Submitted:
26 July 2023
Posted:
28 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Result
2.1. Identification and sequence feature
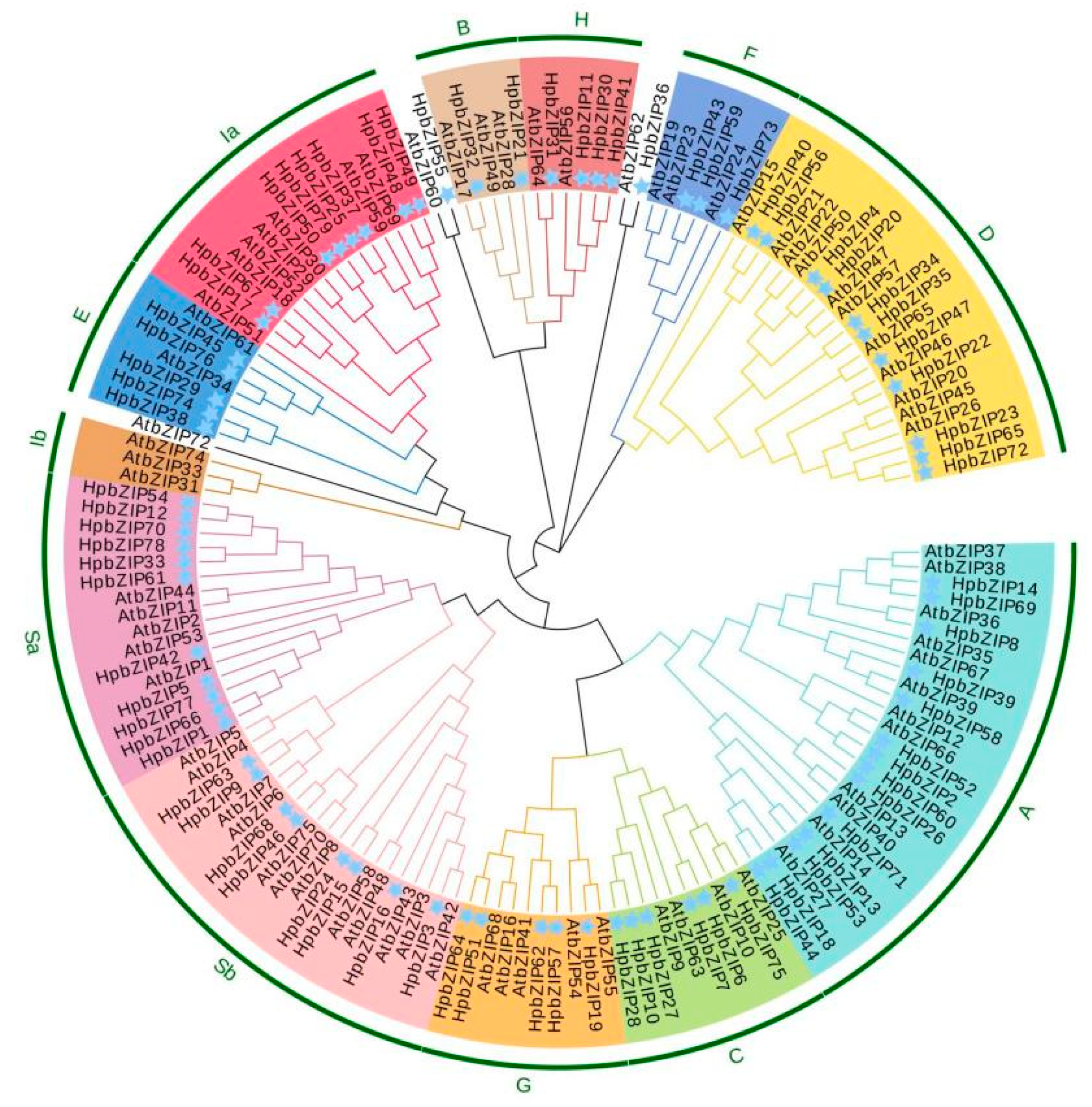
2.2. Phylogenetic analysis of HpbZIPs
