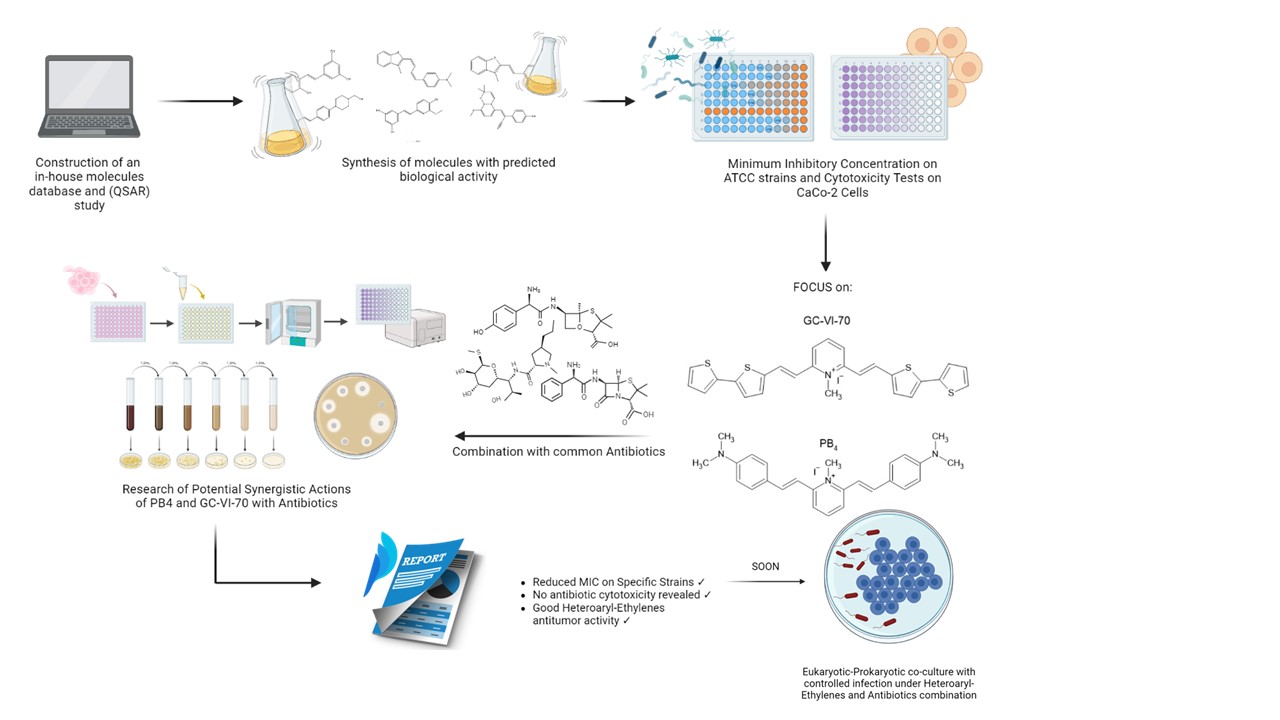
1. Introduction
ESKAPE bacteria, comprising
Enterococcus faecalis,
Staphylococcus aureus,
Klebsiella pneumoniae,
Acinetobacter baumannii,
Pseudomonas aeruginosa,
Salmonella enterica and
Escherichia coli, are notorious pathogens known for their ability to escape the effects of multiple antibiotics. The rising tide of antibiotic resistance among ESKAPE bacteria poses a significant challenge to modern healthcare [
1],demanding urgent efforts to develop innovative strategies for the discovery of new antibacterial compounds as fundamental tool to combat this growing threat [
2].
Multidrug-resistant bacteria (MDR), such as methicillin-resistant
Staphylococcus aureus (MRSA) [
3]and carbapenem-resistant Enterobacterales (CRE) [
4] are responsible for a significant number of infections worldwide [
5]. The overuse and misuse of antibiotics [
6], horizontal gene transfer [
7], and inadequate infection control measures have contributed to the emergence and spread of resistance mechanisms among bacterial populations [
8]. We recently reported the molecular modelling study and the versatile synthetic procedure that allowed us to identify the heteroaromatic stilbene derivatives as a new class of antimicrobial compounds. [
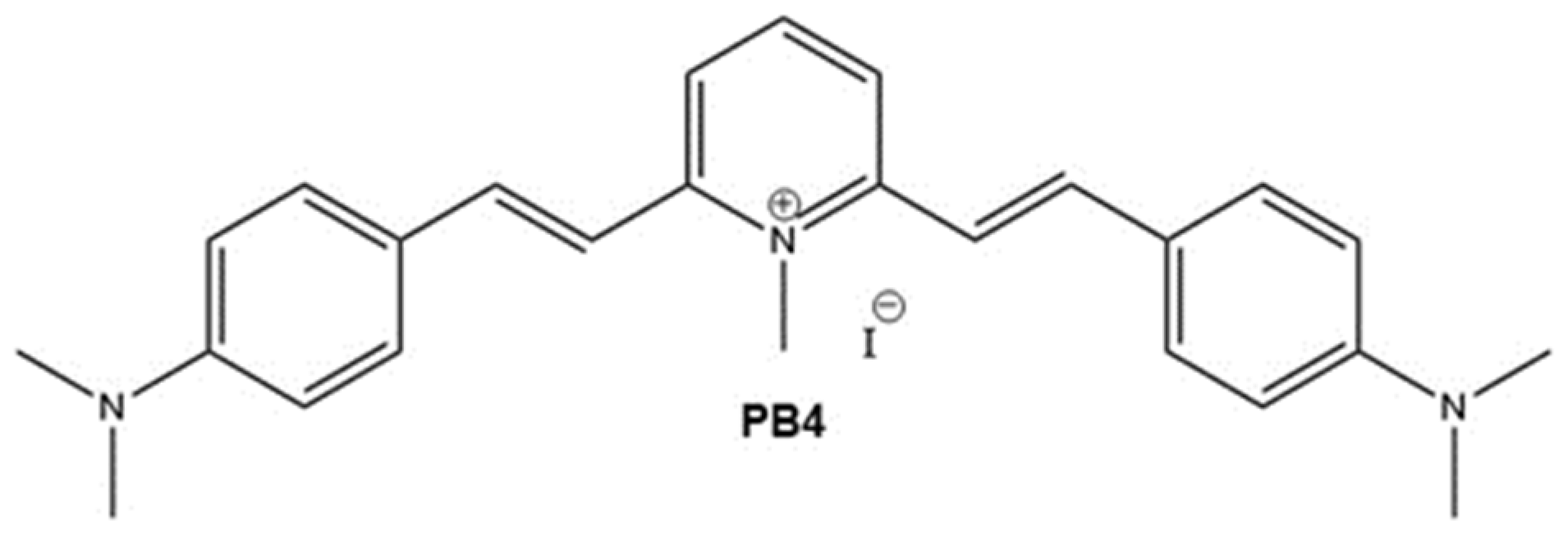
9] The most promising compound, namely PB4 (see
Scheme 1), was able to counteract the growth of Gram-positive and Gram-negative selected ATCC strains and, most notably, of high-priority clinical strains. [
10]
Scheme 1.
Structure of the lead compound PB4.
Scheme 1.
Structure of the lead compound PB4.
Beside the multidrug resistance phenomenon, we have also focused our attention on the development of new antineoplastic drugs (both cytotoxic and adjuvant medicines [
11,
12,
13,
14] Some of the Heteroaryl Ethylenes previously reported were highly cytotoxic also against CaCo-2 colon-rectal cancer cell line and these data have been used for the construction of an in-house database and perform a Structure–Activity Relationship (QSAR) study. The structural design approach has high predictability and, when coupled with a set of Partial Least Squares (PLS) analyses to predict some relevant absorption, distribution, metabolism and excretion (ADME) and biological properties, can contribute to a comprehensive evaluation of the in vitro - in vivo efficacy, both for antimicrobial and antiproliferative activities. [
15,
16] Then, the next step of this multidisciplinary ongoing project is a combined in silico approach based on: i) the extension of the QSAR study to identify new compounds from our in-house chemical database able to act antimicrobial agents; ii) the development of a new QSAR model for the prediction of the cytotoxic activity towards CaCo-2 colon-rectal cancer cell line and iii) the selection of new promising scaffold with sufficient potential to progress to a full drug development program. Through multiple in vitro assays, we evaluated the antibacterial activity, toxicity properties of these compounds, revealing their effectiveness against a range of ATCC bacterial strains. Finally, we tested these compounds, alone and in combination with some recurrent antibiotics in clinical practice (linezolid, gentamycin, ampicillin, erythromycin, rifampin and imipenem) on CaCo-2 cells to evaluate cytotoxicity. This study is the third of a series dealing with heteroaryl-ethylene compounds with antibacterial activity and it could have significant implications for the future of antibiotic development and the fight against MDR bacterial infections [
17,
18].
2. Results
2.1. Structural Design and Synthesis of Heteroaryl Ethylenes
It is well established that the inflammation derived from chronic bacterial infections or an abnormal gastrointestinal colonization, is associated with colorectal cancer. [
19,
20,
21] Understanding these interactions will yield future opportunities concerning prevention and treatment of both bacterial infection and colorectal cancer.
