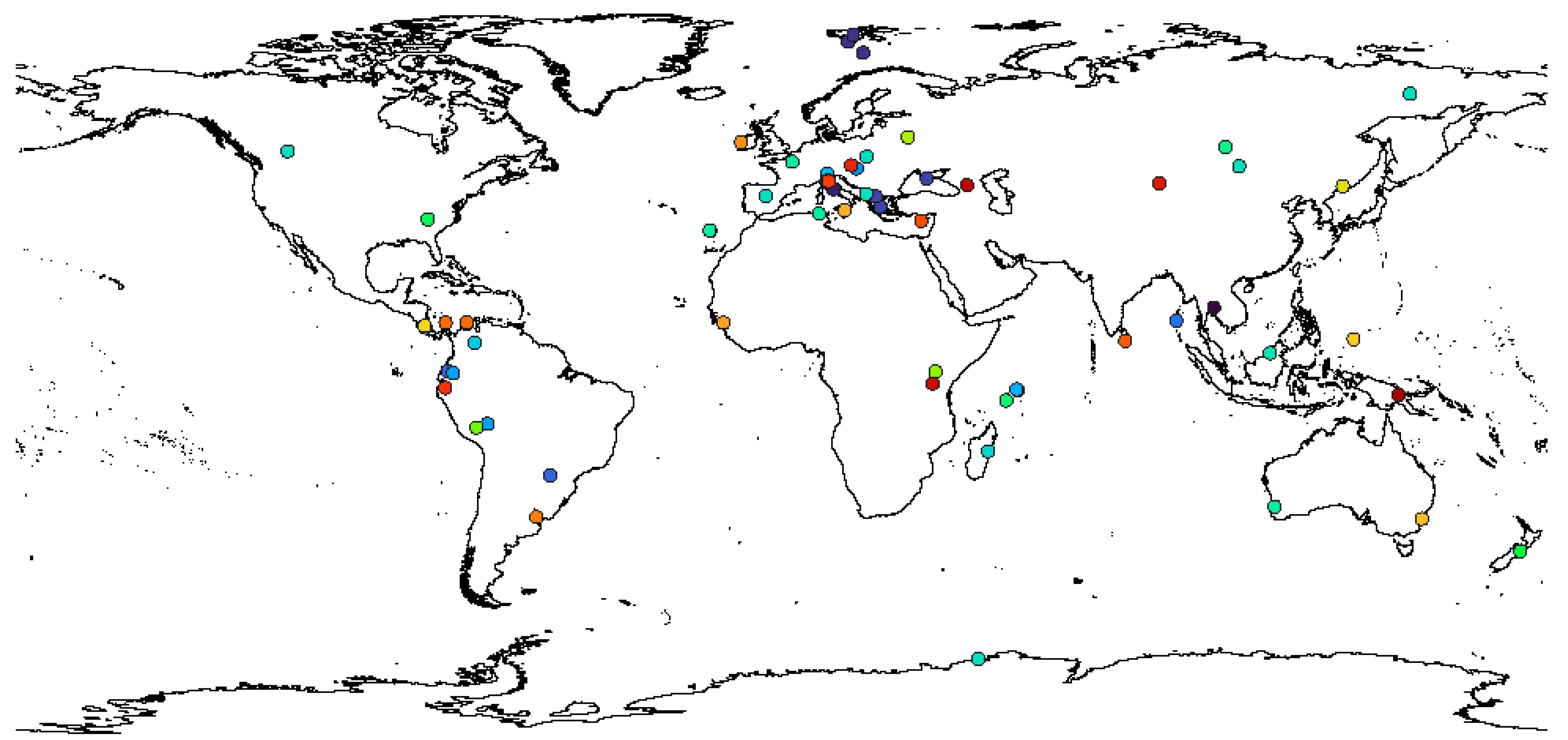
The distribution of species from this genus shows worldwide distribution. The distribution of all 45 species in genus
Paramacrobiotus till date is presented in SM.01 and
Figure 1.
4. Feeding behaviour
The tardigrade species Paramacrobiotus are omnivorous, consumes a variety of organisms, including certain cyanobacteria, algae, and fungi, as well as the rotifer Lecane, the nematode Caenorhabditis, and small juvenile tardigrades. Additionally, the diet of adults and juveniles eat is different: adults favour rotifers and nematodes, whereas juveniles favour unicellular green algae. Moreover, juveniles suck out all of them, including algal cells, animal food, and fungal cells, in contrast to adults who only consume entire fungal and algal cells58.
5. Life history
Life history refers to total life span, development, reproduction and death of an organism59. The life history list in case of tardigrades consists of age at first oviposition, clutch size, fecundity, hatching percentage, hatching success, lifespan, moulting number and total number of ovipositions60,61. The lifespan differs from species to species in case of tardigrades62. The life history of only a few Paramacrobiotus species have been reported till date. Namely, Pam. fairbanski with an average lifespan of 137.3 ± 136.4 days and 194.9 ± 164.4 days respectively and age at first oviposition of 70.7 ± 19.4 days and 76.9 ± 16.4 days respectively63; Pam. kenianus with average lifespan of 125 ± 35 days and 141 ± 54 days respectively, maximum life span of 204 days and 212 days respectively and age at first oviposition of 10 days and10 days respectively60; Pam. metropolitanus with juveniles hatching in 12–20 days, first oviposition in 11–13 days after hatching64; Pam. palaui with average lifespan of 97 ± 31 days, maximum life span of 187 days and age at first oviposition of 10 days60; Pam. richtersi with age at first oviposition of 64.2 ± 1.7 days65; Pam. tonollii with average lifespan of 69.0 ± 45.1 days and maximum life span of 237 days and age at first oviposition 24.4 ± 4.4 days62.
6. Microbiome
The microbiome represents the entire community of microorganisms, including fungi, protists, bacteria, archaea, as well as viruses, that inhabit all known metazoan species. The bacterial component of the microbiome community plays crucial roles in multiple aspects of ecdysozoan host life, such as behavior, metabolism, development, immunity, or pathogen defense, thereby regulating the functioning of the entire organism66,67. Conversely, it has also been demonstrated that the host's phylogeny68 and diet69 have significant impacts on the overall microbial composition. Indeed, many metazoan species appear to harbor their own specific microbiome community70. However, our understanding of the microbiome composition of Tardigrada, based on next-generation sequencing methods (NGS) targeting the standard 16S rRNA bacterial barcoding gene fragment, is limited to a very small number of published articles71–77.
