3.2. Global Remarks on the Spectra Acquisition and Preprocessing
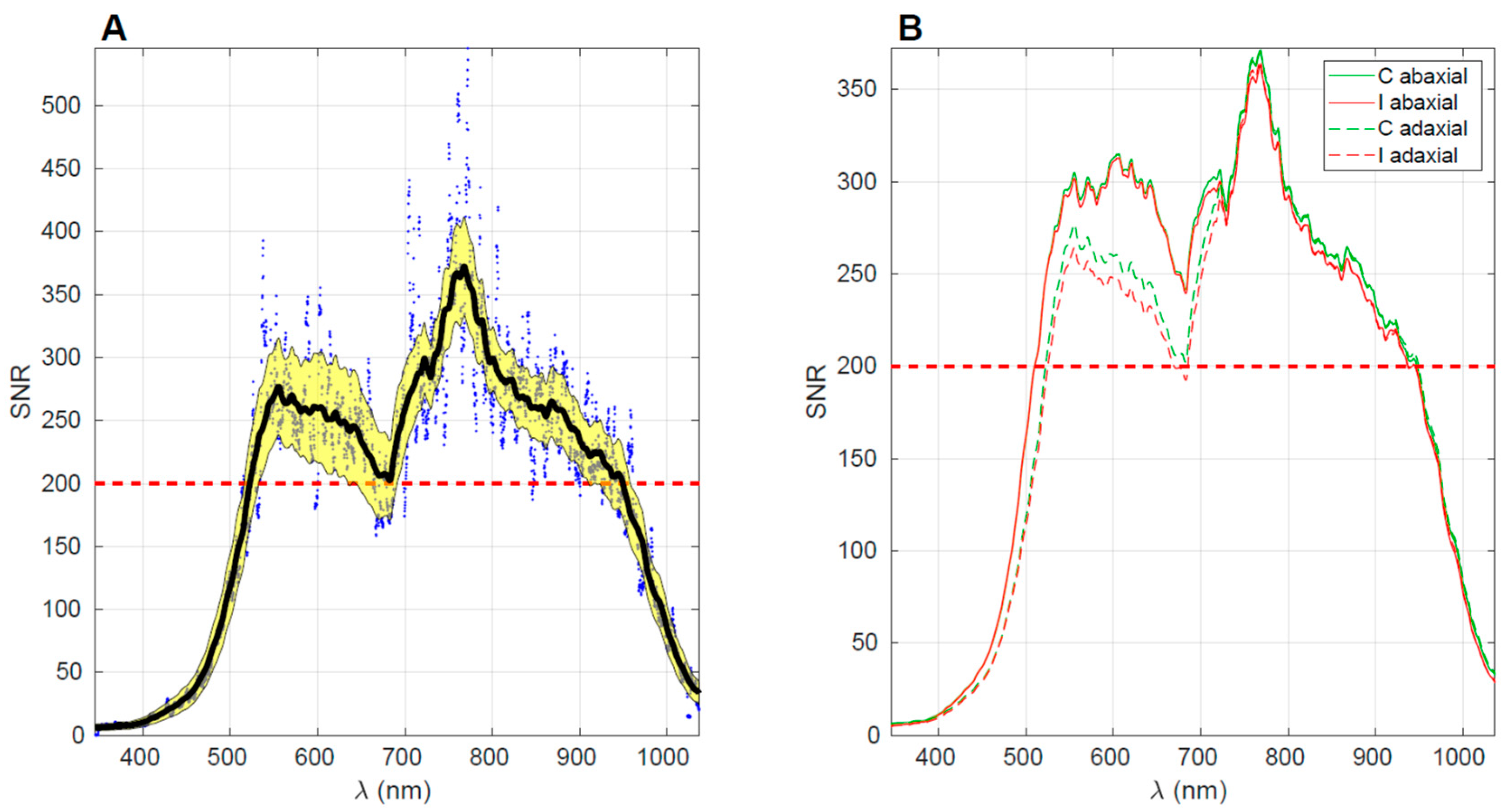
Since the aim of the spectral measurements was to detect potential variations between the control and inoculated plant groups, it is crucial to ensure that the spectra exhibit a suitable signal-to-noise ratio (SNR) across the entire range. Given that these differences might be subtle, it is essential to assess the SNR for each wavelength. SNR is computed by dividing the average signal by its standard deviation, obtained from a series of measurements. In a previous study, the same spectrometer was employed for non-invasive prediction of the internal quality of ’Jintao’ kiwifruit [
29]. It was determined that the spectral ranges suitable for calibration exhibited an SNR greater than 200. Following a similar procedure, the SNR was calculated in this study, and the results are illustrated in
Figure 2.
In
Figure 2A, the signal-to-noise ratio (SNR) is depicted for the spectra obtained from the adaxial side of plant leaves in the control group across the five evaluation days, at 63, 78, 91, 126 and 248 days after inoculation (DAI). Each point represents the SNR value for a specific leaf at each wavelength. These individual values are then averaged and smoothed using a Savitzky-Golay filter with 200 points and a polynomial order of 2, resulting in the smooth dark curve. The yellow band indicates one standard deviation around this average. The dashed red line represents the threshold SNR value of 200, which serves as a criterion for identifying the optimal spectral range in terms of SNR. Based on this criterion, the range from 530 to 920 nm is determined to be the optimal spectral range. It is evident that beyond this range, the SNR curve rapidly declines. This decline can be attributed to two factors: (i) the spectral response of the spectrometer’s detector and (ii) the absorption caused by the pigments present in the leaves. For instance, in the range of 400 to 500 nm, both the low spectral response and the significant absorption by chlorophyll and carotenoids contribute to decreased SNR. The absorption by these pigments substantially reduces the reflected signal, leading to lower SNR values. Similarly, the drop observed above 900 nm is associated with the low quantum efficiency of silicon (which falls below 10% beyond 1000 nm) and the absorption caused by water around 960-980 nm. In
Figure 2B, the average SNR curves are presented for the four datasets: control abaxial, control adaxial, inoculated abaxial, and inoculated adaxial. Interestingly, the smaller spectral features in the abaxial SNR curves closely resemble each other, as do the two adaxial SNR curves. This suggests that the fine details of the SNR are primarily determined by the spectral response of the spectrometer itself, while the larger features are influenced by the characteristics of the samples being measured. The distinction between the abaxial and adaxial leave sides becomes apparent in the 500-700 nm range, which can be attributed to the presence of pigments. On the other hand, the differences between the inoculated and control groups are not clearly evident in this overall plot combining data from all days. However, further analysis conducted separately for each day reveals discernible differences, as described below.
The spectral acquisition was conducted under sunlight and challenging conditions for the equipment. For instance, on hot days, the cooling of the light source was less effective, leading to gradual fluctuations in light intensity. Similarly, the spectrometer’s response and dark noise counts exhibited slow drifts over time. Consequently, using absolute reflectance values for comparing the two groups is not recommended. Instead, a preprocessing technique is necessary to mitigate the additive and multiplicative effects induced by environmental changes. Therefore, the spectra underwent Multiplicative Scatter Correction (MSC) as a preprocessing step. MSC was calculated from the average of the spectra from all days and all plants (i.e., including both groups). Prior to applying MSC, the spectral data was limited to the range of 530-920 nm, as determined by the SNR analysis. It is important to note that MSC causes a loss of direct interpretability for the absolute values of reflectance. However, the overall shape of the spectrum is preserved, allowing for the identification of local variations within specific spectral wavebands through changes in the spectrum shape and its relation to other bands.