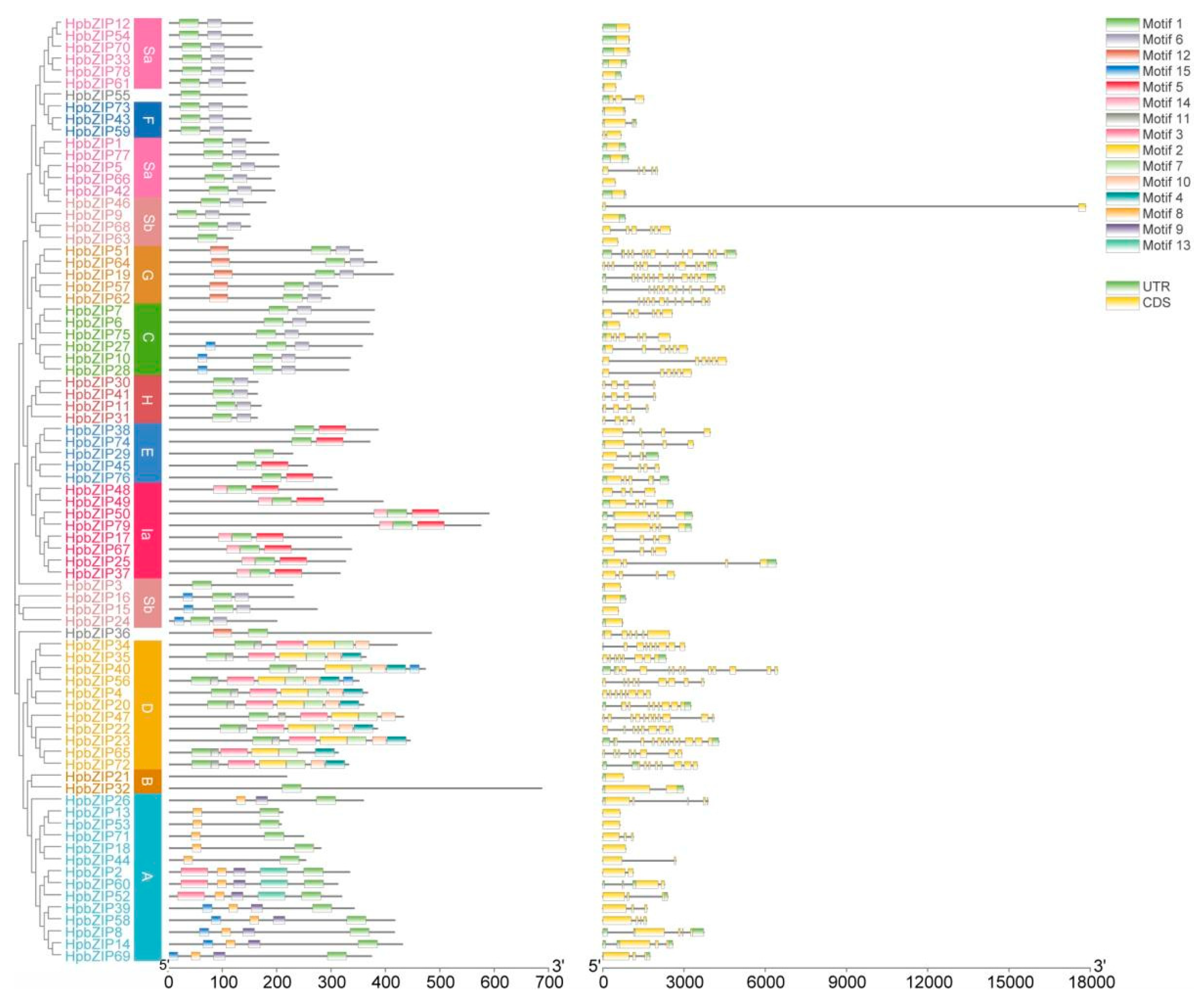
2.3. Gene structure and conserved motif analyses
2.4. Transcript abundance profiling