In the case of species from the genus Paramacrobiotus, the microbiomes of a few species have been studied to date. In 2018, Vecchi et al.71 described the bacterial communities associated with six limno-terrestrial tardigrade taxa, one of which was Pam. areolatus. The study revealed that the microbial community was mainly composed of Proteobacteria and Bacteroidetes. Interestingly, certain classified Operational Taxonomic Units (OTUs) showed variations among species from geographically distant samples, indicating the presence of specific bacterial communities in each species. However, in all the investigated species' microbiome profiles, the order Rickettsiales was consistently identified. This order belongs to the class Alphaproteobacteria and is characterized by both pathogens and intracellular mutualists78. There were two distinct patterns in the diversity observed between tardigrades and their substrates, indicating significantly less microbial diversity in tardigrades compared to their substrates. This phenomenon may be attributed to tardigrades selectively associating with specific microbial communities that promote the growth of certain bacterial species while inhibiting others. Another hypothesis suggests that substrates, being complex matrices with wide surface areas and volumes, can support a high bacterial biomass, resulting in a vast and complex microbial community.
Similarly, Kaczmarek et al.21 conducted a microbiome analysis on two populations of Pam. experimentalis from Madagascar and their laboratory culture environment. These populations of Pam. experimentalis had been maintained in laboratory culture for two years. The most abundant phylum in all samples was Proteobacteria. Firmicutes was the second most dominant phylum in both Pam. experimentalis populations, while Bacteroides was the second most dominant phylum in the laboratory habitat. With the exception of the phyla Verrucomicrobia and Saccharibacteria, which were not found in the tardigrade microbiome, all identified taxa in the Pam. experimentalis microbiome community and laboratory culture environment were widespread and had comparable abundances. This confirms that the tardigrade microbiome significantly differs in composition from the bacteria inhabiting their environment. Moreover, within the microbiome of Pam. experimentalis, Operational Taxonomic Units (OTUs) classified as potential endosymbionts belonging to the order Rickettsiales were identified. The absence of Rickettsiales OTUs in the environment of the studied species further supports the close association of these bacteria with their host.
Furthermore, Mioduchowska et al.73 conducted a study to investigate whether tardigrade species are infected with bacterial endosymbionts belonging to the genus Wolbachia. The analysis included Pam. fairbanksi and Paramacrobiotus sp. In the study Proteobacteria, Firmicutes, and Actinobacteria as the three most prevalent phyla among the analyzed tardigrades, including species outside the genus Paramacrobiotus, were identified. However, the focus of the study was on potential tardigrade endosymbionts, particularly Operational Taxonomic Units (OTUs) from the order Rickettsiales and the genus Wolbachia. Both Rickettsiales and Wolbachia were detected in adult Paramacrobiotus sp., while only Rickettsiales were found in Pam. fairbanksi eggs. Adult Pam. fairbanksi did not have either Wolbachia or Rickettsiales infections. The genus Wolbachia is an intracellular bacterium belonging to the order Rickettsiales and it infects various invertebrates, particularly terrestrial insects79. However, recent studies have identified infections of this bacterial endosymbiont in various freshwater invertebrate species77,80,81. Generally, this bacterium is transmitted vertically from mother to offspring and/or through horizontal transfer directly from the environment or between different hosts82. Subsequently, Wolbachia manipulates host reproduction by inducing parthenogenesis, feminization, male killing, or cytoplasmic incompatibility83,84.
In 2023, Mioduchowska et al.77 described new molecular and bioinformatic tools for detecting Wolbachia in freshwater invertebrates. In this study, Wolbachia was detected in Pam. experimentalis, which were the same isolates analysed by Kaczmarek et al.72. Phylogenetic analysis of the obtained bacterial sequences allowed for their classification within the differentiated supergroup A of the genus Wolbachia. The discovery of Wolbachia in tardigrades opens new frontiers in understanding the Wolbachia-driven biology and ecology of Tardigrada.
7. Reproduction
Reproduction refers to the process where every organism known produces offspring either sexually or asexually. In case of tardigrades, they reproduce only through gametes via many different patterns i.e. dioecious (separate male and female), hermaphroditic (single animal with both male and female reproductive parts) or parthenogenetic (form of asexual reproduction)85. The genus Paramacrobiotus consists of both bisexual and unisexual species/populations. Pam. richtersi is both bisexual and unisexual from Italy; according to modern taxonomy they probably constitute a distinct species, Pam. areolatus population from Italy is bisexual and population from Svalbard is unisexual, Pam. tonolli from the USA is bisexual, Pam. fairbanksi is unisexual from various locations as Antarctic, Italy, Poland, Spain and USA, Pam. kenianus from Kenya is unisexual and Pam. palaui from Micronesia is unisexual, Pam. depressus from Italy is bisexual, Pam. celsus from Italy is bisexual, Pam. spatialis from Italy is bisexual, Pam. arduus from Italy is bisexual, Pam. experimentalis from Madagascar is bisexual and Pam. gadabouti is unisexual from various locations in Portugal, Australia, France and Tunisia. Out of 45, mode of reproduction for only 18 species are known (SM.01). Also, Guidetti et al.11 suggests the mode of reproduction being related to constrained or wide distribution of the species. The amphimictic species displays a very constrained or punctiform distribution, in contrast to the parthenogenetic species' extremely extensive spread and presence over multiple continents. The difference in the ability for dispersal linked to the two modes of reproduction can be used to explain why apomictic and amphimictic populations are distributed differently.
8. Morphological taxonomy
The genus Paramacrobiotus is divided into two morphologically distinct species groups: areolatus (species without a microplacoid or with rudimentary structures in the place of microplacoid in the pharynx) and richtersi (species with a microplacoid in the pharynx)( e.g.23,28). It was suggested that initially the microplacoid was present, however it was lost in some species from the areolatus group. But, the opposite situation, in which the microplacoid gradually appears, is also possible41. For example, in Pam vanescens the microplacoid suggests a gradual reduction. In turn, in Pam. areolatus and Pam. centesimus the microplacoid is generally absent, but a thin cuticular thickening is present in the place where microplacoid should be normally present and can be considered as rudimentary microplacoid14,47. Although, the presence or absence of microplacoid seems to be a clear morphological character dividing genus Paramacrobiotus into two separate phylogenetic lineages (which was suggested by Kaczmarek et al.41) the genetic studies did not confirm this11,47.
At present 45 species are formally attributed to the genus Paramacrobiotus and 13 belong to areolatus group and 32 richtersi group. They can be further divided into smaller groups based on egg types. In total, seven types of eggs were identified. However, two of them (areolatus and richtersi types) are the most common and occur in 37 species (ca. 82%). In the next two species huziori type of eggs are present (ca. 5%). The other types of eggs (i.e. beotiae, chieregoi, csotiensis, tonollii and submorulatus) were identified only in single taxa (for details of egg morphology see Kaczmarek et al.41). What is more, eggs are unknown for one species i.e. Pam. wauensis.