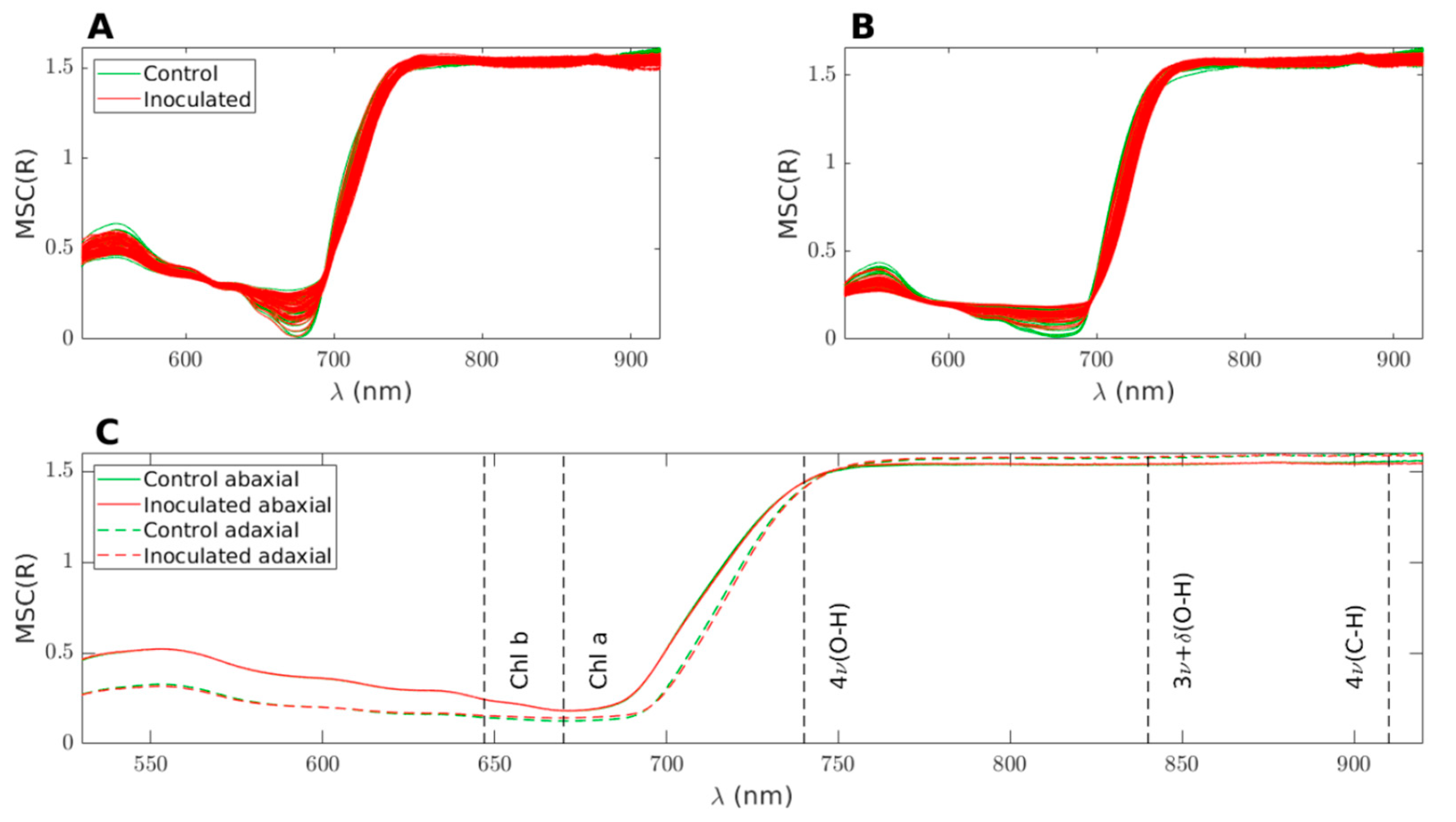
Figure 3 displays the superimposed spectra after applying MSC. The inoculated samples are shown in red, while the control samples are depicted in green.
Figure 3A illustrates the reflectance on the abaxial side of the leaves,
Figure 3B represents the reflectance on the adaxial side, and
Figure 3C displays the average of the four datasets presented in the top figures.
In
Figure 3C, distinct reflectance patterns are observed for the abaxial and adaxial sides. However, when considering the average spectra, it becomes challenging to differentiate between the control and inoculated groups. For the purpose of the following discussion, the spectra can be divided into three main bands. The first band corresponds to the visible range, spanning from 400 to 700 nm, which is influenced by the presence of pigments such as chlorophyll and carotenoids. The second band encompasses the near-infrared (NIR) range, ranging from 750 to 1000 nm, characterized by a plateau. The height of this plateau is primarily determined by the leaf structure, specifically the epidermal layers and the mesophyll [
34]. Finally, there is a transition zone between the visible and NIR ranges, often referred to as the red edge. This region exhibits a sharp slope, connecting the low reflectance values in the visible range to higher reflectance levels in the NIR.
The observed phenomenon where the abaxial side reflects more in the visible range and slightly less in the NIR range compared to the adaxial side can be attributed to the optical characteristics of leaf tissues, which have naturally evolved to optimize their interaction with light from above. This optical engineering of the leaf tissues ensures that light is effectively channeled to the chloroplasts for optimal absorption. The palisade parenchyma, with its channellike structure, plays a key role in facilitating the conduction of light to the chloroplasts. In contrast, the spongy parenchyma, located beneath the palisade parenchyma, induces light scattering, which is particularly important for protecting the leaf from excessive infrared heating. However, when light is incident from below, the order of interaction with the two parenchyma layers is reversed. As a result, the optimal conduction of light to the chloroplasts is compromised, leading to lower absorption in the visible range and, consequently, higher reflection values. Additionally, the reflection of NIR light is also less efficient on the abaxial side, possibly due to the abaxial epidermis being less reflective than the adaxial epidermis.