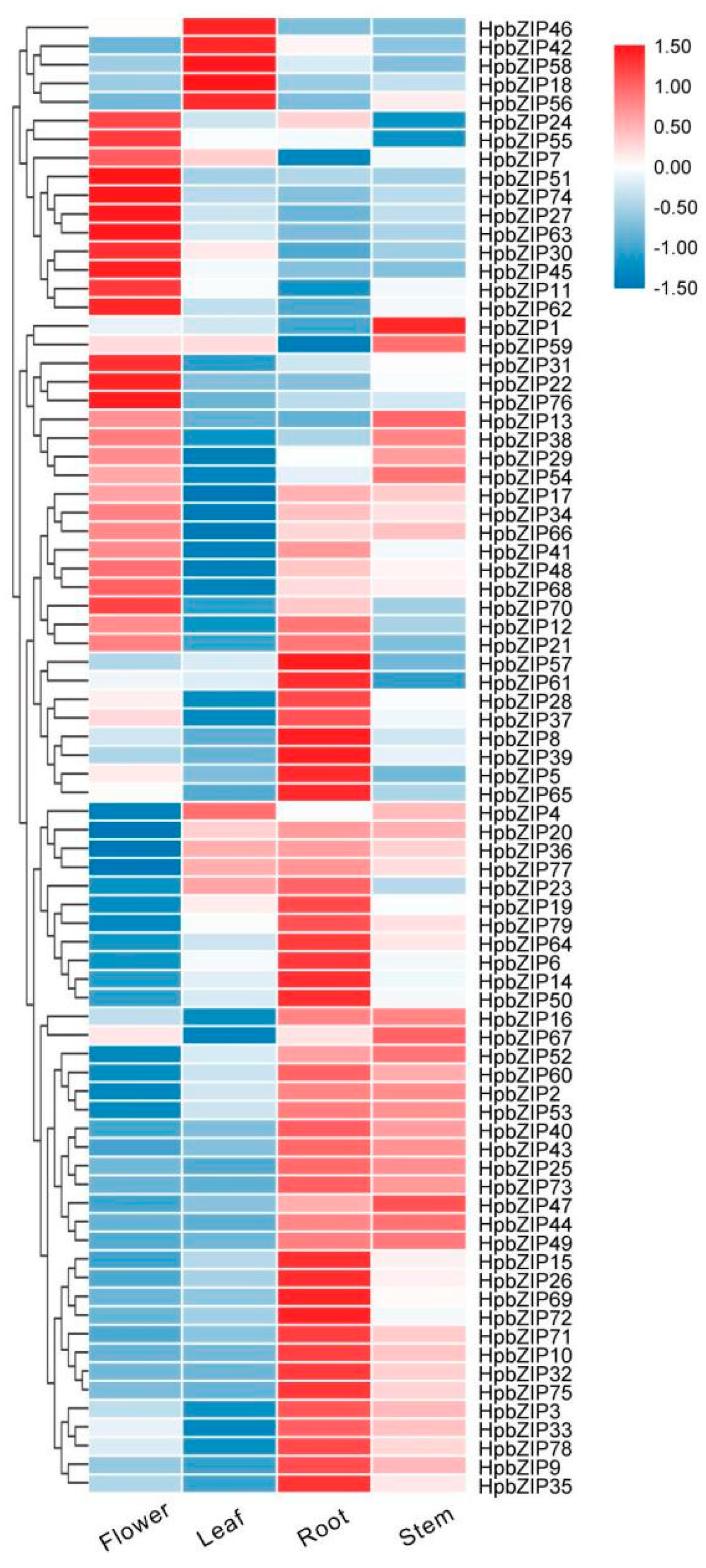
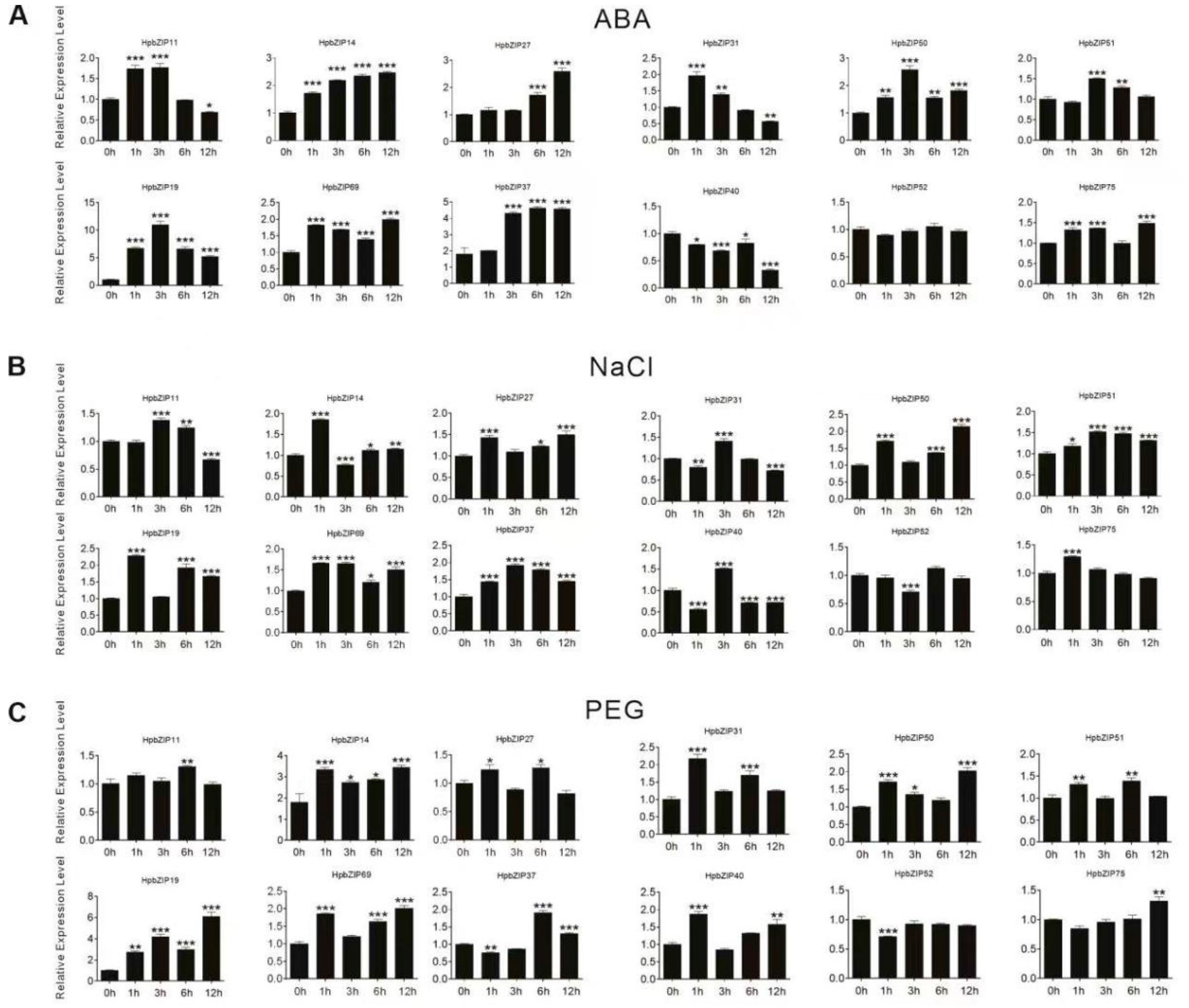
2.5. Expression patterns analysis