Heteroaryl ethylene compounds occupy a relative niche position in the fields of electronic materials thanks to their electrochemical and optoelectronic properties and, although their precise mechanism of action is not known, our recent studies are attempting to widen their fields of interest towards biomedical applications [
9,
10,
14]. This prompted us to perform a systematic investigation on the relationships between molecular structure and their biological activity. We have now extended our multidisciplinary study starting from an in-silico selection of new Heteroaryl Ethylenes as antimicrobial and antitumor agents.
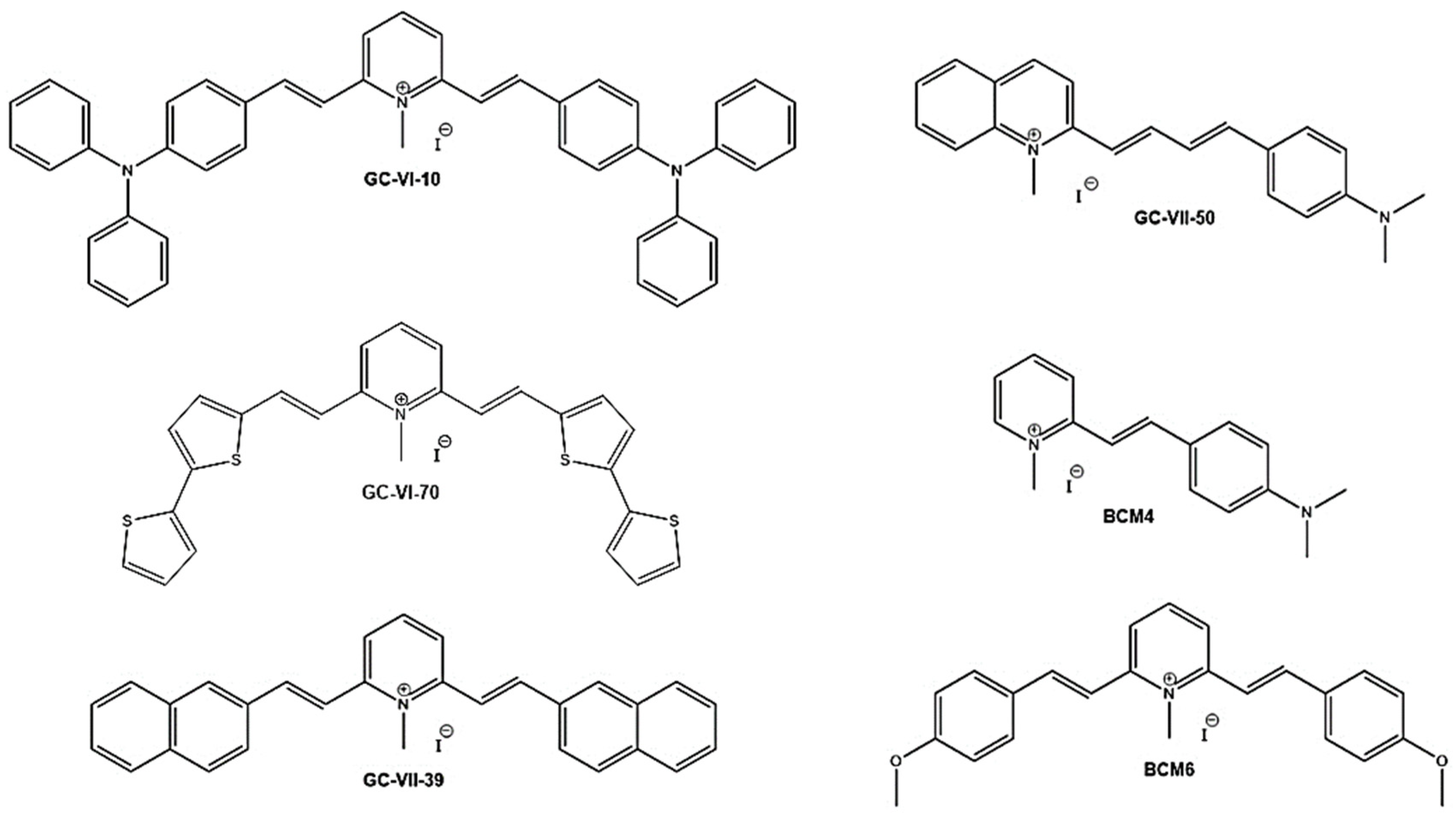
Predicting the biological properties of the Heteroaryl ethylene derivatives is not a standard procedure due to a limited number of research results, thus our QSAR models were developed using literary few data for the inhibitors of S. aureus and an in-house dataset of compounds for the antitumor activity against the colon-rectal cancer cell line. The new drug candidates have been selected to retain some structural figures of the lead compound PB4, namely the double branched D-π-A-π-D motif, through the insertion of different donor units (GC-VI-10, GC-VI-70, GC-VII-39 and BCM6), or the 4-(dimethylamino) styryl unit through the variation of the acceptor moiety (GC-VII-50 and BCM4).
2.1.1. QSAR model for the screening of the in vitro activity against S. aureus ATCC29213
The data regarding the antimicrobial activity, expressed as MIC (mg/L), of 53 Heteroaromatic compounds tested against
S. aureus ATCC29213 define our library for the prediction of the antibacterial activity; this dataset includes 40 compounds employed in our previous QSAR study and the 8 Heteroaryl Ethylenes tested in the same study [
9], 3 compounds of our in-house database already tested against
S. aureus ATCC29213 and some recently reported styryl derivatives [
22].
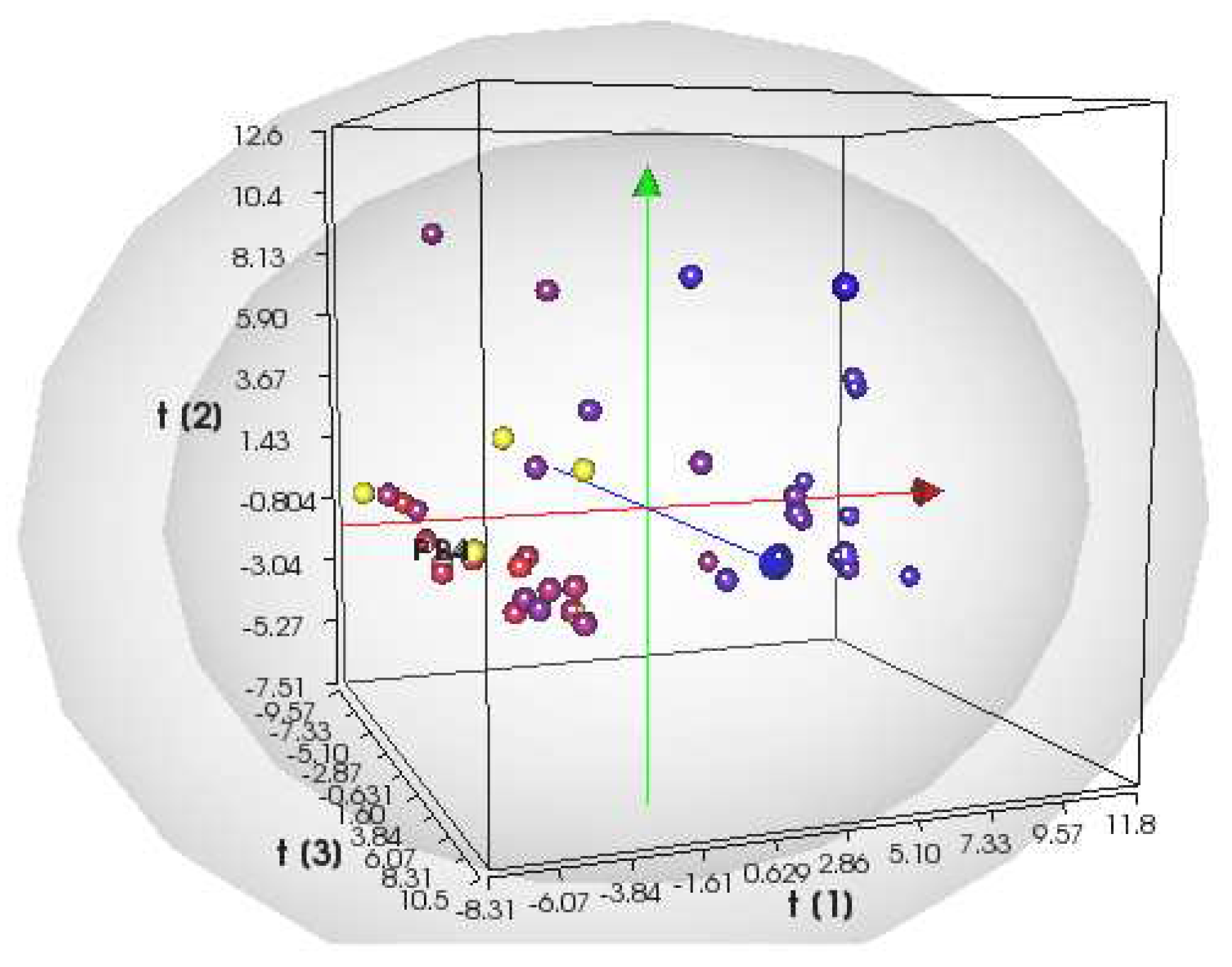
Overall, the PLS model has been improved by the insertion of the new 13 compounds, mainly Heteroaryl Ethylenes molecules with different donor-bridge-acceptor (D-π-A) motif (see Table S1). The PLS 3D score plot (
Figure 1) shows that the most active compounds are located on the left-hand side of the plot with negative LV1 scores and, overall, the model has a good correlation between experimental and predicted activity (R2=0.88). The Leave One Out (LOO) procedure allowed the internal validation of the model: the predictive ability has significantly increased up to a Q2 value of 0.59 (see Figure S2 for R2 vs Q2 plot).