In recent years two very important for taxonomy of the entire genus, species Pam. areolatus and Pam. richtersi were integratively redescribed11,47. Another species Pam. fairbanksi described based, mostly, on genetic data was also morphometrically well characterized few years ago21. However, a few Paramacrobiotus species still need a redescription based on type material or on additional material from type localities. Descriptions of Pam. beotiae, Pam. chieregoi, Pam. csotiensis, Pam. rioplatensis, Pam. submorulatus, Pam. tonollii and Pam. wauensis are inaccurate and some important morphological information are lacking.
Another two species, i.e., Pam. kenianus and Pam. palaui are cryptic taxa described mostly based on genetic data without morphological differential diagnosis22.
Descriptions of the other
Paramacrobiotus species more or less complete, but in most of them exact morphometric data of claws, buccal tubes placoids and above all genetic data are lacking (see
Table 1 and SM.01). Based on all the abovementioned doubts, 3 species, i.e.,
Pam. kenianus,
Pam. palaui and
Pam. wauensis were not included to the key.
9. Molecular taxonomy
Molecular markers serve as valuable tools for species identification. In the integrative taxonomy of Tardigrada, four DNA fragments with different mutation rates are commonly used: two conservative nuclear ribosomal subunit genes, namely 18S rRNA (the small ribosome subunit) and 28S rRNA (the large ribosome subunit), the noncoding nuclear ITS2 fragment (the internal transcribed spacer-2) with high evolution rates, and the protein coding mitochondrial COI barcode gene (the cytochrome oxidase subunit I) with an intermediate effective mutation rate (e.g., Kaczmarek
et al.72). The COI mtDNA molecular marker, in particular, has been recommended for DNA barcoding purposes (
http://www.barcodinglife.org), such as rapid species identification, discrimination between cryptic species, and resolving phylogenetic relationships among closely related species
86,87. To gain additional insights into the phylogenetic relationships within the genus
Paramacrobiotus, an analysis based on COI mtDNA was conducted. This analysis was performed to supplement the information obtained from previous studies using four molecular markers
24.
Due to ongoing revisions and redescriptions of Paramacrobiotus species, studies are becoming more accessible, leading us to anticipate that the species diversity within the genus is greatly underestimated11,23. One significant challenge that needs to be addressed in future studies is the lack of available barcodes. Despite the designation of 45 species to the genus Paramacrobiotus, not all species have available barcode sequences. In this study, we aimed to estimate the phylogenetic relationships among all Paramacrobiotus species (including taxa designated as "cf." – meaning "compare with" and "aff." – meaning "similar to") for which COI barcode sequences are available in the GenBank database. We used the COI sequence of Milnesium berladnicorum Ciobanu, Zawierucha, Moglan & Kaczmarek, 201488 as outgroups to construct the most reliable evolutionary tree. To determine the most appropriate model of sequence evolution, we applied jModelTest v. 2.1.489 with both the Bayesian Information Criterion (BIC) and the Akaike Information Criterion (AIC)90. The GTR + G (Time Reversible model with gamma distributed rate heterogeneity) was selected as the best-fit evolutionary model. The phylogenetic tree was constructed using Bayesian inference (BI) analysis with the program MrBayes 391, following the settings described by Mioduchowska et al.92. The alignment of COI barcode sequences resulted in 574 characters, with 270 variable sites and 241 parsimony informative sites. Uncorrected pairwise distances (p-distances) were calculated using MEGA X93.
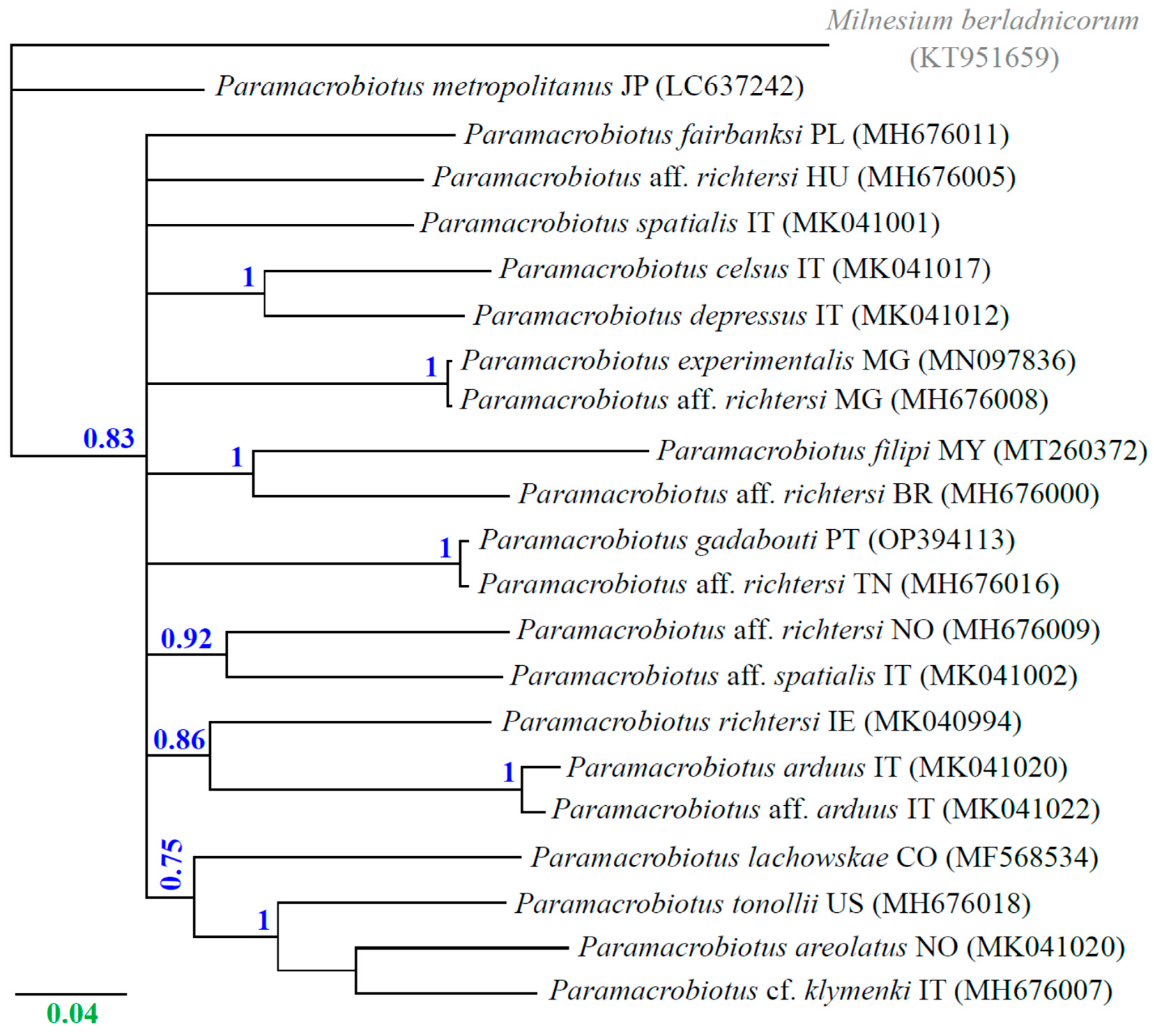
The binary model of phylogenetic relationships, which involves reconstructing gene trees from sequence data, allows us to gain insights into the speciation history of species
94. However, in our analysis of barcode sequences, we observed speciation events that resulted in polytomies within the phylogeny of the genus
Paramacrobiotus (
Figure 2). This means that more than two descendants were observed from certain nodes
95. The presence of unresolved nodes in a polytomic multifurcating tree indicates a lack of signal in the data to resolve relationships within the genus
Paramacrobiotus. This observation is partially consistent with previous studies, where both groups of
richtersi and
areolatus were described as polyphyletic
11,47. However, in the work by Kayastha
et al.24, the interrelationships of the genus
Paramacrobiotus were not depicted as a polytomy when two conservative coding nuclear molecular markers (18S rRNA and 28S rRNA) and a noncoding nuclear marker with high evolution rates (ITS2) were included in the analysis. As a result, the phylogenetic relationships within the genus
Paramacrobiotus were resolved. Interestingly, other examples of polytomies in Tardigrada gene trees based on nuclear molecular markers have also been observed
96.
The genetic p-distances between the analyzed COI barcode sequences of
Paramacrobiotus species ranged from 16% to 27%, indicating different species (
Table 2). However, it was shown that there are very low genetic differences, i.e., a p-distance of 0.3%, between
Pam. aff.
richtersii from Tunisia (GenBank: MH676016) and
Pam. gadabouti from Portugal (GenBank: OP394113), suggesting they belong to the same species (
Table 2). This finding is consistent with the work by Kayastha
et al. 24, where both species were described as
Pam. gadabouti. No genetic differences were found between
Pam. aff.
richtersi from Madagascar (GenBank: MH676008) and
Pam. experimentalis from Madagascar (GenBank: MN097836) (
Table 2). Both sequences represented
Pam. experimentalis, which is also consistent with the previous study (Kayastha
et al.24). Moreover, we found very low genetic differences, i.e., a p-distance of 2.1%, between
Pam. arduus from Italy (GenBank: MK041020) and
Pam. aff.
arduus from Italy (GenBank: MK041022), indicating the same species (
Table 2).
10. Key for species identification
1. Microplacoid present (richtersi group) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2
–. Microplacoid absent (areolatus group) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31
2. Cuticular pattern on dorsal side of the body present and visible in LM (PCM and/or DIC) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
–. Cuticle on dorsal side of the body smooth or cuticular pattern not visible in LM (PCM and/or DIC) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