Although the overall comparison of spectra collected across all measurement days does not reveal distinct differences between the control and inoculated groups, conducting a specific analysis for each measurement day uncovers noticeable distinctions. These differences have been verified using the Wilcoxon Rank Sum Test (also known as the Mann Whitney U-test), which assesses whether the data in the control and inoculated groups originate from continuous distributions with equal medians. The test is performed for each wavelength individually. The application of the classical t-test across the entire spectral range was not feasible due to a significant portion of wavelengths not meeting the assumptions of normality and homogeneity of variances. Surprisingly, even with these violated assumptions, running the t-test produced results that were highly similar to those obtained with the nonparametric test. In
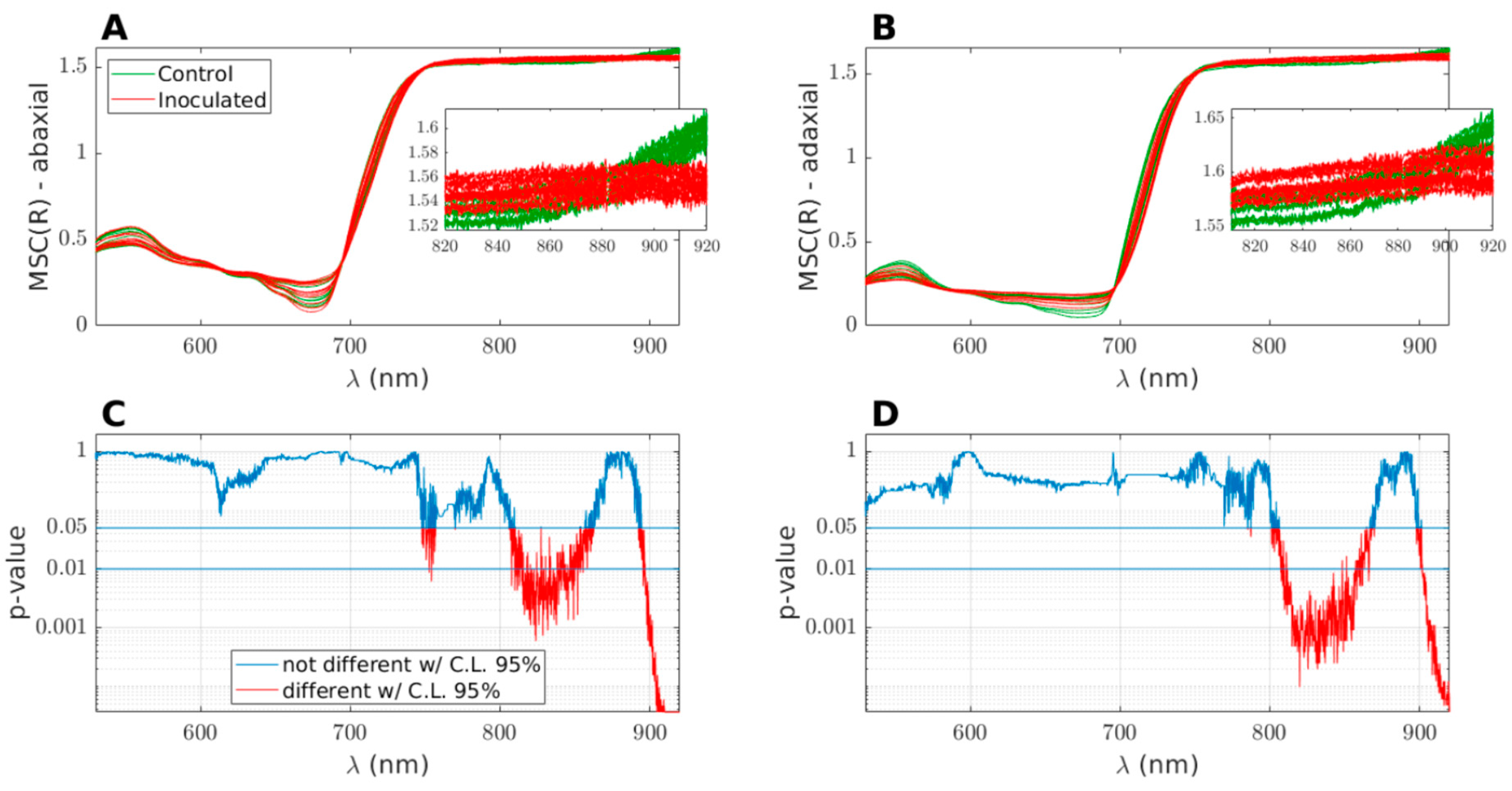
Figure 4, the results for the second measurement day (78 DAI) are presented. The top plots, A and B, depict the MSC-transformed spectra for the abaxial and adaxial sides, respectively. The figure insets provide a closer view of a restricted range within the NIR plateau, specifically between 820 and 920 nm, where the differences between the control and inoculated groups are prominently visible. The bottom plots, C and D, illustrate the
p-values obtained from the Wilcoxon Rank Sum Test for the data in plots A and B, respectively, as a function of wavelength. The
p-values indicate the statistical significance of the differences observed between the two groups. The horizontal lines represent the threshold values of
p = 0.05 (95% confidence level) and
p = 0.01 (99% confidence level). It is important to note that the scale of the
p-value axis is logarithmic. Regions of the curves below the
p = 0.05 threshold are highlighted in red, indicating the wavelengths where the medians of the MSC reflectance for the control and inoculated groups significantly differ.
The differences observed in the spectra can be interpreted as follows:
1. The visible band (up to 700 nm), which is primarily influenced by pigments, does not exhibit significant differences between the control and inoculated groups. This suggests that there were no significant variations in the content of pigments between the two groups.
2. The regions highlighted in red, which correspond to the following spectral bands previously shown in
Figure 3, are where notable differences between the control and inoculated groups can be observed:
(a) Around 750 nm (only on the abaxial side), coinciding with the presence of 4v (O − H), which represents the third overtone of the O–H stretching vibration.
(b) Around 830 nm, close to 3v + δ (O − H), which corresponds to the O–H combination band of the second overtone of stretching with the fundamental bending vibration.
(c) Around 920 nm, aligning with the presence of 4v (C −H), which represents the third overtone of the C–H stretching vibration.
These specific spectral bands indicate distinct molecular vibrations or combinations that may vary between the control and inoculated groups, suggesting differences in the chemical composition or molecular structures of the samples. The first two bands are related with water, while the third may reflect changes in, basically, any organic molecule(s).
However, there is an alternative interpretation that is both more plausible and straightforward. The broad absorption peak of 3v (O−H) at 960 nm is outside of the considered spectral range, but its effects may still be felt above 900 nm. As a result, the observed differences above 900 nm could also be attributed to variations in water content. Hence, adhering to the principle of Occam’s razor, the simplest and most likely explanation is that the spectral differences are indeed influenced by changes in water content.
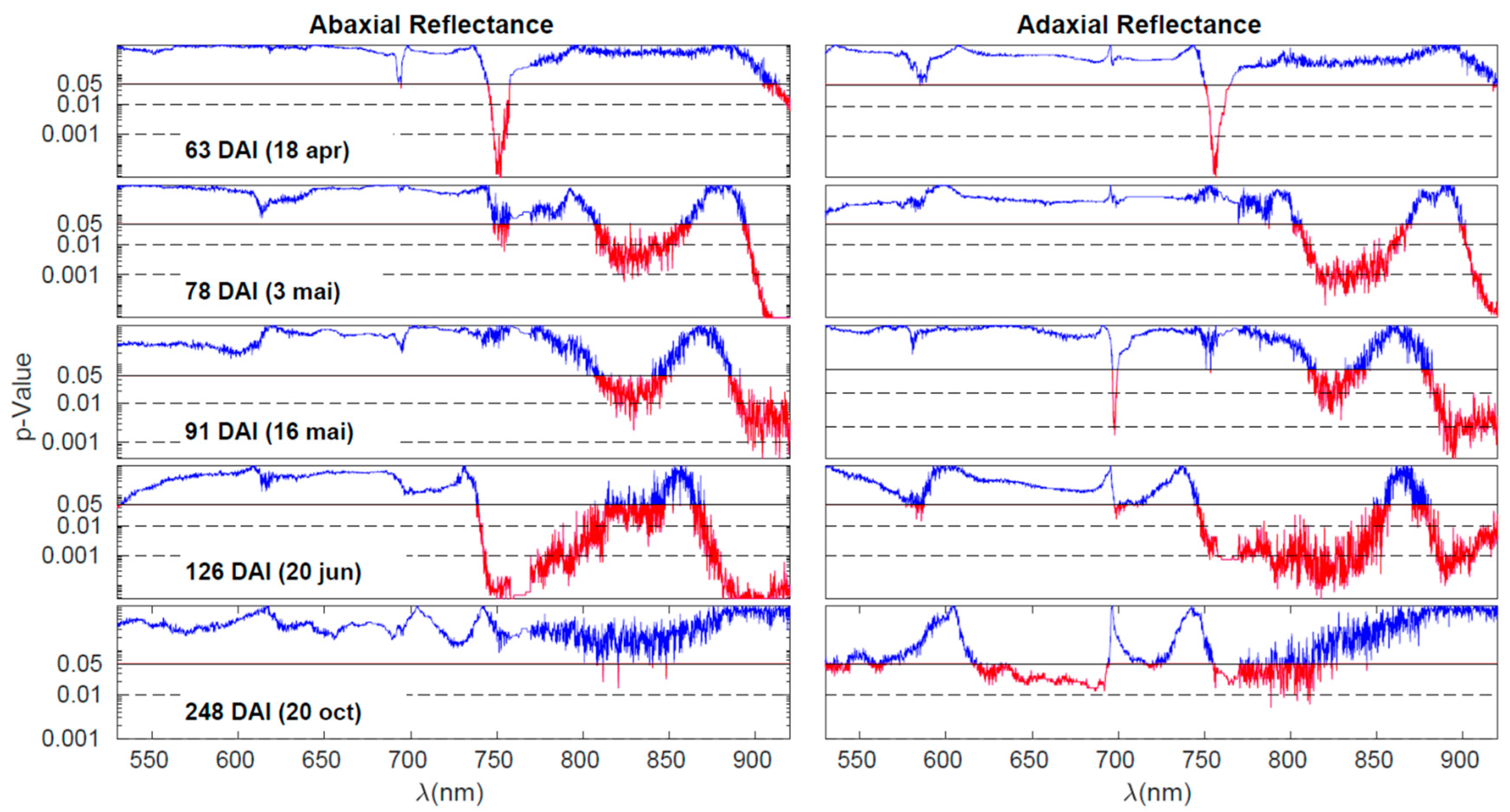
The analysis presented in
Figure 4 can be extended to all five days of measurement, as demonstrated in
Figure 5. In this figure, the
p-values resulting from the Wilcoxon Rank Sum Test, which compares the MSC reflectances of the control and inoculated groups for each measurement day, are depicted. Each line in the figure corresponds to one of the measurement days (63, 78, 91, 126, and 248 DAI), while the columns represent the abaxial and adaxial sides, respectively. The
p-values are plotted as a function of wavelength. The interpretation of the horizontal lines and colour codes remains consistent with
Figure 4.