Figure 1.
Scores plot at the third latent variable (LV1 vs LV2 vs LV3) of the PLS S. aureus -MIC activity model. Compounds are color-coded by their activity values, using a scale from red (actives) to blue (inactives), according to the experimental MIC values. The projections of the new compounds are indicated by the yellow circles.
Figure 1.
Scores plot at the third latent variable (LV1 vs LV2 vs LV3) of the PLS S. aureus -MIC activity model. Compounds are color-coded by their activity values, using a scale from red (actives) to blue (inactives), according to the experimental MIC values. The projections of the new compounds are indicated by the yellow circles.
This molecular modelling approach provide information about key molecular properties, as molecular interaction fields (MIFs)-based descriptors, that correlate with the biological activity (see Variable Influence on Projection VIP and Weights plot in Figure S3 and S4). Amongst the 128 VS+ descriptors, the bacteriostatic activity against S. aureus ATCC 29213 is highly influenced by the percentage of unionized species at higher pH (%FU), 3D pharmacophoric descriptors for H-bond donor regions (DODODO, ACDODO, and DRDODO), H-bond donors volume (WO), physicochemical properties such as distance of the hydrophobic volume from the center of mass (ID), the partition coefficient water/cyclohexane (LogPc-Hex) as well as ADME properties such as skin and CACO-2 cells permeability (SKIN and CACO-2). The improved PLS model highlights also the correlation of the activity with the compound’s solubilities computed at various pH and the corresponding shape of the solubility profile curve (LnLgS and LgS).To highlight new promising scaffold, we used this model for the external prediction of the bacteriostatic activity of the new six structures (
Scheme 2) from our in-house database of heteroaryl-ethylenes (yellow circles in
Figure 1 inset indicate the predicted compounds). Compound GC-VI-10 is located out of the 95% degree of confidence of the Scores plot while the remaining candidates are located within the 99% degree of confidence, moreover the most significant figure is their projections in the region of active antimicrobial compounds active against
S. aureus ATCC 29213.
Scheme 2.
Structures of the candidate antimicrobial/antitumor compounds.
Scheme 2.
Structures of the candidate antimicrobial/antitumor compounds.
2.1.2. QSAR model for cytotoxic activity towards CaCo2 colon-rectal cancer cell line
For the development of the QSAR model to evaluate the cytotoxic activity, we resorted to an in-house dataset of compounds previously synthesized and tested by our groups for their potential antitumor activity against the CaCo-2 colon-rectal cancer cell line. Based on these results, we create a database composed by 38 heteroaryl ethylene compounds and their antiproliferative effect against CaCo-2 (Table S2).
A preliminary Principal Components Analysis (PCA) applied to the X-matrix of the VolSurf+ descriptors allowed us an overview of the extended dataset. The first three principal components explained 66.9% of variance and the PCA score plot is shown in Figure S5. Only three compounds are located at the border of the 95% degree of confidence of the plot, while the remaining 35 compounds are located within the 99% degree of confidence. Neither classification of compounds nor any training information is given to the PCA model but, interestingly, active and inactive compounds can be nicely discriminated by the first two PC.
Then we perform the PLS regression using VolSurf+: the PLS 3D score plot (
Figure 2) shows that the chemical space is properly explored and provides a good discrimination between inactive (blue circles) and active compounds (red circles), with the latter being positioned in the lower-left side of the 3D plot.
Figure 2.
Scores plot at the third latent variable (LV1 vs LV2 vs LV3) of the PLS CaCo-2 activity model. Compounds are color-coded by their activity values, using a scale from red (actives) to blue (inactives), according to the experimental logIC50 values. The projections of the new compounds are indicated by the yellow circles.
Figure 2.
Scores plot at the third latent variable (LV1 vs LV2 vs LV3) of the PLS CaCo-2 activity model. Compounds are color-coded by their activity values, using a scale from red (actives) to blue (inactives), according to the experimental logIC50 values. The projections of the new compounds are indicated by the yellow circles.
The PLS model for cytotoxic activity towards CaCo-2 colon-rectal cancer cell line was validated using the Leave One Out (LOO) method and presented the following values of statistical coefficients (see Figure S6): R2 = 0.86 and Q2 = 0.56. The analysis of the Weights and VIP plots (Figure S7 and S8) revealed that descriptors for H-bond acceptor regions (WN and DRACACAC), CaCo-2 cells permeability (CACO-2), physicochemical properties such as distance of the hydrophobic volume from the center of mass (ID) and hydrophobic volume (CW) are directly related with activity; descriptors for ADME properties such as skin permeability and volume of distribution (SKIN and VD), physicochemical properties used for hydrophobic volumes (CD and DD) or Polar Surface Area (PSAR) and the ratio between the hydrophilic and lipophilic part of a molecule (CP) are inversely correlated with the cytotoxic activity towards CaCo-2 colon-rectal cancer cell line.
Since the model seems to clearly separate active from inactive heteroaryl ethylene compounds, we projected the six compounds previously selected into the model for cytotoxic activity prediction. Surprisingly, the picture that emerges follow the results given in the previous PLS model: compound GC-VI-10 is located out of the 95% degree of confidence of the Scores plot while the remaining candidates projected in the region of the most active antimicrobial compounds active towards CaCo-2 colon-rectal cancer cell line.
2.2. Impact on Biological Activity and Antimicrobial Susceptibility Test
The compounds selected through the QSAR procedure have been tested to determine the Minimum Inhibitory Concentration (MIC). Our results, reported in
Table 1, shown that heteroaryl-ethylenes are active against
S. aureus (ATCC 29213, ATCC 12598, ATCC BAA-1556),
E. faecalis (ATCC 29212),
E. coli (ATCC 25922), and
A. baumannii (ATCC 17978). The two heteroaryl-ethylene molecules that reported the best results are PB4 and GC-VI-70: the former is our ‘lead’ compounds [
9], the latter, GC-VI-70, possess the same double branched D-π-A-π-D structure but two bis-thiophene heteroaryl donor groups.
Surprisingly, BCM4 and GC-VII-39, showed MIC levels ranging from 16 mg/L to ≥ 128 mg/L for all tested strains. BCM6 was ineffective against Gram-negative bacteria (MIC values from 64 to ≥ 128 mg/L) whereas showed lower MIC values for Gram-positives (from 4 to 8 mg/L), except for
E. faecalis ATCC 29212 which reported a MIC value of 32 mg/L. The same trend was also reported for GC-VII-50 (MIC values from 0,25 to 8 mg/L). GC-VI-70 displayed high MIC values for Gram-negative bacteria and a better antibacterial activity against Gram-positive with MIC values ranging from 2 up to 16 mg/L. Moreover, all tested strains showed high MIC levels for GC-VI-10, except for
E. faecalis ATCC 29212 whose MIC value is 4 mg/L. All MIC values are listed in
Table 1.
2.2.1. Impact on Biological Activity and Antimicrobial Susceptibility Test
Due to its promising antimicrobial activity as well as its previously determined cytotoxic profile [
9,
10], PB4 was selected to perform Broth Microdilution (BMD) in combination with commonly used antibiotics such as linezolid, rifampin and erythromycin for Gram positive, imipenem for Gram negative and gentamycin and ampicillin for both, in order to examine a potential synergism.
Data reported in
Table 2 evidenced that PB4 led to a significant rifampin MIC reduction for S. aureus USA 300 (4-fold log reduction) and for E. faecalis ATCC 29212 (2-fold log reduction). Moreover, the combination with gentamycin led to 2-fold log MIC reduction for
K. pneumoniae ATCC 700603,
P. aeruginosa ATCC 27853 and
E. faecalis ATCC 29212. No significant MIC reductions have been reported for the combinations with linezolid, erythromycin, ampicillin, and imipenem, for all tested bacterial strains. Specifically, when combined with ampicillin, PB4 demonstrated a two-dilution decrease in MIC for several bacterial strains, including
P. aeruginosa ATCC 27853,
K. pneumoniae ATCC 700603, and
E. faecalis ATCC 29212. Particularly, in association with rifampicin, PB4 decreased the MIC of E. faecalis ATCC 29212 by 5 dilutions (from 0.125 mg/L to 0.006 mg/L); in combination with ampicillin, decreased MICs by two dilutions on
P. aeruginosa ATCC 27853 (2 mg/L to 0.5 mg/L),
K. pneumoniae ATCC 700603 (8 mg/L to 2 mg/L) and
E. faecalis ATCC 29212 (4 mg/L to 1 mg/L). Additionally, a synergism between PB4 and the newly tested GC-VI-70 was demonstrated, with a three-dilution decrease in the MIC of PB4 against
E. faecalis ATCC 29212 (from 0.5 mg/L to 0.06 mg/L) and a two-dilution decrease against
S. aureus ATCC 12598 (from 0.25 mg/L to 0.06 mg/L). Furthermore, since GC-VI-70 displayed the best MIC/cytotoxicity profile (see Section 3.4), with the lowest MIC value (2 mg/L) reported for
E. faecalis ATCC 29212 and an IC50 value on Caco-2 cells of 0.32 µM, it was selected to evaluate a potential synergism in combination with PB4 against Gram-positive bacteria. The combination of the two molecules revealed a significant synergistic effect, reducing PB4 MIC values by 2 logs for
E. faecalis ATCC 29212 and for
S. aureus ATCC 12598 (see
Table 3).