3. Eggs of areolatus type. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. danielae
–. Eggs of richtersi type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . 4
4. Eyes present, lunules under claws IV dentate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. corgatensis
–. Eyes absent, lunules under claws IV smooth . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 5
5. Dorsal cuticle covered with very small circular or elongated tubercles, egg processes less than 14.5 μm height . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. halei
–. Dorsal cuticle covered with small dots (granules) or small polygons, egg processes more than 15.5 μm height . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
6. Dorsal cuticle covered with small dots (granules) . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. vanescens
–. Dorsal cuticle covered with small polygons . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. danielisae
7. Areolation between egg processes absent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
–. Areolation between egg processes present . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
8. Lunules under claws IV dentate, eggs of beotiae type . . . . . . . . . . .. . . . . .. . . . . . . . . . . . . Pam. beotiae
–. Lunules under claws IV smooth, egg of chieregoi type . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. chieregoi
9. Eggs of submorulatus type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . Pam. submorulatus
–. Eggs of richtersi or areolatus type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
10. Eggs of richtersi type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .11
–. Eggs of areolatus type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 25
11. Only five or six areoles present around each egg process . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
–. The number of areoles around each egg process larger than six . . . . . . . .. . . . . . . . . . . . . . . . . . . . . 18
12. Eyes present. . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. priviterae
–. Eyes absent. . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .13
13. Granulation on leg I-III present. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . .14
–. Granulation on legs I-III absent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. depressus
14. The pt values of the macroplacoid length less than 43.5. . .. . . .. . . . . . . . . . . . . . . . . . . . . . . Pam. pius
–. The pt values of the macroplacoid length more than 49.0. . . .. . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . 15
15. Egg process jagged . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . .. . . . . .. . . .. . . . . . . . . . . . . . . . . . . . . . 16
–. Egg process not jagged . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
16. Egg processes height less than 15.0 μm and parthenogenetic mode of reproduction. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. fairbanksi
–. Egg processes height more than 15.1 μm and bisexual mode of reproduction. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. celsus
17. Egg diameter without processes less than 62.5 μm. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. arduus
–. Egg diameter without processes more than 65.0 μm. . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. spatialis
18. Eyes present . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 19
–. Eyes absent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 21
19. Granulation on legs I-III present . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20
–. Granulation on legs I-III absent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. magdalenae
20. Egg bare diameter less than 87.9 μm, egg process height more than 15 μm, egg processes hemispherical with blunt terminal part . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. sklodowskae
–. Egg bare diameter more than 92.0 μm, egg process height less than 13.5 μm, egg processes hemispherical with cylindrical indented apices . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. sagani
21. Lunules under claws IV dentate. . . . . . . . . .. . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . .Pam. alekseevi
–. Lunules under claws IV smooth. . . . . . . . . .. . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . .22
22. Egg processes with cap-like vesicular structures on the top . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 23
– Egg processes without cap-like vesicular structures on the top. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 24
23. Egg processes with elongated terminal portion, second macroplacoid length less than 6.5 μm, pt values of second macroplacoid length less than 14.0, pt values of macroplacoid row length less than 59.0, placoid row length less than 34.5 μm and pt values of placoid row length less than 74.0. . .. ... . . . . . . . . . . . . . . . .. ... . . . . . ... .. . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. filipi
– Egg processes without elongated terminal portion, second macroplacoid length 7.0 μm or more, pt values of second macroplacoid length more than 15.0, pt values of macroplacoid row length more than 60.0, placoid row length more than 34.9 μm and pt values of placoid row length more than 77.5. . . . . .. . . . . . . . . . . . .. .. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. gadabouti
24. Egg processes with long, thin and flexible terminal portion and egg process height more than 24.5 μm . . . . . . .. . . . . . .. . . . . . .. . . . . . .. . . . . . .. . . . . . .. . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. lorenae
– Egg processes without long, thin and flexible terminal portions and egg process height less than 22.5 μm . . . . . . . .. . . . . . .. . . . .. . . . . . .. . . . . . .. . . . .. . . . . .. . . . .. . . . . . . . . . . . . . . . . . . . . . . Pam. richtersi
25. Cuticle with oval pores, egg processes with cap-like structure on the top and clearly narrower under caps . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. garynahi
–. Cuticle without oval pores, egg processes without cap-like structure on the top and without narrowing at the top . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .26
26. Egg processes hemispherical with blunt apex not divided and without elongated terminal part . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. savai
–. Egg processes different . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . 27
27. Egg processes with long flexible spines on the top . . . . . . . . . . . . . . . .. . . . . . . . .Pam. rioplatensis