On the first day of measurement (63 DAI), there is a noticeable difference primarily around 750 nm. This difference can be attributed to variations in water content (4v (O−H)) and/or a shift in the red edge. The red edge is a sensitive region characterized by a steep slope, marking the transition from low reflectance in the visible range to high reflectance in the NIR plateau. Any alteration in pigments or the leaf structure that affects NIR scattering can lead to changes in the slope of the red edge, making it particularly useful for detecting NIR variations. Examining the p-values curve, we observe an additional region marked in red on the abaxial side, above 900 nm. This can also be influenced by changes in the off-range water absorption peak at 960 nm. In summary, the most plausible explanation for the differences observed on the first day is as follows: there are initial changes occurring in water levels (indicated by the variations above 900 nm), which slightly impact the NIR plateau. This change is further amplified in the slope of the red edge, making it appear as the primary source of differentiation between the control and inoculated groups.
The patterns observed on days 78, 91, and 126 DAI exhibit similarities that have been previously examined in the example of the analysis on one of the days. These patterns align with variations in water content, reflecting distinctions in the 3v+δ (O−H) combination band and the O–H combination band at 840 nm and 960 nm, respectively. Additionally, the 4v (C − H) band within the 910-930 nm range may contribute to these observations, although distinguishing the individual contributions of these bands can be challenging.
By the fourth day (126 DAI), differences attributable to water remain, but the distinctions across the near-infrared (NIR) plateau become more pronounced. With the exception of a band centred around 870 nm, all regions within the NIR plateau exhibit variations between healthy and inoculated plants. This suggests a significant disruption in the physical structure of the leaves. As previously mentioned, the palisade and spongy parenchyma tissues play a crucial role in light scattering within the NIR plateau. The observed differences across the entire spectral range of the plateau may indicate alterations in the structure of the parenchyma tissues, likely stemming from disparities in the initial water content. The final day of observation reveals a reversal of the observed patterns. On the abaxial side of the leaves, no notable differences are present except for occasional spikes of noise. On the adaxial side, most of the differences within the NIR plateau diminish, with the exception of the band ranging from 750 nm to 820 nm. However, a new development arises in the range of pigments, specifically between 620 nm and 700 nm, where differences between the two groups are apparent. The similarity observed in the NIR plateau between the inoculated and control plants could stem from two possible causes:
the inoculated plants have regained their initial structural integrity, or
the control plants have also experienced structural change.
The latter scenario is plausible considering that a summer season elapsed between the fourth and final measurements. On the other hand, the differences observed in the pigment range may also be attributed to two potential factors:
variations in pigment content between the two groups, or
structural changes in leaf anatomy altering the light path to and from the chloroplasts, thereby impacting reflectance levels.
In the latter hypothesis, the pigment content may be similar in both groups, but the reflectance levels differ. An important clue for discerning which scenario occurred lies in the absence of visible range differences on the abaxial side. This suggests that the pigment content remains unchanged, as any divergence would affect abaxial reflectance. Therefore, the most plausible explanation for the observations on the final day points to structural changes. The leaf structure of the control plants underwent alterations that made their NIR response resemble that of the infected plants. However, these same structural modifications resulted in visible reflectance differences between the two groups. How is this possible?
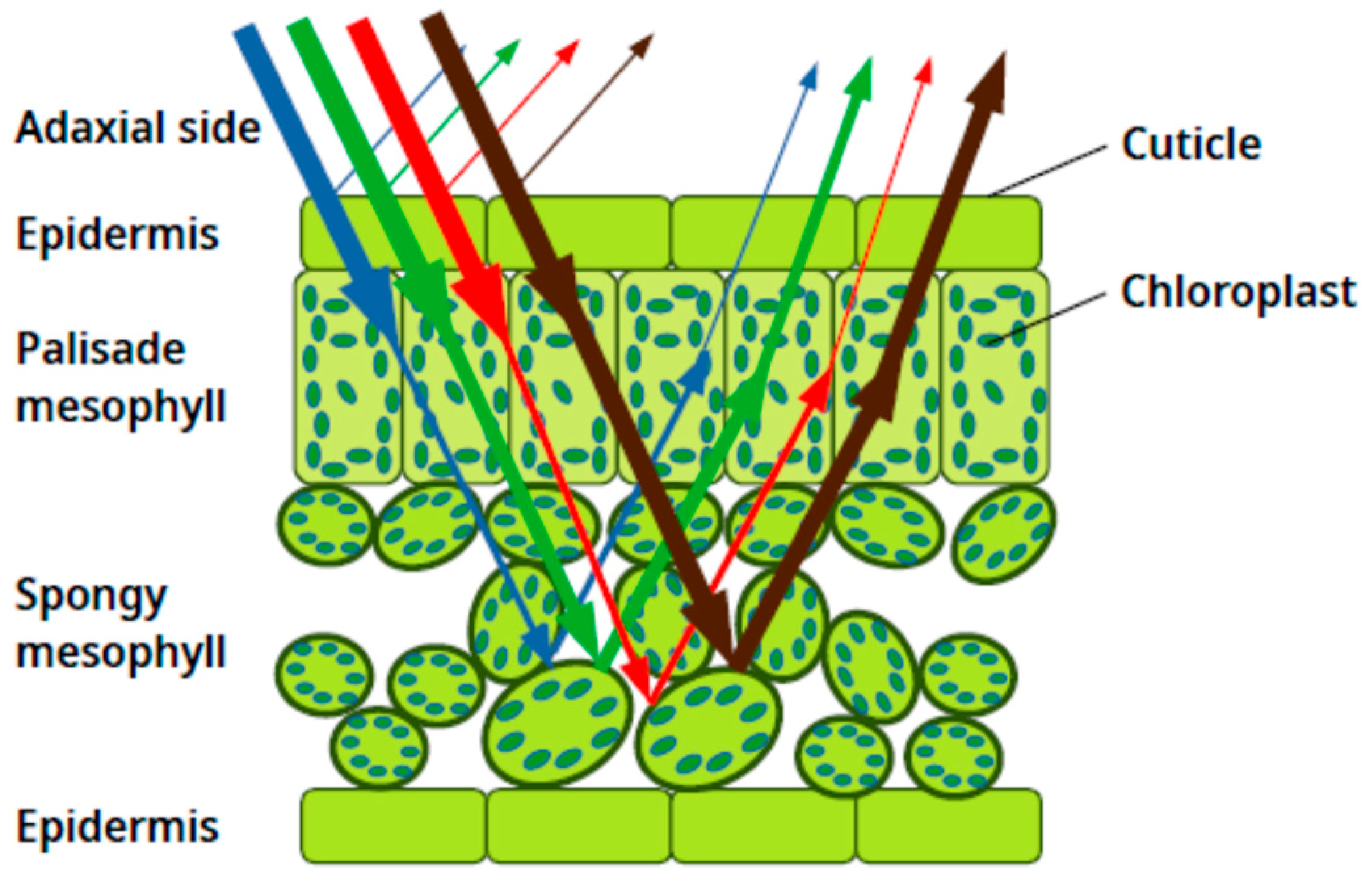
To comprehend why structural changes can lead to distinct alterations in visible and NIR reflectance patterns,
Figure 6 provides valuable insights. In this figure, it is evident that visible light primarily interacts with the upper portion of the leaf, including the chloroplasts and extending down to the palisade mesophyll. Conversely, NIR light primarily interacts with the spongy mesophyll, where it is scattered. Based on the observations from the final day, the following conclusions can be drawn:
The pigments’ content is similar in both the control and inoculated groups.