2.3. Evaluation of Heteroaryl-Ethylenes compounds Cell Cytotoxicity
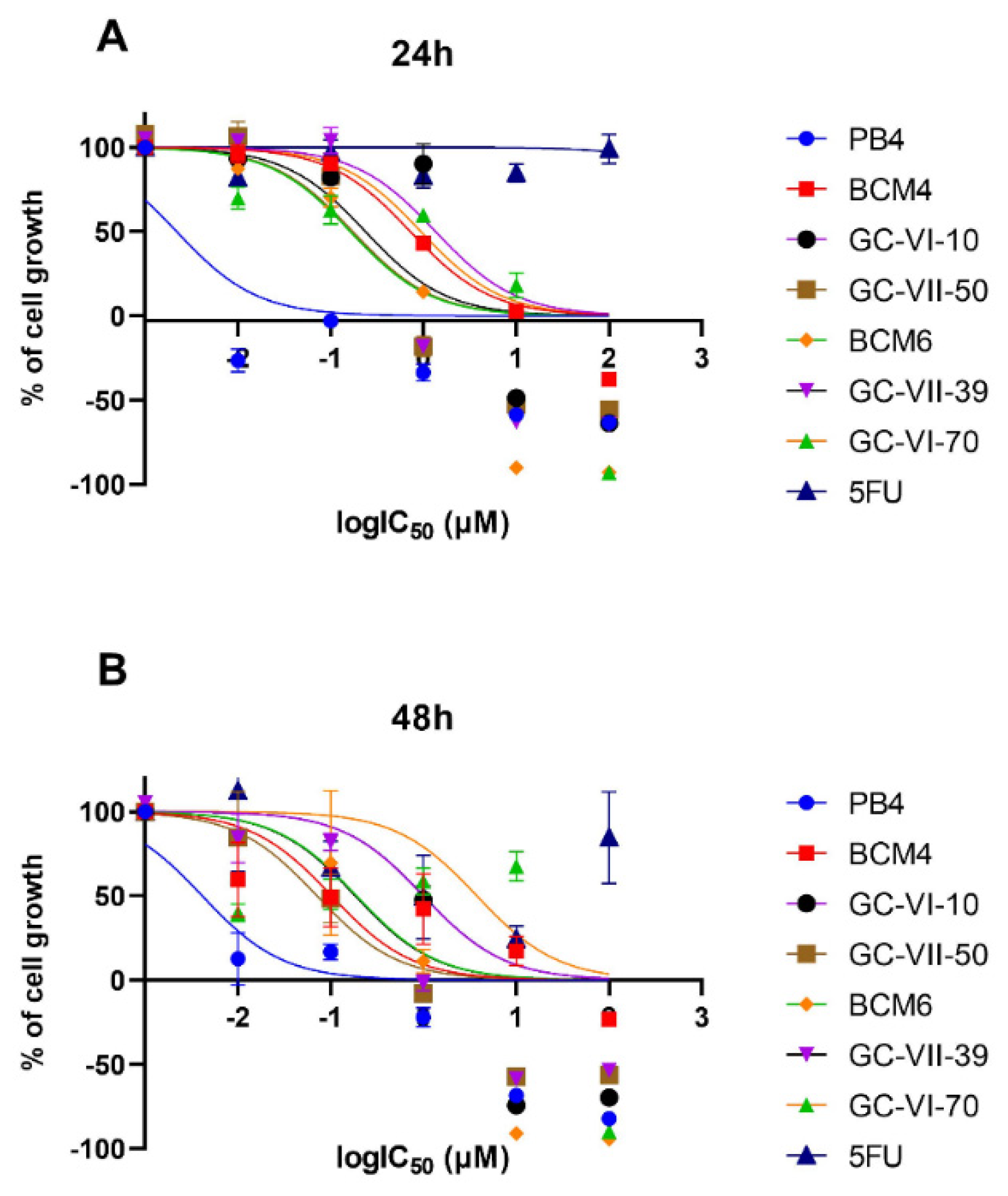
To fully assess the biological activity of the new molecules, they were tested at different concentration on CaCo-2 (ATCC HTB-37) colon-rectal cancer cell line. These human cancer cells were treated with compound solutions ranging in concentration from 100 µM to 0.01 µM. The reference compound for these assays was 5-Fluorouracil (5-FU), a pyrimidine analogue from the antimetabolite family. After 24 and 48 hours of incubation, the antiproliferative activity was evaluated by MTT assays to obtain cell growth curves and half maximal inhibitory concentration (IC50) valuesand the results are shown in Figure 1 and in
Table 4, respectively.
Figure 1.
Dose-response curve for each compound tested in the evaluation of cytotoxicity at 24 (A) and 48 (B) hours.
Figure 1.
Dose-response curve for each compound tested in the evaluation of cytotoxicity at 24 (A) and 48 (B) hours.
The results of the MTT assay showed that all heteroaryl-ethylene compounds are significantly more cytotoxic than 5-FU [
23], which is used in the assay as a reference antitumor compound. The compounds that reported the highest activity at 24 hours were BCM6, GC-VII-39 and GC-VII-50 with IC50 of 0.16, 0.23 and 0.17 μM, respectively. For comparison, 5-FU reported a 24-hour IC50 of about 27 μM. Due to the low cytotoxicity, we have evaluated the synergistic effects, as antimicrobial, when combining GC-VI-70 with PB4 (see
Section 2.2.1 and
Table 3)
2.3.1. Evaluation of Antibiotics Cytotoxicity
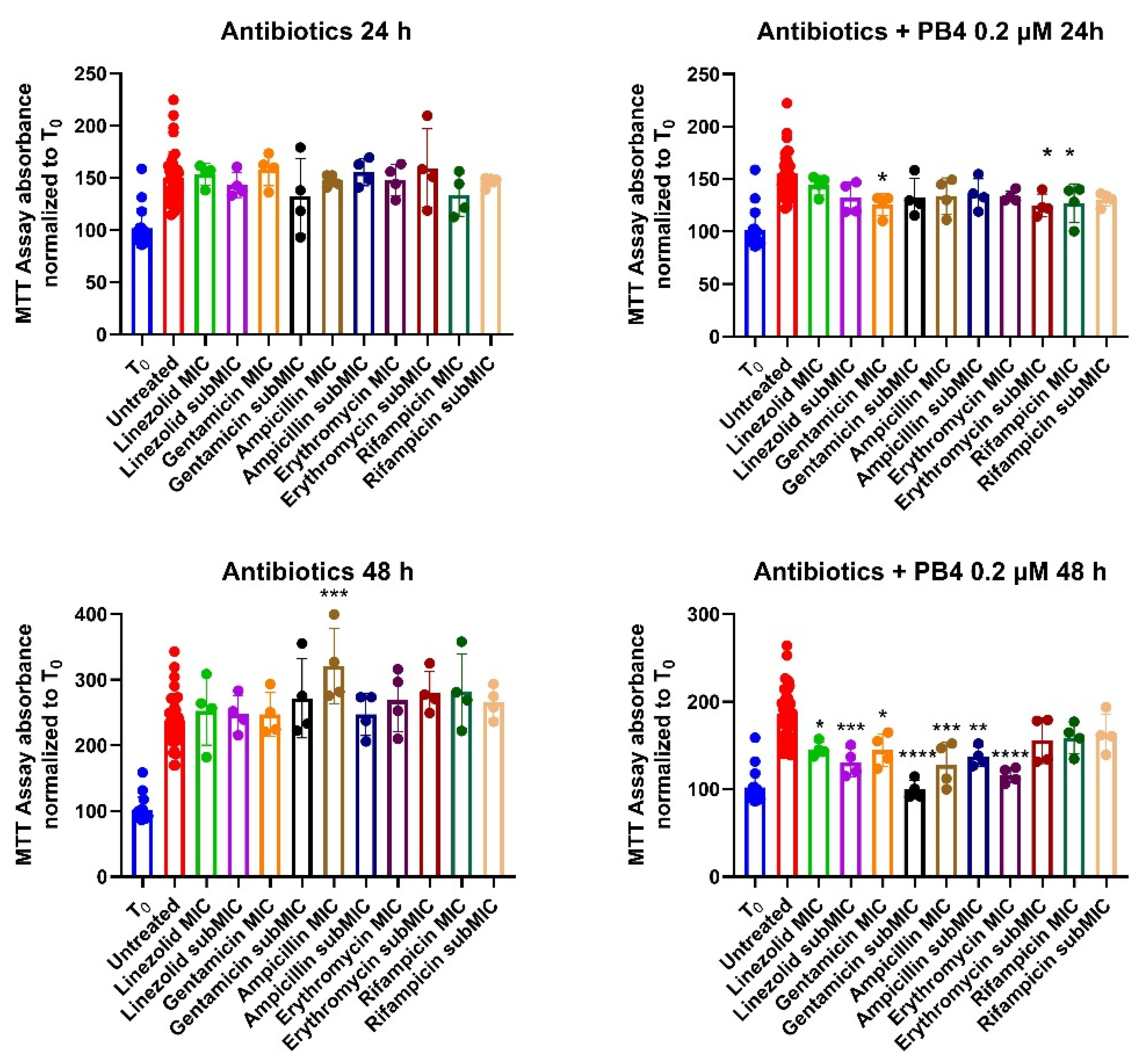
The MTT assay was performed also for the selected antibiotics to evaluate the effect of a combined treatment with PB4. The results are expressed as a percentage of cell growth compared to control, and not as IC50 (μM) values since only two concentrations have been tested rather than a defined range as in the case of heteroaryl-ethylene compounds. The results are shown in Figure 2 and
Table 3. Since we are setting up a co-culture model employing E. faecalis ATCC 29212 and CaCo-2 cells, the antibiotic concentrations chosen for cytotoxicity tests were only two: MIC and sub-MIC concentrations reported for E. faecalis ATCC 29212 (
Table 1). The use of PB4 as adjuvant to standard antibiotics has a dual purpose: i) contain bacterial proliferation, trying to use concentrations of both compounds that do not exert cytotoxic activity on the cells and ii) propose a new solution to emerging antibiotic resistance.