–. Egg processes without long flexible spines on the top . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .28
28. Egg processes base width less than 12.5 μm. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. peteri
–. Egg processes base width more than 13.0 μm. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29
29. Granulation on IVth pair of legs absent and egg processes height more than 15.5 μm . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. hapukuensis
–. Granulation on IVth pair of legs present and egg processes height less than 15.0 μm . 30
30. Presence of wrinkled surface on the egg areolae and the absence of cuticular bulge on inner surface of claws I–III. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ..Pam. experimentalis
–. Lack of wrinkled surface on the egg areolae and the presence of cuticular bulge on inner surface of claws I–III. . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . .. . . . . .. . .. . . . . . . . . . Pam. metropolitanus
31. Egg of csotiensis type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. csotiensis
–. Eggs of areolatus, huziori, tonollii or richtersi type . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . 32
32. Eggs of tonollii type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. tonollii
–. Eggs of areolatus, huziori or richtersi type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . .33
33. The egg areolation of the huziori type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .34
–. Eggs of richtersi or areolatus type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .35
34. Only one row of larger teeth present in the second band in the oral cavity, the distances between all macroplacoids are approximately the same, accessory points well developed but not protruding high above the primary branch, diameter of bases of egg processes approximately equal to or slightly smaller than their height, 9–11 processes on egg circumference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . Pam. huziori
–. A row of larger teeth and a posterior band of small granules/conical teeth present in the second band of teeth in the oral cavity, the second macroplacoid situated closer to the first than to the third macroplacoid, accessory points extremely well developed, protruding high above the primary branch, diameter of bases of egg processes greater than their height, 12–16 processes on egg circumference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. derkai
35. Eggs of richtersi type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. spinosus
–. Eggs of areolatus type . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 36
36. The first/anterior band of teeth visible under PCM . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 37
–. The first/anterior band of teeth absent or not visible under PCM . . . . . . . . . . . . . . . . . . . . .Pam. intii
37. Lunules under claws IV smooth. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .38
–. Lunules under claws IV dentate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .39
38. Eyes present, macroplacoid length sequence 2<3<1, full egg diameter more than 93.0 μm and egg process height more than 17.5 μm. . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . . . . . . . .Pam. lachowskae
–. Eyes absent, macroplacoid length sequence 2<1<3, full egg diameter less than 92.0 μm and egg process height less than 11.5 μm . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ..Pam. centesimus
39. Eyes present, macroplacoid length sequence 2<1<3 and egg processes elongated . . . . . . . . . . . . . .40
–. Eyes absent, macroplacoid length sequence 2<3<1 and egg processes short. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. klymenki
40. Egg process height more than 26.5 μm and egg process surface smooth . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . Pam. areolatus
–. Egg process height less than 17.5 μm and egg process surface apically covered by irregular granulation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . . . . . . . . . . . .. . . . . . . . . . . . Pam. walteri
5. Conclusions
The genus Paramacrobiotus shows cosmopolitan distribution with presence of both bisexual and parthenogenetic species. Although the integrative descriptions and redescriptions are improving the overall situation and allowing for fresh opportunities for detailed study, the phylogeny of the genus Paramacrobiotus seems to be unresolved. Also, there are many other studies regarding life-history, cryptobiotic abilities and microbiome community as well as bacterial endosymbiont infections identification, which are lacking, and such studies are required for the advancement of tardigrade knowledge in general.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. SM.01 Locations, mode of reproduction and presence of genetic data for all the
Paramacrobiotus species.
Author Contributions
Conceptualization, P.K. and Ł.K.; methodology, P.K. and M.M.; formal analysis, P.K. and M.M.; investigation, P.K.; data curation, P.K.; writing—original draft preparation, P.K.; writing—review and editing, P.K., M.M. and Ł.K; visualization, P.K. and M.M.; supervision, Ł.K. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All the DNA sequences data are from GenBank.
Acknowledgments
Studies have been partially conducted in the framework of activities of BARg (Biodiversity and Astrobiology Research group). PK is scholarship holder of Passport to the future - Interdisci-plinary doctoral studies at the Faculty of Biology, Adam Mickiewicz University, Poznań POWR.03.02.00-00-I006/17. The work of MM was supported by National Science Centre, Poland, grant no. 2021/43/D/NZ8/00344 and grant no. 1220/146/2021 from the Small Grants Pro-gramme of the University of Gdansk (i.e., Ugrants-first competition). Tomasz Bartylak for helping with QGIS.
Conflicts of Interest
The authors declare no competing interests.
References
- Guidetti R, Bertolani RB. Tardigrade taxonomy: an updated check list of the taxa and a list of characters for their identification. Zootaxa. 2005;845(1):1. [CrossRef]
- Degma P, Guidetti R. Notes to the current checklist of Tardigrada. Zootaxa. 2007;1579(1):41-53. [CrossRef]
- Vicente F, Bertolani R. Considerations on the taxonomy of the Phylum Tardigrada. Zootaxa. 2013;3626(2):245-248. [CrossRef]
- Degma P, Guidetti R. Actual checklist of Tardigrada species. 2009-2023. [CrossRef]
- Nelson DR, Guidetti R, Rebecchi L, Kaczmarek Ł, McInnes S. Phylum Tardigrada. In: Thorp and Covich’s Freshwater Invertebrates. Elsevier; 2020:505-522. [CrossRef]
- Ramazzotti G, Maucci W. II Phylum Tardigrada. III. Edizione Riveduta e Aggiornata.(II Phylum Tardigrada. 3rd ed.; 1983.