The spongy mesophyll of the control group underwent alterations during the summer and acquired a structure similar to that of the inoculated group.
The upper epidermal layer and/or the palisade mesophyll experienced differential changes between the control and inoculated groups, resulting in distinct responses in the visible range.
These findings suggest that while the pigments’ content remains consistent, structural modifications in the spongy mesophyll account for the similarity in NIR reflectance between the two groups. Conversely, differential changes in the upper epidermal layer and/or the palisade mesophyll contribute to the divergent responses observed in the visible range. Indeed, the explanation provided aligns with the observed similarity in abaxial reflectances as well. In this scenario, where reflected light predominantly interacts with the spongy mesophyll, the comparable structure of this mesophyll layer in both the control and inoculated groups explains the absence of discernible differences in abaxial reflectances.
Vegetation Indices (VI) are commonly employed to assess vegetation health through remote sensing. Therefore, it is important to explore whether VI can effectively detect distinctions between the two groups. The indices considered in this study are presented in
Table 1.
It is crucial to note that the vegetation indices (VI) were derived from the raw reflectance data rather than the MSC-transformed data. Calculating the VIs directly from the raw data ensures that they are not distorted. This approach poses no issue since any potential spectrum fluctuations, which necessitated the application of MSC as a compensation transformation, are naturally accounted for by the nature of the VIs themselves. VIs are primarily defined by ratios, enabling them to compensate for such fluctuations. Relatively to the contents of
Table 1, Normalized Difference Vegetation Index (NDVI) is the most widely adopted VI and with the greenness (health) of vegetation [
35]. NDVI values range from 1 to -1. Healthy NDVI values are greater than 0.3 and lower NDVI values represent barren or drought-tolerant plants. The Green Normalized Difference Vegetation Index (GNDVI) method is used for estimating photosynthetic activity and is a commonly used vegetation index to determine water and nitrogen uptake into the plant canopy [
36]. The Water Index (WI) is sensitive to changes in canopy water status. As the water content of vegetation canopies increases, the strength of the absorption around 970 nm increases relative to that of 900 nm [
37]. We have included a similar index, calculated from the reflectance around 950 nm instead. This is called here Water Band Index (WBI). The Soil-adjusted vegetation index (SAVI) accounts for the differential red and near-infrared extinction through the vegetation canopy. The index is a transformation technique that minimizes soil brightness influences from spectral vegetation indices involving red and NIR wavelengths [
38]. The indices O1 – O4 are simple ratio and/or differences between two bands, less used in the literature and that do not have a uniformly adopted nomenclature. O1 is a simple version of the Weighted Difference Vegetation Index (WDVI) [
39] with the a coefficient taken as 1. Since it measures the difference in height between the red and NIR plateaus, and the later depends on the scattering by the leaf structure, it may be related with changes in leaf structure. O2 is the simple red/NIR ratio [
40], while O4 is simply its inverse and usually called Simple Ratio NIR/RED Difference Vegetation Index (DVI). Finally, O3 is the green/red ratio [