After 48 hours of treatment, no antibiotic alters the physiological cell growth (
Table 5), which indeed exceeds T0 and in some cases even doubles (Ampicillin-MIC 48 hours, 189,3%). Regarding the combination of antibiotics and PB4 0.2 μM, no significant difference was noted compared to cells treated with just PB4, and growth is greatly inhibited compared to solutions containing only antibiotic (mean of 77,7% and 80,7% at 24 and 48 hours compared to 87,3% and 158,5%).
A Dunnett's multiple comparisons test was performed to highlight any statistical differences between treated and untreated cells (Table S3). No remarkable statistical differences emerged for the cells treated with antibiotics alone, the only exception being observed between untreated cells and cells treated with Ampicillin at MIC concentration after 48 h. When cells are treated with antibiotics and PB4, after 24 hours, there is a statistical difference only with cells treated with gentamicin MIC, erythromycin subMIC and rifampicin MIC. All these differences, however, have a barely satisfactory P value. Nevertheless, the cells treated with Antibiotic and PB4 after 48 hours exhibited significant statistical differences with untreated cells, except rifampicin-treated cells at MIC and subMIC concentrations, and erythromycin-treated cells at subMIC concentrations.
Figure 2.
Percentage of cell growth compared to T0 with antibiotics and cells treated with antibiotics and 0.2 μM of PB4 at 24 (above) and 48 (under) hours.
Figure 2.
Percentage of cell growth compared to T0 with antibiotics and cells treated with antibiotics and 0.2 μM of PB4 at 24 (above) and 48 (under) hours.
3. Discussion
One approach to address antimicrobial resistance relay on the design and synthesis of novel molecules with antimicrobial properties exploring novel approaches based on rational in-silico design [
24,
25]. Taking advantage of a large in-house database of heteroaryl-ethylene molecule, we developed two QSAR models to selected six compounds and started the experimental study by testing their antibacterial activity against a range of Gram-positive and Gram-negative bacterial strains. The results revealed promising antimicrobial activity for these molecules and a good agreement with the QSAR model for antimicrobial activity against
S. aureus ATCC 29213. Furthermore, their effectiveness in combination therapy with commonly used antibiotics was investigated to explore possible synergistic interactions. Combination therapy, involving these molecules as adjuvants to existing antibiotics showed interesting results: the addition of PB4 significantly reduced the MIC of different antibiotic acting on the protein synthesis such as Rifampin in
S. aureus and
E. faecalis, gentamicin against
E. faecalis,
K. pneumoniae and
P. aeruginosa.
Overall, these results provide us important preliminary data to set up a eukaryotic-prokaryotic co-culture employing
E. faecalis ATCC 29212 and CaCo-2 cells.
E. faecalis co-culture with human intestinal epithelial cells has been used in several studies to investigate the bacterial ability to adhere to, penetrate, and destroy the host epithelium [
26,
27]. In the context of this infection model, the use of molecules such as PB4 or GC-VI-70 in combination with antibiotics may allow us to prolong the co-culture model avoiding both uncontrolled bacterial proliferation and cytotoxic outcomes. As shown, the combination of antibiotics such as gentamicin and rifampicin with PB4 can lead to significant decreases in MIC values, leaving cellular metabolism almost unchanged. Combinations like this will be crucial for setting up the coculture experiments as all heteroaryl compounds presented in this study exhibit high cytotoxic activity against colorectal cancer cells.
Taken together, these results encouraged us to focus our research on heteroaryl-ethylene structures for the development of drugs with multitarget biological activity and particular attention has to be paid to the design of compounds active against Gram negative strains [
11,
28]. The biological activity of PB4, will be the main focus of the studies of this multidisciplinary research group, to understand the mechanisms by which this substance proves to be antimicrobial and cytotoxic. Despite their cytotoxicity, this compound showed to be promising for combination therapies with common antibiotics, in order to enhance their effectiveness also when used at very low concentrations that do not show cytotoxic effects [
29,
30,
31]
The development of new synthetic molecules with antibacterial activity is of paramount importance, as they offer the potential to overcome existing resistance mechanisms, provide broad-spectrum activity, and reduce toxicity [
32]. At the same time, is noteworthy that the mechanisms of action should be evaluated in terms of the observed synergistic effects. In addition, in vivo studies will be useful to assess the safety, pharmacokinetics, and efficacy of these molecules in more complex biological systems. It becomes so undeniable how further research and investment in the discovery and development of such molecules are crucial to combating the threat of antibiotic resistance and ensuring effective treatment options for multidrug-resistant bacterial infections [
33,
34]. This work highlights an important awareness: the design of new chemical compounds is of dominant importance for medicine [
35]. It enables the development of innovative drugs with enhanced efficacy and reduced side effects.
4. Materials and Methods
4.1. Dataset of the QSAR models
The Principal Component Analysis (PCA) and a PLS models were performed through the Volsurf+ (VS+) software, developed by Molecular Discovery (see
Supplementary Materials for further details). [
36,
37]
The dataset of the QSAR model for antimicrobial activity comprised 53 Heteroaromatic compounds (
Table S1) with experimental values for antimicrobial activity against
S. aureus ATCC29213. The antimicrobial activity was evaluated by means of standard MIC (minimal inhibition concentration, mg/L).
For the development of the QSAR model for cytotoxic activity, the in-house dataset comprised 38 Heteroaryl Ethylenes (
Table S2) with experimental in-vitro activity values, expressed as logGI50, against CaCo-2 colon-rectal cancer cell line.
4.2. Compounds Synthesis
Compounds BCM4, BCM6, GC-VII-39, GC-VII-50, GC-VI-70, GC-VI-10 were synthesized via base-catalysed Knoevenagel condensation between the proper pyridinium/quinolinium ions, as their iodide salts, and the corresponding aldehyde following previously reported procedures. [
11,
12,
38,
39,
40,
41]
4.3. Bacterial Strains
To assess the antibacterial properties and the spectrum of activity of the six new synthetic chemical compounds as well as our lead compound, PB4, nine ATCC bacterial strains, both Gram-positive and Gram-negative, were selected (
Table 6). For S. aureus were selected two methicillin-susceptible
Staphylococcus aureus (MSSA) and 1 MRSA (ATCC BAA-1556). All the control strains were provided by the American Type Culture Collection (Manassas, VA, USA). All tests were conducted in duplicate and both replicates showed the same MIC values.
4.4. Bacterial Growth Conditions
S. aureus and E. faecalis strains were cultivated respectively on Mannitol Salt Agar (Cat. No. CM0085B) and Bile Aesculin Agar (Cat. No. CM0888), whereas E. coli, A. baumannii, P. aeruginosa, K. pneumoniae and S. enterica on MacConkey Agar (Cat. No. CM0007). All bacterial strains were incubated O/N at 37°C. All culture media were purchased from Thermo ScientificTM OxoidTM, Basingstoke, UK.
4.5. Antimicrobial Susceptibility Test
The Antimicrobial Susceptibility Test (AST) was conducted performing Broth Microdilution (BMD) to determine the Minimum Inhibitory Concentration (MIC) of the selected compounds and PB4, as well as of some well-known antibiotics such as linezolid (Cat. No. 460592500), gentamicin (Cat. No. 15750037), ampicillin (Cat. No. J60977.06), erythromycin (Cat. No. J62279.09), and rifampin (Cat. No. J60836.03). All antibiotics were provided by Thermo ScientificTM OxoidTM, Basingstoke, UK. The tested concentrations range from 128 to 0,125 mg/L.