- Beasley CW. The Phylum Tardigrada. Third Edition by G. Ramazzotti and W. Maucci, English Translation. P. Abilene, USA; 1995.
- Thulin G. Über die phylogenie und das system der. Hereditas. 1928;11(2-3):207-266. [CrossRef]
- Guidetti R, Schill RO, Bertolani R, Dandekar T, Wolf M. New molecular data for tardigrade phylogeny, with the erection of Paramacrobiotus gen. nov. J Zool Syst Evol Res. 2009;47(4):315-321. [CrossRef]
- Tumanov DV. Notes on the tardigrada of Thailand, with a description of Macrobiotus alekseevi sp. nov. (Eutardigrada, Macrobiotidae). Zootaxa. 2005;999(1):1. [CrossRef]
- Guidetti R, Cesari M, Bertolani R, Altiero T, Rebecchi L. High diversity in species, reproductive modes and distribution within the Paramacrobiotus richtersi complex (Eutardigrada, Macrobiotidae). Zool Lett. 2019;5(1):1. [CrossRef]
- Murray J. XXV.—Arctic Tardigrada, collected by Wm. S. Bruce. Earth Environ Sci Trans R Soc Edinb. 1907;45(3):669-681. [CrossRef]
- Durante Pasa M, Maucci W. Moss Tardigrada from the Scandinavian Peninsula. Zesz Nauk Uniw Jagiell Pr Zool Kraków. 1979;79(25):47-85.
- Pilato G. Macrobiotus centesimus, new species of eutardigrade from the South America. Boll Delle Sedute Della Accad Gioenia Sci Nat Catania. 2000;33:97-101.
- Maucci W, Durante Pasa MV. Tardigradi muscicoli delle isole Andamane. Bollettino del Museo Civico di Storia Naturale di Verona. Published online 1980:281-291.
- Pilato G, Binda MG, Lisi O. Notes on tardigrades of the Seychelles with the description of three new species. Ital J Zool. 2004;71(2):171-178. [CrossRef]
- Iharos G. Neue Tardigraden-arten aus Ungarn. Acta Zool Acad Sci Hung. 1966;12(1-2):111-122.
- Pilato G, Binda MG, Napolitano A, Moncada E. Notes on South American tardigrades with the description of two new species: Pseudechiniscus spinerectus and Macrobiotus danielae. Trop Zool. 2001;14(2):223-231. [CrossRef]
- Pilato G, Binda MG, Lisi O. Three new species of eutardigrades from the Seychelles. N Z J Zool. 2006;33(1):39-48. [CrossRef]
- Degma P, Michalczyk Ł, Kaczmarek Ł. Macrobiotus derkai, a new species of Tardigrada (Eutardigrada, Macrobiotidae, huziori group) from the Colombian Andes (South America). Zootaxa. 2008;1731(1):1. [CrossRef]
- Kaczmarek Ł, Mioduchowska M, Kačarević U, et al. New records of Antarctic tardigrada with comments on interpopulation variability of the Paramacrobiotus fairbanksi Schill, Förster, Dandekar and Wolf, 2010. Diversity. 2020;12(3):108. [CrossRef]
- Schill RO, Förster F, Dandekar T, Wolf M. Using compensatory base change analysis of internal transcribed spacer 2 secondary structures to identify three new species in Paramacrobiotus (Tardigrada). Org Divers Evol. 2010;10(4):287-296. [CrossRef]
- Stec D, Dudziak M, Michalczyk Ł. Integrative descriptions of two new Macrobiotidae species (Tardigrada: Eutardigrada: Macrobiotoidea) from French Guiana and Malaysian Borneo. Zool Stud. 2020;59:e23. [CrossRef]
- Kayastha P, Stec D, Sługocki Ł, Gawlak M, Mioduchowska M, Kaczmarek Ł. Integrative taxonomy reveals new, widely distributed tardigrade species of the genus Paramacrobiotus (Eutardigrada: Macrobiotidae). Sci Rep. 2023;13(1):2196. [CrossRef]
- Kaczmarek Ł, Michalczyk Ł, Diduszko D. Some tardigrades from Siberia (Russia, Baikal region) with a description of Macrobiotus garynahi sp. nov. (Eutardigrada: Macrobiotidae: richtersi. Zootaxa. 2005;1053(1):35-45. [CrossRef]
- Bartels PJ, Pilato G, Lisi O, Nelson DR. Macrobiotus (Eutardigrada, Macrobiotidae) from the Great Smoky Mountains National Park, Tennessee/North Carolina, USA (North America): two new species and six new records. Zootaxa. 2009;2022(1):45-57. [CrossRef]
- Michalczyk L, Kaczmarek L. A new species Macrobiotus magdalenae (Tardigrada: Eutardigrada: Macrobiotidae, richtersi group) from Costa Rican rain forest (Central America). N Z J Zool. 2006;33(3):189-196. [CrossRef]
- Kaczmarek Ł, Cytan J, Zawierucha K, Diduszko D, Michalczyk Ł. Tardigrades from Peru (South America), with descriptions of three new species of Parachela. Zootaxa. 2014;3790(2):357. [CrossRef]
- Pilato G, Kiosya Y, Lisi O, Sabella G. New records of Eutardigrada from Belarus with the description of three new species. Zootaxa. 2012;3179(1):39. [CrossRef]
- Stec D, Roszkowska M, Kaczmarek Ł, Michalczyk Ł. Paramacrobiotus lachowskae, a new species of Tardigrada from Colombia (Eutardigrada: Parachela: Macrobiotidae). N Z J Zool. 2018;45(1):43-60. [CrossRef]
- Biserov VI. Macrobiotus lorenae sp. n., a new species of Tardigrada (Eutardigrada Macrobiotidae) from the Russian Far East. Arthr Sel. 1996;5:145-149.
- Sugiura K, Matsumoto M, Kunieda T. Description of a model tardigrade Paramacrobiotus metropolitanus sp. nov. (Eutardigrada) from Japan with a summary of its life history, reproduction and genomics. Zootaxa. 2022;5134(1):92-112. [CrossRef]
- Pilato G, Claxton S, Binda M. Tardigrades from Australia. III. Echiniscus marcusi and Macrobiotus peteri, new species of tardigrades from New South Wales. Animalia. 1989;16:43-48.