41]. The Photochemical Reflectance Index (PRI) is sensitive to changes in carotenoid pigments (e.g. xanthophyll pigments) in live foliage [
42]. The Rededge Vegetation Stress Index (RVSI) identifies spectral changes in the red edge, which may also be traced to changes in leaf structure [
43]. The Modified Chlorophyll Absorption in Reflectance Index (MCARI) minimizes the effects of nonphotosynthetic materials on spectral estimates of absorbed photosynthetically active radiation [
44]. The Visible Atmospherically Resistant Index (VARI) is designed to emphasize vegetation in the visible portion of the spectrum, while mitigating illumination differences and atmospheric effects [
45]. The Browning Reflectance Index (BRI) was for estimation of superficial scald development in anthocyanin-free fruit upon a background of Chl and Car absorption [
46], but we have included it in the attempt of finding browning in the leafs.
Table 1.
Vegetation Indices (VI) used in this work.
Table 1.
Vegetation Indices (VI) used in this work.
| Name |
Formula* |
Reference |
% Detection |
| NDVI |
(N - R)/(N + R) |
[35] |
10 |
| GNDVI |
(G - R)/(G + R) |
[36] |
20 |
| WBI |
R950/R900 |
[37] |
80 |
| SAVI |
[(1 + L)(N - R)]/(N + R + L), L = 0.5 |
[38] |
40 |
| O1 |
N - aR, a = 1 |
[39] |
70 |
| O2 |
R/N |
[40] |
10 |
| O3 |
G/R |
[41] |
20 |
| O4 |
N/R |
[47] |
10 |
| PRI |
(R531 - R570)/(R531 + R570) |
[42] |
20 |
| RSVI |
(R714 + R752)/2 - R733
|
[43] |
40 |
| MCARI |
[(R700 - R670)-0.2(R700 - R500)] X R700/R670
|
[44] |
10 |
| VARI |
(G - R)/(G + R - B) |
[45] |
30 |
| WI |
R900/R970 |
[37] |
100 |
| BRI |
(1/R550 - 1/R700)/N
|
[46] |
30 |
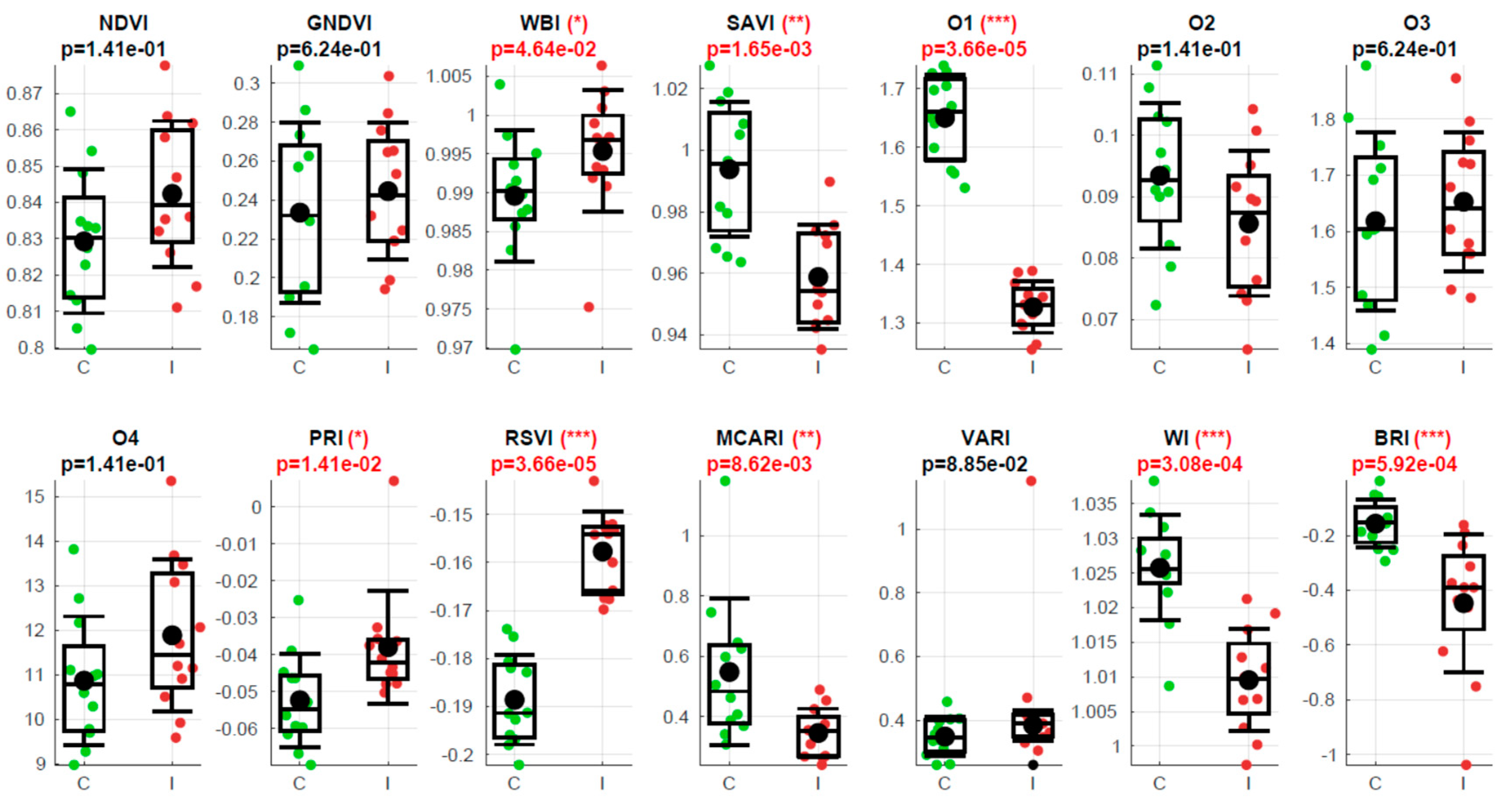
Figure 7 illustrates the results of the Vegetation Indices (VI) obtained from the adaxial side on the fourth day of measurements, corresponding to 126 DAI. The figure also displays the outcomes of the Wilcoxon Rank Sum Test comparing the two groups. Among the 14 VIs examined, eight of them demonstrate the ability to differentiate between the two groups. Notably, the fourth day exhibits the highest number of significant differences, which aligns with the observations depicted in
Figure 5, where the fourth day exhibits more bands shown in red.
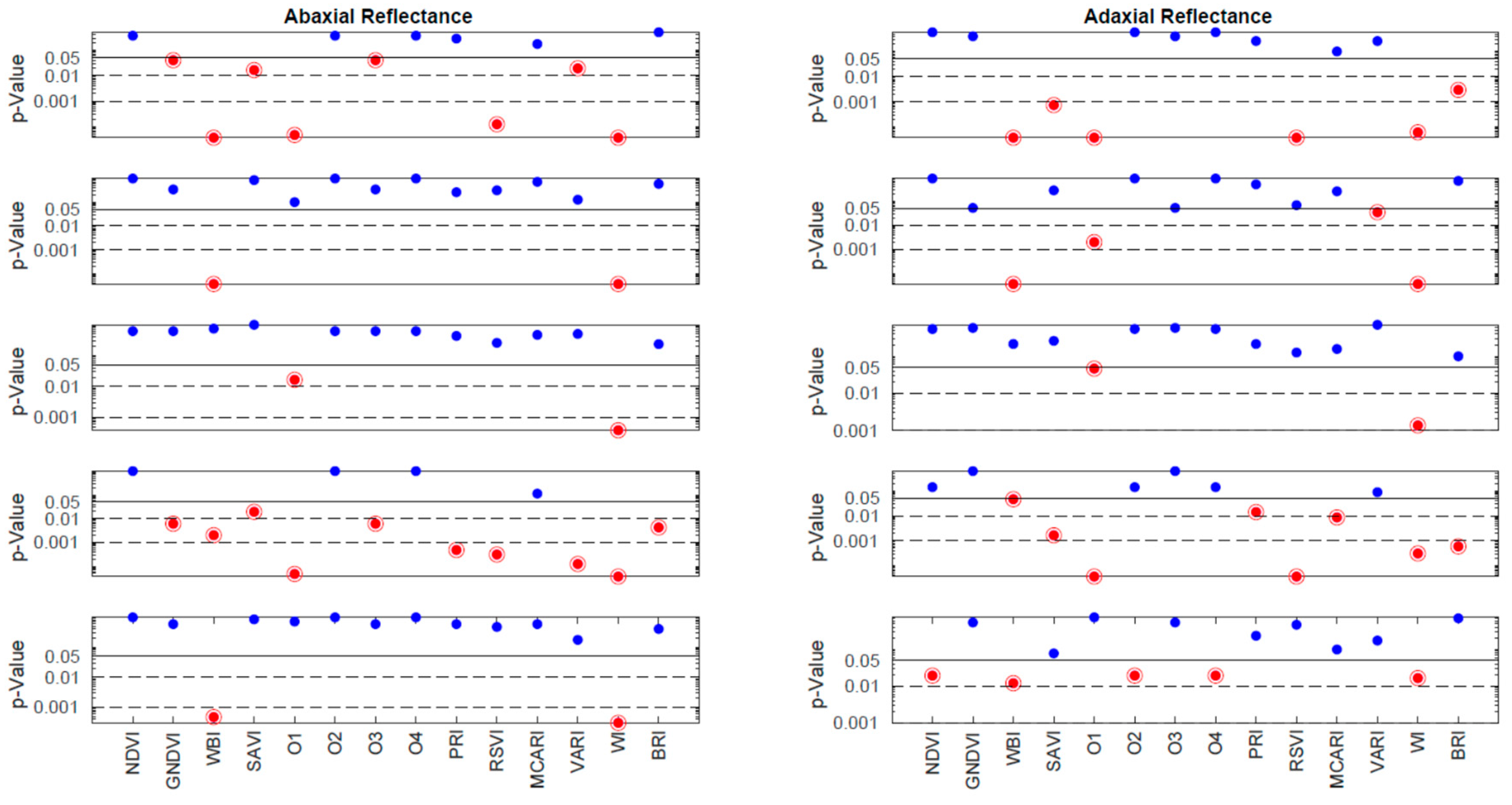
Figure 8 is analogous to
Figure 5, but it focuses on the VIs. Each row in the figure corresponds to one of the measurement days (63, 78, 91, 126, and 248 DAI), while the columns represent the abaxial and adaxial sides. The p-values are plotted against the VIs, which are shown along the x-axis of the bottom row. The interpretation of the horizontal lines and color codes remains consistent with
Figure 5, where p-values below 0.05 are indicated by a red point encircled by a circumference.
Figure 7.
Vegetation Indices (VI) acquired from the adaxial side on the fourth day of measurements, or 126 DAI. Each subplot represents a specific VI, where the control values are represented in green and the inoculated values in red. The name of the parameter is displayed at the top of each subplot, and below it is the p-value resulting from the Wilcoxon Rank Sum Test, which compares the medians of the two groups. Significant p-values are highlighted in red. The significance levels are denoted by a star code: (*) indicates p < 0.05 (95% confidence level), (**) indicates p < 0.01 (99% C.L.), and (***) indicates p < 0.001 (99.9% C.L.). The boxes and error bars are defined by five horizontal segments, which, from top to bottom, represent the mean plus standard deviation, the third quartile, the median, the first quartile, and the mean minus standard deviation, respectively.
Figure 7.