We tested the combination of PB4 and the five antibiotics to examine their synergic effects on the ATCC 29212
E. faecalis strain. PB4 was used at fixed concentration of 0,125 μg/mL (0,24 μM), corresponding to a quarter of MIC value, whereas antibiotics concentrations range from 128 to 0,125 mg/L. BMD was performed using Cation-Adjusted Müeller Hinton Broth (CA-MHB) (Cat. No. 212322, BD BBLTM, Franklin Lakes, NJ, USA) according to standard methods. [
42] All antibiotics were solved in appropriate solvent according to CLSI guidelines. [
43] All heteroaryl-ethylenes compounds were solved in 100% DimethylSulfoxide (DMSO) (Cat. No. 85190, Thermo ScientificTM OxoidTM, Basingstoke, UK) obtaining a starting concentration of 8000 mg/L. Intermediate dilutions were carried out using CA-MHB in order to reduce final DMSO concentration. The highest concentration of tested compounds was 128 mg/L, with a final DMSO concentration in the well of almost 1,6 %. As described in literature, this DMSO concentration was shown to be well-tolerated by bacteria and it is comparable to the concentration of 1% recommended by CLSI guidelines [
42]. Additionally, a synergic assay was also performed to assess the effectiveness of PB4/GC-VI-70 combination which are, respectively, a heteroaryl-ethylene compound, previously tested in published studies [
9], and one of the six tested compounds that showed the best cytotoxicity/MIC profile. The synergism was assessed by BMD on Gram-positive ATCC strains: ATCC 29231, ATCC 12598, ATCC BAA-1556 (
S. aureus) and ATCC 29212 (
E. faecalis). PB4 was used at fixed concentration equal to the MIC value for the specific strain, whereas GC-VI-70 tested concentrations range from 128 to 0,125 mg/L.
4.6. Evaluation of the Cytotoxic Activity of the compounds on Human Colorectal Adenocarcinoma Cells
To evaluate the effect of the Heteroaryl-Ethylenes compounds and the antibiotics, an MTT ([3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]) assay was performed as previously described [
10]. Briefly, human colorectal adenocarcinoma cells (CaCo-2 HTB-37TM, American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s MEM (DMEM) with 10% heat-inactivated fetal bovine serum, 2 mM L-Alanyl-L-Glutamine, penicillin-streptomycin (50 units-50 μg for mL, just for cells treated with Heteroaryl-Ethylenes compounds) and incubated at 37 °C in a humidified atmosphere of 5% CO2, 95% air. CaCo-2 cells were plated in 96 well pates and incubated at 37 °C. The Heteroaryl-Ethylenes solutions and 5-FU (Cat. No. F6627, Merck KGaA, Darmstadt, Germany) were prepared as a 1 mM solution in 10 mL with 0.01% DMSO. In addition to this, the cytotoxic activity of five common antibiotics, linezolid (Cat. No. 460592500), gentamicin (Cat. No. 15750037), ampicillin (Cat. No. J60977.06), erythromycin (Cat. No. J62279.09) and rifampin (Cat. No. J60836.03), was also tested, treating cells at two concentrations corresponding to MIC and half-MIC values obtained for ATCC 29212
E. faecalis strain. Both concentrations were also tested in combination with PB4 0,2 µM. All antibiotics were provided by Thermo ScientificTM OxoidTM, Basingstoke, UK. A summary of solutions tested, and their relatives’ concentrations are reported in
Table 7. Twenty-four hours after plating, cells were treated with 20 μL of each solution. Untreated cells were used as controls. Microplates were incubated at 37 °C in a humidified atmosphere of 5% CO2, 95% air for 24 h, and then cytotoxicity was measured with colorimetric assay based on the use of tetrazolium salt MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide. IC50: this parameter expresses the concentration of the tested compound necessary to kill half of the cell population after 24 and 48 h of incubation relative to untreated controls. The absorbance values at 569 nm were obtained using multiwell plate reader (Synergy H1, Biotek, Via Rodolfo Farneti, 8, 20129 Milano MI). Each value stems from the average of four wells. The IC50 values were calculated by nonlinear regression analysis using the GraphPad Prism 6.0 software.
5. Conclusions
These compounds serve as building blocks for designing novel therapeutic agents [
44], targeting previously untreatable diseases. The easy end versatile synthetic process allows for optimization of drug properties, such as solubility [
45], stability [
46], and bioavailability [
47], leading to better drug formulations. Additionally, new chemical compounds offer opportunities for discovering new mechanisms of action, expanding the understanding of disease biology, and ultimately advancing medical treatment [
24]. The application of combination therapy employing novel molecules as adjuvants to existing antibiotics offers several advantages. Firstly, it may restore or enhance the effectiveness of existing antibiotics that have lost their efficacy against resistant strains. Secondly, it can help reducing the overall dosage of antibiotic compounds, potentially minimizing the risk of adverse side effects and decreasing the selective pressure for resistance development. Moreover, the use of synergistic combinations can potentially overcome existing resistance mechanisms, providing an effective treatment option against multidrug-resistant bacteria.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
For research articles with several authors, a short paragraph specifying their individual contributions must be provided. The following statements should be used “Conceptualization, N.M., A.M.; methodology, C.B., P.G.B., E.N.; software, C.Bona.; validation, D.B., A.Ma. and G.C.; formal analysis, N.M, P.G.B., and C. Bona.; investigation, D.A.B., A.M.; resources, G.C., C.G.F and S.S.; data curation, C.B., P.G.B., C. Bona. and N.M.; writing—original draft preparation, C.B., P.G.B., C. Bona. and N.M.; writing—review and editing, D.B. D.A.B., and A.M.; visualization, A.M.; supervision, D.B., S.S. and N.M.; project administration, D.B., S.S. and N.M.; funding acquisition, C.G.F. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
We would like to thank the Bio-nanotech Research and Innovation Tower (BRIT) service center belonging to the University of Catania (Italy) for the use of its facility and instruments as well as for providing us a valuable technical assistance. This work has been partially funded by European Union (NextGeneration EU), through the MUR-PNRR project SAMOTHRACE (ECS00000022) and through the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT), and by Italian Ministry of Health Research Program (grant number RC2022-N4).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ventola, C.L. The Antibiotic Resistance Crisis. P T 2015, 40, 277–283. [Google Scholar] [PubMed]
- Chinemerem Nwobodo, D.; Ugwu, M.C.; Oliseloke Anie, C.; Al-Ouqaili, M.T.S.; Chinedu Ikem, J.; Victor Chigozie, U.; Saki, M. Antibiotic Resistance: The Challenges and Some Emerging Strategies for Tackling a Global Menace. Journal of Clinical Laboratory Analysis 2022, 36, e24655. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus Aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.Z.; Kendall, B. Carbapenem Resistant Enterobacteriaceae. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2023. [Google Scholar]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog Glob Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [CrossRef]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Frontiers in Microbiology 2019, 10. [Google Scholar] [CrossRef]
- Schramm, L.; Byrne, M.K.; Sweetnam, T. Antibiotic Misuse Behaviours of Older People: Confirmation of the Factor Structure of the Antibiotic Use Questionnaire. Antibiotics 2023, 12, 718. [Google Scholar] [CrossRef]
- Bongiorno, D.; Musso, N.; Bonacci, P.G.; Bivona, D.A.; Massimino, M.; Stracquadanio, S.; Bonaccorso, C.; Fortuna, C.G.; Stefani, S. Heteroaryl-Ethylenes as New Lead Compounds in the Fight against High Priority Bacterial Strains. Antibiotics 2021, 10, 1034. [Google Scholar] [CrossRef]
- Bivona, D.A.; Mirabile, A.; Bonomo, C.; Bonacci, P.G.; Stracquadanio, S.; Marino, A.; Campanile, F.; Bonaccorso, C.; Fortuna, C.G.; Stefani, S.; et al. Heteroaryl-Ethylenes as New Effective Agents for High Priority Gram-Positive and Gram-Negative Bacterial Clinical Isolates. Antibiotics 2022, 11, 767. [Google Scholar] [CrossRef]