- Lisi O, Binda MG, Pilato G. Eremobiotus ginevrae sp. nov. and Paramacrobiotus pius sp. nov., two new species of Eutardigrada. Zootaxa. 2016;4103(4):344. [CrossRef]
- Binda MG, Pilato G, Moncada E, Napolitano A. Some tardigrades from Central Africa with the description of two new species: Macrobiotus ragonesei and M. priviterae (Eutardigrada Macrobiotidae). Trop Zool. 2001;14(2):233-242. [CrossRef]
- Murray J. Scottish Tardigrada. A review of our present knowledge. Ann Scott Nat Hist. 1911;78:88-95.
- Claps M, Rossi G. Tardígrados de Uruguay, con descripción de dos nuevas especies (Echiniscidae, Macrobiotidae). Iheringia Sér Zool. 1997;83:17-22.
- Daza A, Caicedo M, Lisi O, Quiroga S. New records of tardigrades from Colombia with the description of Paramacrobiotus sagani sp. nov. and Doryphoribius rosanae sp. nov. Zootaxa. 2017;4362(1). [CrossRef]
- Binda MG, Pilato G. Macrobiotus savai and Macrobiotus humilis, two new species of tardigrades from Sri Lanka. Boll DellAccademia Gioenia Sci Nat. 2001;34:101-111.
- Michalczyk Ł, Kaczmarek Ł, Węglarska B. Macrobiotus sklodowskae sp. nov. (Tardigrada: Eutardigrada: Macrobiotidae, richtersi group) from Cyprus. Zootaxa. 2006;1371(1):45. [CrossRef]
- Kaczmarek Ł, Gawlak M, Bartels PJ, Nelson DR, Roszkowska M. Revision of the Genus Paramacrobiotus Guidetti et al ., 2009 with the description of a new species, re-descriptions and a key. Ann Zool. 2017;67(4):627-656. [CrossRef]
- Ramazzotti G. Tre nouve specie di Tardigradi ed altre specie poco comuni. Atti Soc Nat Milano. 1956;10:284-291.
- Pilato G, Binda MG, Catanzaro R. Remarks on some tardigrades of the African fauna with the description of three new species of Macrobiotus Schultze 1834. Trop Zool. 1991;4(2):167-178. [CrossRef]
- Biserov VI. Tardigrades of the Caucasus with a taxonomic analysis of the genus Ramazzottius (Parachela: Hypsibiidae). Zool Anz. 1998;236(1997):139-159.
- Iharos G. Neuere Daten zur Kenntnis der Tardigraden-Fauna von Neuguinea. Opusc Zool Bp. 1973;11:65-73.
- Marley NJ, Kaczmarek Ł, Gawlak M, et al. A clarification for the subgenera of Paramacrobiotus Guidetti, Schill, Bertolani, Dandekar and Wolf, 2009, with respect to the International Code of Zoological Nomenclature. Zootaxa. 2018;4407(1). [CrossRef]
- Stec D, Krzywański Ł, Zawierucha K, Michalczyk Ł. Untangling systematics of the Paramacrobiotus areolatus species complex by an integrative redescription of the nominal species for the group, with multilocus phylogeny and species delineation in the genus Paramacrobiotus. Zool J Linn Soc. 2020;188(3):694-716. [CrossRef]
- Keilin D. The Leeuwenhoek Lecture - The problem of anabiosis or latent life: history and current concept. Proc R Soc Lond Ser B - Biol Sci. 1959;150(939):149-191. [CrossRef]
- Rebecchi L, Altiero T, Guidetti R. Anhydrobiosis: the extreme limit of desiccation tolerance. Invertebr Surviv J. 2007;4(2):65-81.
- Guidetti R, Altiero T, Rebecchi L. On dormancy strategies in tardigrades. J Insect Physiol. 2011;57(5):567-576. [CrossRef]
- Møbjerg N, Halberg KA, Jørgensen A, et al. Survival in extreme environments - on the current knowledge of adaptations in tardigrades: Adaptation to extreme environments in tardigrades. Acta Physiol. 2011;202(3):409-420. [CrossRef]
- Greven H. From johann August Ephraim Goeze to Ernst Marcus: A ramble through the history of early tardigrade research (1773 Until 1929). In: Schill RO, ed. Water Bears: The Biology of Tardigrades. Vol 2. Zoological Monographs. Springer International Publishing; 2018:1-55. [CrossRef]
- Reuner A, Hengherr S, Brümmer F, Schill RO. Comparative studies on storage cells in tardigrades during starvation and anhydrobiosis. Curr Zool. 2010;56(2):259-263. [CrossRef]
- Rizzo AM, Negroni M, Altiero T, et al. Antioxidant defences in hydrated and desiccated states of the tardigrade Paramacrobiotus richtersi. Comp Biochem Physiol B Biochem Mol Biol. 2010;156(2):115-121. [CrossRef]
- Tsujimoto M, Imura S, Kanda H. Recovery and reproduction of an Antarctic tardigrade retrieved from a moss sample frozen for over 30 years. Cryobiology. 2016;72(1):78-81. [CrossRef]
- Giovannini I, Boothby TC, Cesari M, Goldstein B, Guidetti R, Rebecchi L. Production of reactive oxygen species and involvement of bioprotectants during anhydrobiosis in the tardigrade Paramacrobiotus spatialis. Sci Rep. 2022;12(1):1938. [CrossRef]
- Roszkowska M, Gołdyn B, Wojciechowska D, et al. How long can tardigrades survive in the anhydrobiotic state? A search for tardigrade anhydrobiosis patterns. Klymkowsky M, ed. PLOS ONE. 2023;18(1):e0270386. [CrossRef]
- Bryndová M, Stec D, Schill RO, Michalczyk Ł, Devetter M. Dietary preferences and diet effects on life-history traits of tardigrades. Zool J Linn Soc. 2020;188(3):865-877. [CrossRef]
- Nylin S, Gotthard K. Plasticity in life-history traits. Annu Rev Entomol. 1998;43(1):63-83. [CrossRef]
- Schill RO. Life-history traits in the tardigrade species Paramacrobiotus kenianus and Paramacrobiotus palaui. J Limnol. 2013;72(1s):e20. [CrossRef]
- Ito M, Saigo T, Abe W, Kubo T, Kunieda T. Establishment of an isogenic strain of the desiccation-sensitive tardigrade Isohypsibius myrops (Parachela, Eutardigrada) and its life history traits. Zool J Linn Soc. 2016;178(4):863-870. [CrossRef]
- Lemloh M, Brümmer F, Schill RO. Life-history traits of the bisexual tardigrades Paramacrobiotus tonollii and Macrobiotus sapiens. J Zool Syst Evol Res. 2011;49(s1):58-61. [CrossRef]
- Altiero T, Rebecchi L, Bertolani R. Phenotypic variations in the life History of two clones of Macrobiotus richtersi (Eutardigrada, Macrobiotidae). Hydrobiologia. 2006;558(1):33-40. [CrossRef]