Vegetation Indices (VI) acquired from the adaxial side on the fourth day of measurements, or 126 DAI. Each subplot represents a specific VI, where the control values are represented in green and the inoculated values in red. The name of the parameter is displayed at the top of each subplot, and below it is the p-value resulting from the Wilcoxon Rank Sum Test, which compares the medians of the two groups. Significant p-values are highlighted in red. The significance levels are denoted by a star code: (*) indicates p < 0.05 (95% confidence level), (**) indicates p < 0.01 (99% C.L.), and (***) indicates p < 0.001 (99.9% C.L.). The boxes and error bars are defined by five horizontal segments, which, from top to bottom, represent the mean plus standard deviation, the third quartile, the median, the first quartile, and the mean minus standard deviation, respectively.
The distribution of the red points in
Figure 8 does not exhibit an obvious pattern. It does not align precisely with the distribution of the red and blue components in
Figure 5. This discrepancy arises because the vegetation indices (VIs) convey nonlinear information that may not directly mirror the spectral information. However, upon a 8 more detailed analysis, the two figures do agree in the conclusions. As indicated in
Table 1, the VIs that are most successful in discriminating between the two groups are WI (100%) and WBI (80%), both of which are associated with water content. The third most successful parameter is O1 or WDVI, which relates to changes in leaf structure. The success rates of all the other VIs are below 50%. Notably, SAVI and RSVI achieve a 40% success rate. While SAVI is a modified version of NDVI, RSVI targets the red edge, which, as mentioned earlier, may also be influenced by structural changes. In conclusion, the analysis of the VIs confirms the findings of the spectral analysis, highlighting that water content and leaf structural changes appear to be the primary distinguishing factors between the two groups.
Figure 8.
The p-values obtained by the Wilcoxon Rank Sum Test, on the comparison between the Vegetation Indices (VI) of control and inoculated groups from the data obtained in each measurement day. Each row corresponds to one of these days (63, 78, 91, 126 and 248 DAI), the left and right column correspond to the abaxial and adaxial sides, respectively. The p-values are shown as a function of the wavelength. The horizontal lines are at p = 0.05 (95% CL), at p = 0.01 (99% CL) and at p = 0.001 (99.9% CL). Note the scale is logarithmic. The points below the threshold p = 0.05 are represented in encircled red points, meaning that control and inoculated groups have different medians for that specific VI.
Figure 8.
The p-values obtained by the Wilcoxon Rank Sum Test, on the comparison between the Vegetation Indices (VI) of control and inoculated groups from the data obtained in each measurement day. Each row corresponds to one of these days (63, 78, 91, 126 and 248 DAI), the left and right column correspond to the abaxial and adaxial sides, respectively. The p-values are shown as a function of the wavelength. The horizontal lines are at p = 0.05 (95% CL), at p = 0.01 (99% CL) and at p = 0.001 (99.9% CL). Note the scale is logarithmic. The points below the threshold p = 0.05 are represented in encircled red points, meaning that control and inoculated groups have different medians for that specific VI.
Before delving into this section, it is important to mention that a Principal Component Analysis (PCA) was initially conducted on the spectroscopic results for each day. The objective was to identify two distinct clusters in the scores plot, which would effectively separate the two groups. Additionally, the loadings plot was examined to identify the most significant bands contributing to group separation. While this approach yielded successful results for some days, it was not consistently effective across all days. Furthermore, the information obtained from the PCA analysis did not provide substantial clarity or offer substantially distinct insights compared to those derived from
Figure 5. Consequently, an alternative feature extraction technique was employed. Exploiting the fact that the data is organized in the form of a three-dimensional array (rows = samples) × (columns = wavelengths) × (sheets = days), a method called PARAFAC (Parallel Factor Analysis) was adopted [
48]. PARAFAC facilitates a trilinear decomposition directly on the three dimensions of the data, providing a comprehensive overview of the entire process across the five days, rather than isolated analyses for each individual day. Furthermore, each dimension undergoes compression, similar to PCA. For instance, in the wavelength dimension, the more than 2000 variables were condensed to just three factors, effectively explaining the majority of data variability. Ultimately, each dimension is characterized by a reduced number of factors through the application of the PARAFAC technique. This reduction in dimensionality allows for a more concise representation of the data while still capturing the essential information and variability present in each dimension. After applying a