- Fortuna, C.G.; Barresi, V.; Bonaccorso, C.; Consiglio, G.; Failla, S.; Trovato-Salinaro, A.; Musumarra, G. Design, Synthesis and in Vitro Antitumour Activity of New Heteroaryl Ethylenes. Eur J Med Chem 2012, 47, 221–227. [Google Scholar] [CrossRef]
- Barresi, V.; Bonaccorso, C.; Consiglio, G.; Goracci, L.; Musso, N.; Musumarra, G.; Satriano, C.; Fortuna, C.G. Modeling, Design and Synthesis of New Heteroaryl Ethylenes Active against the MCF-7 Breast Cancer Cell-Line. Mol. BioSyst. 2013, 9, 2426–2429. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorso, C.; Grasso, G.; Musso, N.; Barresi, V.; Condorelli, D.F.; La Mendola, D.; Rizzarelli, E. Water Soluble Glucose Derivative of Thiocarbohydrazone Acts as Ionophore with Cytotoxic Effects on Tumor Cells. Journal of Inorganic Biochemistry 2018, 182, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorso, C.; Naletova, I.; Satriano, C.; Spampinato, G.; Barresi, V.; Fortuna, C.G. New Di(Heteroaryl)Ethenes as Apoptotic Anti-Proliferative Agents Towards Breast Cancer: Design, One-Pot Synthesis and In Vitro Evaluation. ChemistrySelect 2020, 5, 2581–2587. [Google Scholar] [CrossRef]
- Pantaleão, S.Q.; Fernandes, P.O.; Gonçalves, J.E.; Maltarollo, V.G.; Honorio, K.M. Recent Advances in the Prediction of Pharmacokinetics Properties in Drug Design Studies: A Review. ChemMedChem 2022, 17, e202100542. [Google Scholar] [CrossRef] [PubMed]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.M.; Govender, T.; Naicker, T.; Kruger, H.G. Current Trends in Computer Aided Drug Design and a Highlight of Drugs Discovered via Computational Techniques: A Review. Eur J Med Chem 2021, 224, 113705. [Google Scholar] [CrossRef]
- Roman, G. Thiophene-Containing Compounds with Antimicrobial Activity. Archiv der Pharmazie 2022, 355, 2100462. [Google Scholar] [CrossRef] [PubMed]
- Leontiev, R.; Hohaus, N.; Jacob, C.; Gruhlke, M.C.H.; Slusarenko, A.J. A Comparison of the Antibacterial and Antifungal Activities of Thiosulfinate Analogues of Allicin. Sci Rep 2018, 8, 6763. [Google Scholar] [CrossRef]
- Eyvazi, S.; Vostakolaei, M.A.; Dilmaghani, A.; Borumandi, O.; Hejazi, M.S.; Kahroba, H.; Tarhriz, V. The Oncogenic Roles of Bacterial Infections in Development of Cancer. Microb Pathog 2020, 141, 104019. [Google Scholar] [CrossRef]
- Si, H.; Yang, Q.; Hu, H.; Ding, C.; Wang, H.; Lin, X. Colorectal Cancer Occurrence and Treatment Based on Changes in Intestinal Flora. Semin Cancer Biol 2021, 70, 3–10. [Google Scholar] [CrossRef]
- Sun, J.; Kato, I. Gut Microbiota, Inflammation and Colorectal Cancer. Genes Dis 2016, 3, 130–143. [Google Scholar] [CrossRef]
- Wangngae, S.; Ngivprom, U.; Khrootkaew, T.; Worakaensai, S.; Lai, R.-Y.; Kamkaew, A. Cationic Styryl Dyes for DNA Labelling and Selectivity toward Cancer Cells and Gram-Negative Bacteria. RSC Adv. 2023, 13, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Diasio, R.B.; Harris, B.E. Clinical Pharmacology of 5-Fluorouracil. Clin Pharmacokinet 1989, 16, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Koirala, B.; Hernandez, Y.; Zimmerman, M.; Park, S.; Perlin, D.S.; Brady, S.F. A Naturally Inspired Antibiotic to Target Multidrug-Resistant Pathogens. Nature 2022, 601, 606–611. [Google Scholar] [CrossRef]
- Shil, A.; Chichger, H. Artificial Sweeteners Negatively Regulate Pathogenic Characteristics of Two Model Gut Bacteria, E. Coli and E. Faecalis. Int J Mol Sci 2021, 22, 5228. [Google Scholar] [CrossRef]
- Wachsmannova, L.; Stevurkova, V.; Ciernikova, S. Changes in SNAI1 and VIM Gene Expression in Caco2 Cells after Cocultivation with Bacteria from Colorectal Cancer Biopsies. Neoplasma 2019, 66, 271–275. [Google Scholar] [CrossRef]
- Campos, J.; Núñez, C.; Díaz, J.J.; Sánchez, R.M.; Gallo, M.A.; Espinosa, A. Anticancer Bisquaternary Heterocyclic Compounds: A Ras-Ional Design. Farmaco 2003, 58, 221–229. [Google Scholar] [CrossRef]
- Rogers, G.B.; Carroll, M.P.; Bruce, K.D. Enhancing the Utility of Existing Antibiotics by Targeting Bacterial Behaviour? Br J Pharmacol 2012, 165, 845–857. [Google Scholar] [CrossRef]
- Konaklieva, M.I. Addressing Antimicrobial Resistance through New Medicinal and Synthetic Chemistry Strategies. SLAS Discovery 2019, 24, 419–439. [Google Scholar] [CrossRef]
- Marschall, E.; Cryle, M.J.; Tailhades, J. Biological, Chemical, and Biochemical Strategies for Modifying Glycopeptide Antibiotics. Journal of Biological Chemistry 2019, 294, 18769–18783. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the Sustainable Discovery and Development of New Antibiotics. Nat Rev Chem 2021, 5, 726–749. [Google Scholar] [CrossRef] [PubMed]
- Dinicola, S.; De Grazia, S.; Carlomagno, G.; Pintucci, J.P. N-Acetylcysteine as Powerful Molecule to Destroy Bacterial Biofilms. A Systematic Review. Eur Rev Med Pharmacol Sci 2014, 18, 2942–2948. [Google Scholar] [PubMed]
- Leitão, J.H. New Insights into Antibacterial Compounds: From Synthesis and Discovery to Molecular Mechanisms of Action. Antibiotics 2020, 9, 471. [Google Scholar] [CrossRef]
- Nicolaou, K.C. The Emergence and Evolution of Organic Synthesis and Why It Is Important to Sustain It as an Advancing Art and Science for Its Own Sake. Israel Journal of Chemistry 2018, 58, 104–113. [Google Scholar] [CrossRef]
- Cruciani, G.; Crivori, P.; Carrupt, P.-A.; Testa, B. Molecular Fields in Quantitative Structure–Permeation Relationships: The VolSurf Approach. Journal of Molecular Structure: THEOCHEM 2000, 503, 17–30. [Google Scholar] [CrossRef]
- Cruciani, G.; Pastor, M.; Guba, W. VolSurf: A New Tool for the Pharmacokinetic Optimization of Lead Compounds. Eur J Pharm Sci 2000, 11 Suppl 2, S29–39. [Google Scholar] [CrossRef]
- Xu, X.; Qiu, W.; Zhou, Q.; Tang, J.; Yang, F.; Sun, Z.; Audebert, P. Nonlinear Optical Absorption Properties of Two Multisubstituted P-Dimethylaminophenylethenyl Pyridiniums. J Phys Chem B 2008, 112, 4913–4917. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Shewale, D.J.; Sengupta, A.; Soppina, V.; Kanvah, S. Lutidine Derivatives for Live-Cell Imaging of the Mitochondria and Endoplasmic Reticulum. Org. Biomol. Chem. 2022, 20, 7047–7055. [Google Scholar] [CrossRef] [PubMed]
- Fortuna, C.G.; Bonaccorso, C.; Qamar, F.; Anu, A.; Ledoux, I.; Musumarra, G. Synthesis and NLO Properties of New Trans2-(Thiophen-2-Yl)Vinyl Heteroaromatic Iodides. Org. Biomol. Chem. 2011, 9, 1608–1613. [Google Scholar] [CrossRef]
- Carlotti, B.; Benassi, E.; Spalletti, A.; Fortuna, C.G.; Elisei, F.; Barone, V. Photoinduced Symmetry-Breaking Intramolecular Charge Transfer in a Quadrupolar Pyridinium Derivative. Phys. Chem. Chem. Phys. 2014, 16, 13984–13994. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute Performance Standars of Antimicrobial Susceptibility Testing. 2020.
- Weinstein, M.P.; Patel, J.B. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: M07-A11; Documents / Clinical and Laboratory Standards Institute; 11. edition.; Committee for Clinical Laboratory Standards: Wayne, PA, 2018; ISBN 978-1-56238-836-2.
- Yakushiji, F. [Development of Novel Biologically Active Compounds Based on Synthetic Organic Chemistry]. Yakugaku Zasshi 2022, 142, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tehler, U.; Fagerberg, J.H.; Svensson, R.; Larhed, M.; Artursson, P.; Bergström, C.A.S. Optimizing Solubility and Permeability of a Biopharmaceutics Classification System (BCS) Class 4 Antibiotic Drug Using Lipophilic Fragments Disturbing the Crystal Lattice. J. Med. Chem. 2013, 56, 2690–2694. [Google Scholar] [CrossRef] [PubMed]
- Masimirembwa, C.M.; Bredberg, U.; Andersson, T.B. Metabolic Stability for Drug Discovery and Development: Pharmacokinetic and Biochemical Challenges. Clin Pharmacokinet 2003, 42, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Bhalani, D.V.; Nutan, B.; Kumar, A.; Singh Chandel, A.K. Bioavailability Enhancement Techniques for Poorly Aqueous Soluble Drugs and Therapeutics. Biomedicines 2022, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
Table 1.
Activity of all tested molecules on ATCC bacterial strains. All MIC values are expressed in mg/L and µM. The most promising MIC values are highlighted in bold.
Table 1.