- Sugiura K, Minato H, Suzuki AC, Arakawa K, Kunieda T, Matsumoto M. Comparison of sexual reproductive behaviors in two species of Macrobiotidae (Tardigrada: Eutardigrada). Zoolog Sci. 2019;36(2):120. [CrossRef]
- Hohberg K. Tardigrade species composition in young soils and some aspects on life history of Macrobiotus richtersi J. Murray, 1911. Pedobiologia. 2006;50(3):267-274. [CrossRef]
- Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Animal Behavior and the Microbiome. Science. 2012;338(6104):198-199. [CrossRef]
- Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685-690. [CrossRef]
- Ramalho MO, Bueno OC, Moreau CS. Microbial composition of spiny ants (Hymenoptera: Formicidae: Polyrhachis) across their geographic range. BMC Evol Biol. 2017;17(1):96. [CrossRef]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: A Metagenomic analysis in humanized gnotobiotic Mice. Sci Transl Med. 2009;1(6). [CrossRef]
- Derycke S, De Meester N, Rigaux A, et al. Coexisting cryptic species of the Litoditis marina complex (Nematoda) show differential resource use and have distinct microbiomes with high intraspecific variability. Mol Ecol. 2016;25(9):2093-2110. [CrossRef]
- Vecchi M, Newton ILG, Cesari M, Rebecchi L, Guidetti R. The microbial community of tardigrades: Environmental influence and species specificity of microbiome structure and composition. Microb Ecol. 2018;76(2):467-481. [CrossRef]
- Kaczmarek Ł, Roszkowska M, Poprawa I, et al. Integrative description of bisexual Paramacrobiotus experimentalis sp. nov. (Macrobiotidae) from republic of Madagascar (Africa) with microbiome analysis. Mol Phylogenet Evol. 2020;145:106730. [CrossRef]
- Mioduchowska M, Nitkiewicz B, Roszkowska M, et al. Taxonomic classification of the bacterial endosymbiont Wolbachia based on next-generation sequencing: is there molecular evidence for its presence in tardigrades? Genome. 2021;64(10):951-958. [CrossRef]
- McQueen JP, Gattoni K, Gendron EMS, Schmidt SK, Sommers P, Porazinska DL. Host identity is the dominant factor in the assembly of nematode and tardigrade gut microbiomes in Antarctic Dry Valley streams. Sci Rep. 2022;12(1):20118. [CrossRef]
- Tibbs-Cortes LE, Tibbs-Cortes BW, Schmitz-Esser S. Tardigrade community microbiomes in North American orchards include putative endosymbionts and plant pathogens. Front Microbiol. 2022;13:866930. [CrossRef]
- Zawierucha K, Trzebny A, Buda J, et al. Trophic and symbiotic links between obligate-glacier water bears (Tardigrada) and cryoconite microorganisms. Wilson BA, ed. PLOS ONE. 2022;17(1):e0262039. [CrossRef]
- Mioduchowska M, Konecka E, Gołdyn B, et al. Playing peekaboo with a master manipulator: Metagenetic detection and phylogenetic analysis of Wolbachia supergroups in freshwater invertebrates. Int J Mol Sci. 2023;24(11):9400. [CrossRef]
- Yu XJ, Walker DH. The Order Rickettsiales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, eds. The Prokaryotes: Volume 5: Proteobacteria: Alpha and Beta Subclasses. Springer; 2006:493-528. [CrossRef]
- Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9(4):393-405. [CrossRef]
- Mioduchowska M, Czyż MJ, Gołdyn B, et al. Detection of bacterial endosymbionts in freshwater crustaceans: the applicability of non-degenerate primers to amplify the bacterial 16S rRNA gene. PeerJ. 2018;6:e6039. [CrossRef]
- Mioduchowska M, Katarzyna Z, Tadeusz Z, Jerzy S. Wolbachia and Cardinium infection found in threatened unionid species: a new concern for conservation of freshwater mussels? Conserv Genet. 2020;21(2):381-386. [CrossRef]
- Lewis Z, Lizé A. Insect behaviour and the microbiome. Curr Opin Insect Sci. 2015;9:86-90. [CrossRef]
- Engelstädter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst. 2009;40(1):127-149. [CrossRef]
- Ferrari J, Vavre F. Bacterial symbionts in insects or the story of communities affecting communities. Philos Trans R Soc B Biol Sci. 2011;366(1569):1389-1400. [CrossRef]
- Bertolani R. Evolution of the reproductive mechanisms in tardigrades — A review. Zool Anz. 2001;240(3-4):247-252. [CrossRef]
- Hebert PDN, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 2003;270(suppl_1). [CrossRef]
- Lu JM, Li T, Chen HW. Molecular phylogenetic analysis of the Stegana ornatipes species group (Diptera: Drosophilidae) in China, with description of a new species. J Insect Sci. 2011;11(20):1-12. [CrossRef]
- Ciobanu D, Zawierucha K, Moglan I, Kaczmarek Ł. Milnesium berladnicorum sp. n. (Eutardigrada, Apochela, Milnesiidae), a new species of water bear from Romania. ZooKeys. 2014;429:1-11. [CrossRef]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772-772. [CrossRef]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Thorne J, ed. Syst Biol. 2004;53(5):793-808. [CrossRef]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572-1574. [CrossRef]
- Mioduchowska M, Kačarević U, Miamin V, et al. Redescription of Antarctic eutardigrade Dastychius improvisus (Dastych, 1984) and some remarks on phylogenetic relationships within Isohypsibioidea. Eur Zool J. 2021;88(1):117-131. [CrossRef]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Battistuzzi FU, ed. Mol Biol Evol. 2018;35(6):1547-1549. [CrossRef]
- Bapteste E, Van Iersel L, Janke A, et al. Networks: expanding evolutionary thinking. Trends Genet. 2013;29(8):439-441. [CrossRef]
- Suh A. The phylogenomic forest of bird trees contains a hard polytomy at the root of Neoaves. Zool Scr. 2016;45(S1):50-62. [CrossRef]
- Fleming JF, Arakawa K. Systematics of tardigrada: A reanalysis of tardigrade taxonomy with specific reference to Guil et al. (2019). Zool Scr. 2021;50(3):376-382. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).