Savitzky-Golay filter to the MSC-transformed data, the following results were obtained. The filter had a width of 155 points, approximately equivalent to 30 nm, and a polynomial order of 2 with a derivative order of 1. This filtering technique emphasized the shape of the spectra, enabling improved discrimination between the groups. However, as a trade-off, the clarity regarding the specific wavelengths’ contributions was reduced. The adoption of a large filter width was necessary to effectively remove noise from the data. The model presented in this study was ultimately reduced to two factors. The primary criterion for determining the number of factors was the core consistency check, which was performed using the loadings obtained from the PARAFAC model. When three factors were considered, the core consistency check did not pass, leading to the decision to retain only two components. While including more factors could potentially account for additional variability in the data, it would primarily capture noise. The first two components explain 44% of the total data variability on the adaxial side and 43% on the abaxial side. Although there is still unexplained variability, these two components are sufficient to capture the differences between the two groups.
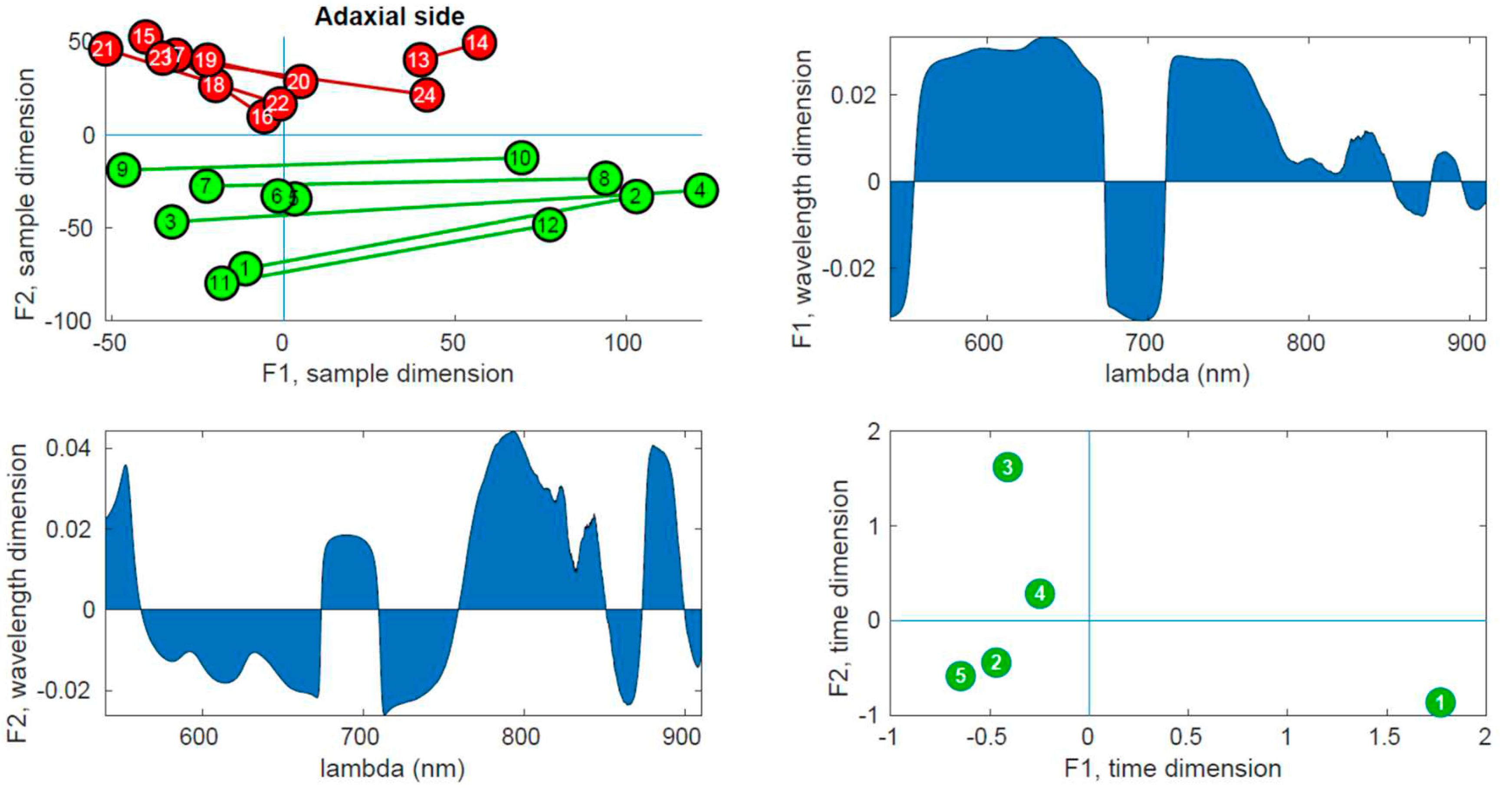
Figure 9 displays the PARAFAC results obtained from the adaxial side data and
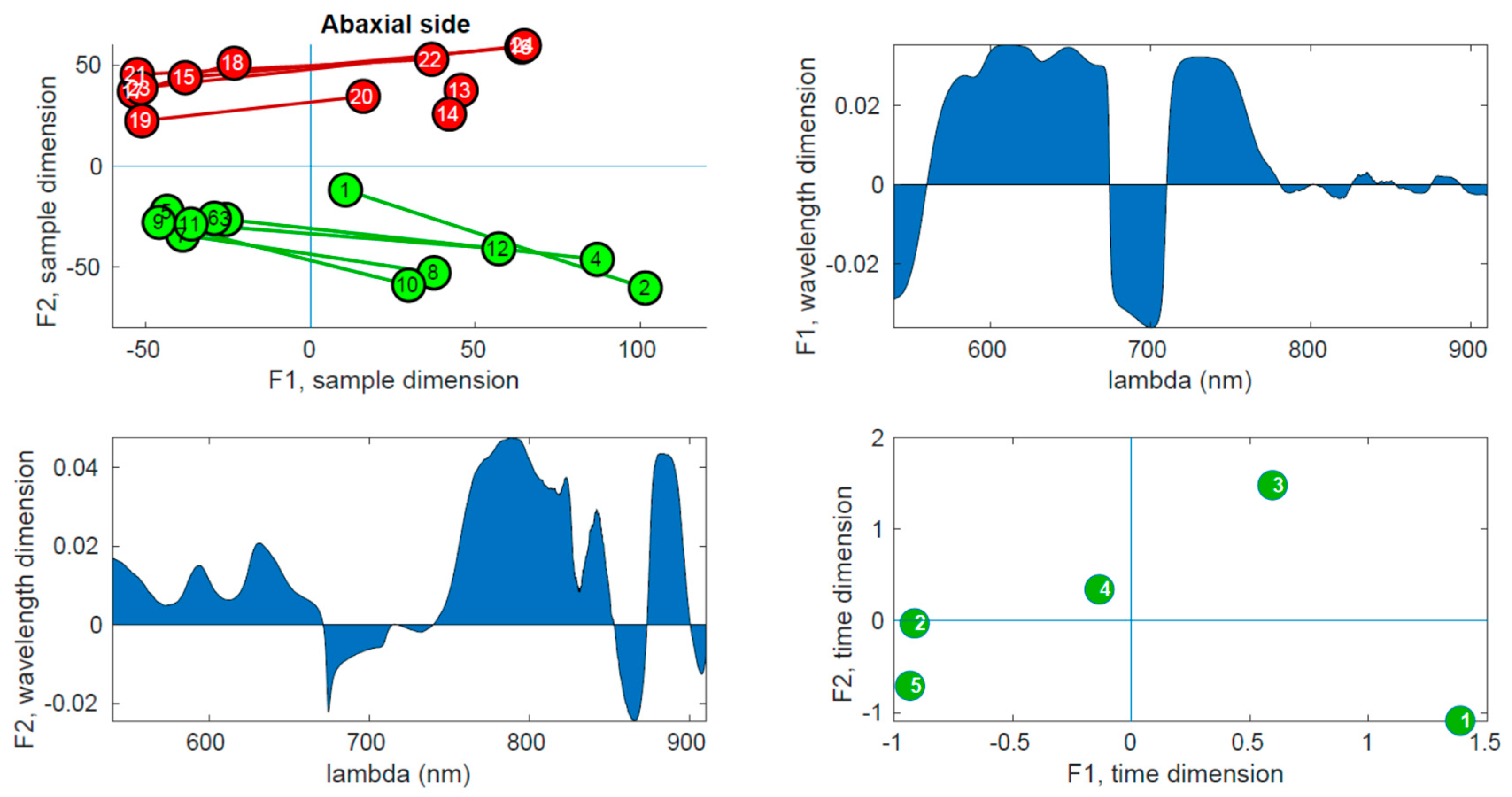
Figure 10 shows the corresponding results for the abaxial side.
In the upper right panel, the scores of the samples are plotted in the first two components, where control leaves are represented by green markers and inoculated leaves by red markers. The upper left panel shows the wavelength loadings in the first component, while the lower left panel illustrates the wavelength loadings in the second component. Finally, the lower right panel exhibits the loadings associated with the third dimension (time) in the first two components. The upper right plots in these figures reveal the most notable observation. A distinct separation between the two groups is evident along the second factor. Generally, the control samples exhibit values of F2 < 0, while the inoculated samples show values of F2 > 0, a trend that holds true for both adaxial and abaxial sides. Additionally, an interesting observation is that the first factor, F1, appears to capture variations within the same tree, while the second factor, F2, captures variations between the trees. This distinction arises because the segments connecting each pair of leaves from the same tree align predominantly with the F1 axis, indicating that tree-related variations are primarily associated with variations in F1. This pattern is more pronounced in the control leaves compared to the inoculated leaves, particularly on the adaxial side. The reduced parallelism observed in the inoculated group suggests internal changes resulting from the infection. The area plots (upper right and lower left) illustrate the loadings across the wavelength dimension. F1, which corresponds to intra-tree variations, exhibits significant contributions primarily below 750 nm. On the other hand, F2, associated with inter-tree variations, demonstrates substantial contributions primarily above 750 nm. Consequently, intra-variations are predominantly linked to changes in pigments, while inter-variations (which enable group differentiation) are primarily associated with water content and leaf structural changes. These findings align with the previous conclusions drawn from the direct spectral analysis as well as the examination of vegetation indices, reinforcing the overall consistency of the results. The final subplot in the lower left show cases the loadings along the time dimension, which corresponds to the order of the measurement days. Its interpretation follows the previously assigned meanings of F1 and F2. For instance, the first and last days exhibit the greatest separation in terms of the first factor, indicating that the leaves from these days are the most dissimilar in terms of pigment content. This can be attributed to the natural aging process of the leaves. Conversely, in terms of water and structural changes (F2), the first and fifth days appear to converge towards a similar state by the end of the experiment. This observation is consistent with the spectral analysis in
Figure 5, where the first and fifth rows exhibit relatively similar patterns, with fewer red bands compared to the other three days. However, caution is required when interpreting other observations due to the derivative spectra used in the analysis performed by PARAFAC. For instance, in both
Figure 9 and
Figure 10, it is the third day that appears to be the furthest from the rest in terms of F2, suggesting it is the day with the most significant differences between the two groups. On the contrary,
Figure 5 and
Figure 8 indicate that the fourth day exhibits more discriminative bands. The resolution to this apparent contradiction lies in recognizing that PARAFAC results are based on derivative data, and it is plausible that the rate of changes is higher on the third day, leading to a more pronounced separation along the F2 axis.