Activity of all tested molecules on ATCC bacterial strains. All MIC values are expressed in mg/L and µM. The most promising MIC values are highlighted in bold.
| |
|
PB4 |
BCM4 |
BCM6 |
GC-VII-39 |
GC-VI-70 |
GC-VII-50 |
GC-VI-10 |
| Species |
Strain |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
| E. faecalis |
ATCC 29212 |
0.5 |
0.98 |
≥128 |
≥349.5 |
32 |
65.93 |
16 |
30.45 |
2*
|
3.32 |
8*
|
18.5 |
4*
|
5.2 |
| S. aureus |
ATCC 29213 |
0.125 |
0.24 |
128 |
349.5 |
4*
|
8.2 |
128 |
243.6 |
16 |
26.6 |
0.5*
|
1.1 |
128 |
168.9 |
| S. aureus |
ATCC 12598 |
0.25 |
0.49 |
64 |
174.7 |
4*
|
8.23 |
32 |
60.9 |
16 |
26.6 |
0.25*
|
0.5 |
64 |
84.4 |
| S. aureus |
ATCC BAA-1556 USA300 |
≤0.125 |
≤0.24 |
64 |
174.7 |
8*
|
16.4 |
>128 |
>243.6 |
4*
|
6.65 |
2*
|
4.6 |
128 |
168.9 |
| K. pneumoniae |
ATCC 700603 |
64 |
125.1 |
>128 |
≥349.5 |
>128 |
>263.7 |
≥128 |
≥243.6 |
>128 |
>212.7 |
>128 |
>297.4 |
>128 |
>168.9 |
| A. baumannii |
ATCC 17978 |
≤0.125 |
≤0.24 |
≥128 |
≥349.5 |
64 |
131.8 |
≥128 |
≥243.6 |
32 |
53.19 |
16 |
37.18 |
>128 |
>168.9 |
| P. aeruginosa |
ATCC 27853 |
>128 |
>250.2 |
>128 |
≥349.5 |
>128 |
>263.7 |
≥128 |
≥243.6 |
>128 |
>212.7 |
>128 |
>297.4 |
>128 |
>168.9 |
| S. enterica |
ATCC 14028 |
64 |
125.1 |
>128 |
≥349.5 |
>128 |
>263.7 |
≥128 |
≥243.6 |
>128 |
>212.7 |
>128 |
>297.4 |
>128 |
>168.9 |
| E. coli |
ATCC 25922 |
2 |
3.91 |
≥128 |
≥349.5 |
128 |
263.7 |
≥128 |
≥128 |
128 |
212.7 |
64 |
148.7 |
>128 |
>168.9 |
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table 2.
Antimicrobial activity of PB4 in combination with commonly used antibiotics on E. faecalis ATCC 29212. MIC values are expressed in mg/L. Data in bold and with asterisks indicate the most significant decreases.
Table 2.
Antimicrobial activity of PB4 in combination with commonly used antibiotics on E. faecalis ATCC 29212. MIC values are expressed in mg/L. Data in bold and with asterisks indicate the most significant decreases.
| Species |
Strain |
LNZ |
LNZ + PB4 |
RD |
RD + PB4 |
E |
E + PB4 |
CN |
CN + PB4 |
AMP |
AMP + PB4 |
IMI |
IMI + PB4 |
| E. faecalis |
ATCC 29212 |
4 |
2 |
0,25* |
0,06* |
2 |
1 |
4* |
1* |
0,25 |
0,125 |
- |
- |
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
| S. aureus |
ATCC 29213 |
4 |
4 |
0,125 |
0,125 |
0,5 |
0,5 |
0,5 |
0,5 |
2 |
8 |
- |
- |
| ATCC 12598 |
4 |
4 |
0,125 |
0,125 |
0,5 |
0,5 |
0,5 |
0,5 |
0,125 |
0,125 |
- |
- |
| USA 300 |
2 |
1 |
0,125* |
0,006* |
32 |
32 |
0,5 |
0,5 |
8 |
8 |
- |
- |
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
| K. pneumoniae |
ATCC 700603 |
- |
- |
- |
- |
- |
- |
8* |
2* |
> 128 |
> 128 |
0,03 |
0,03 |
| A. baumannii |
ATCC 17978 |
- |
- |
- |
- |
- |
- |
2 |
2 |
32 |
32 |
0,06 |
0,06 |
| P. aeruginosa |
ATCC 27853 |
- |
- |
- |
- |
- |
- |
2* |
0,5* |
> 128 |
> 128 |
0,25 |
0,25 |
| S. enterica |
ATCC 14028 |
- |
- |
- |
- |
- |
- |
1 |
1 |
2 |
2 |
0,06 |
0,06 |
| E. coli |
ATCC 25922 |
- |
- |
- |
- |
- |
- |
0,5 |
0,5 |
8 |
8 |
0,06 |
0,06 |
Table 3.
Synergistic activity of PB4 in combination with GC-VI-70 on Gram-positive bacterial strains. All MIC values are expressed in mg/L and µM. In bold and with asterisks the significant decreases.
Table 3.
Synergistic activity of PB4 in combination with GC-VI-70 on Gram-positive bacterial strains. All MIC values are expressed in mg/L and µM. In bold and with asterisks the significant decreases.
| |
|
PB4 |
GC-VI-70 |
PB4 +
GC-VI-70
|
| Species |
Strain |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
| E. faecalis |
ATCC 29212 |
0,5 |
0,98 |
2 |
3,3 |
0,06* |
0,2* |
| S. aureus |
ATCC 29213 |
0,125 |
0,24 |
16 |
26,5 |
1 |
1,66 |
| S. aureus |
ATCC 12598 |
0,25 |
0,49 |
16 |
26,5 |
0,06* |
0,2* |
| S. aureus |
ATCC BAA-1556 USA 300 |
≤0,125 |
≤0,24 |
4 |
6,6 |
2 |
3,3 |
Table 4.
IC50 (µM) for all tested molecules on CaCo-2 cell line after 24 and 48 hours of treatment.
Table 4.
IC50 (µM) for all tested molecules on CaCo-2 cell line after 24 and 48 hours of treatment.
| Molecule |
IC50 at 24 hours (µM) |
IC50 at 48 hours (µM) |
| BCM4 |
0,72 |
0,10 |
| BCM6 |
0,16 |
0,18 |
| GC-VII-39 |
0,23 |
0,18 |
| GC-VII-50 |
0,17 |
0,07 |
| GC-VI-70 |
0,94 |
3,76 |
| GC-VI-10 |
1,35 |
0,99 |
| 5-FU |
27,36 |
12,91 |
| PB4 |
0,31 |
0,3 |
Table 5.
Percentage of growth compared to control of cells treated with antibiotics and with the antibiotic/PB4 combination 0.2 μM after 24 and 48 hours. The same percentage is also reported for the percentage of cell growth at the point of IC50 reported, to demonstrate that the inhibition of growth in antibiotic/PB4 combination is dictated solely by the presence of the heteroaryl-ethylene compound.
Table 5.
Percentage of growth compared to control of cells treated with antibiotics and with the antibiotic/PB4 combination 0.2 μM after 24 and 48 hours. The same percentage is also reported for the percentage of cell growth at the point of IC50 reported, to demonstrate that the inhibition of growth in antibiotic/PB4 combination is dictated solely by the presence of the heteroaryl-ethylene compound.
| |
Antibiotic |
Antibiotic +
PB4 0,2 µM
|
| 24h |
48h |
24h |
48h |
| Linezolid |
MIC |
153,5 |
252,9 |
144,0 |
145,7 |
| subMIC |
143,4 |
247,6 |
132,0 |
130,6 |
| Gentamicin |
MIC |
158,2 |
247,5 |
125,6 |
141,1 |
| subMIC |
132,3 |
271,7 |
132,4 |
99,2 |
| Ampicillin |
MIC |
147,7 |
321,1 |
133,8 |
127,7 |
| subMIC |
155,6 |
247,5 |
135,6 |
137,0 |
| Erythromycin |
MIC |
148,2 |
269,0 |
133,6 |
117,5 |
| subMIC |
159,5 |
280,6 |
124,9 |
155,8 |
| Rifampin |
MIC |
133,7 |
282,6 |
126,9 |
158,9 |
| subMIC |
146,6 |
265,6 |
130,6 |
163,7 |
| Mean |
147,9 |
268,6 |
131,9 |
137,7 |
| PB4 0,2 µM |
| 24h |
48h |
| 68,8 |
72,7 |
Table 6.
ATCC bacterial strains selected for antimicrobial susceptibility tests.
Table 6.
ATCC bacterial strains selected for antimicrobial susceptibility tests.
| |
Strain |
Species |
| Gram + |
ATCC 29212 |
Enterococcus faecalis |
| ATCC 29213 |
Staphylococcus aureus |
| ATCC 12598 |
Staphylococcus aureus |
| ATCC BAA-1556 |
Staphylococcus aureus
sub. Rosenbach (USA300 clone) |
| Gram - |
ATCC 700603 |
Klebsiella pneumoniae |
| ATCC 17978 |
Acinetobacter baumannii |
| ATCC 27853 |
Pseudomonas aeruginosa |
| ATCC 14028 |
Salmonella enterica |
| ATCC 25922 |
Escherichia coli |
Table 7.
Compounds and concentrations used in cytotoxicity tests.
Table 7.
Compounds and concentrations used in cytotoxicity tests.
| Compound |
|
|
| |
PB4 |
BCM4 |
BCM6 |
GC-VII-39 |
GC-VII-50 |
GC-VI-10 |
5-FU |
GC-VI-70 |
|
|
| Concentration Tested |
100 µM, 10 µM, 1 µM, 0.1 µM and 0.01 µM |
|
|
| |
Antibiotic |
| |
Linezolid |
Gentamicin |
Ampicillin |
Erythromycin |
Rifampin |
| |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
mg/L |
µM |
| MIC Concentration Tested |
4 |
11.85 |
4 |
7.74 |
0.25 |
0.72 |
2 |
2.73 |
0.25 |
0.30 |
| Sub-MIC Concentration Tested |
2 |
5.93 |
2 |
3.87 |
0.125 |
0.36 |
1 |
1.36 |
0.125 |
0.15 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).