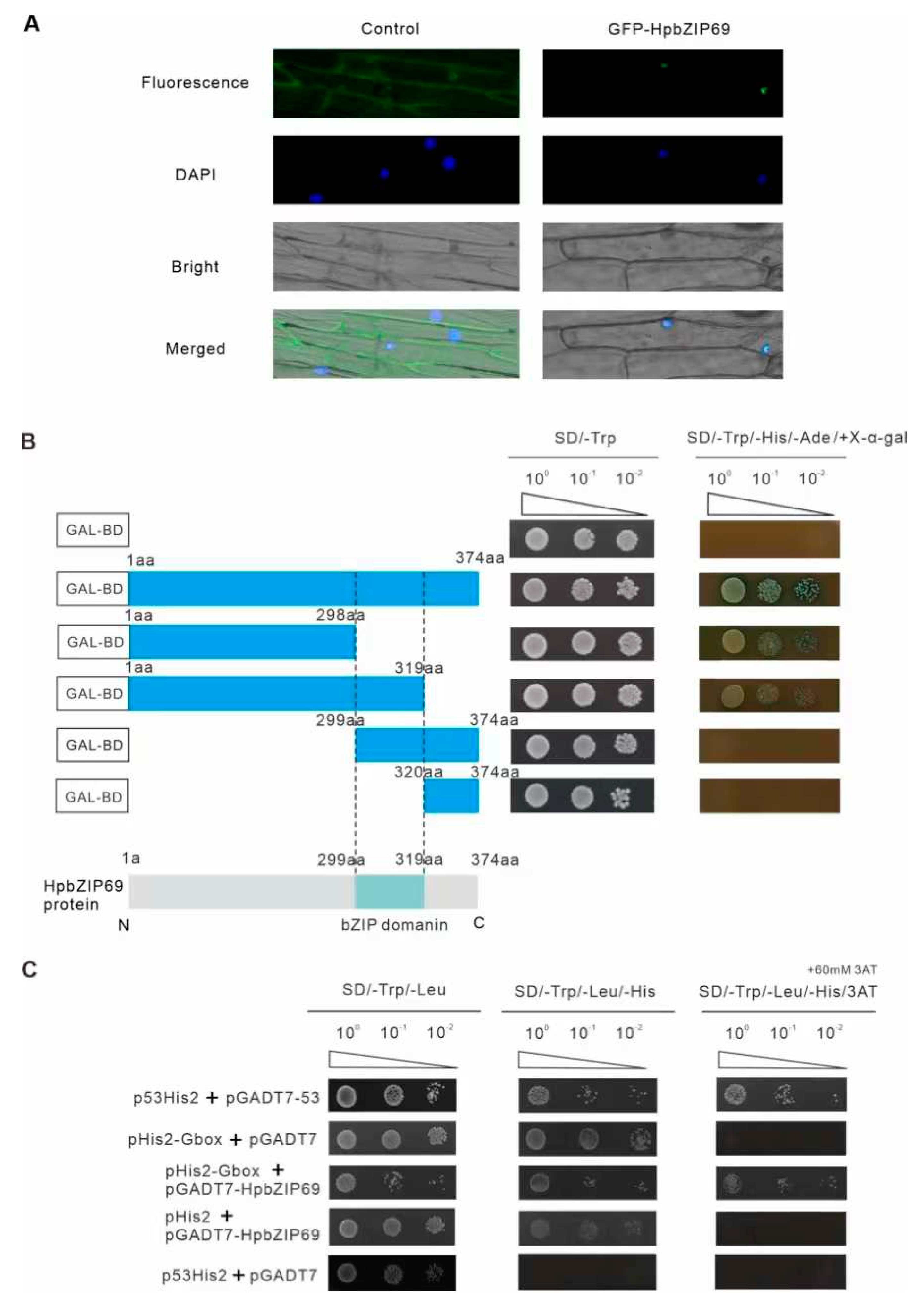
2.6. Characterization of Transcription Activity of HpbZIP69
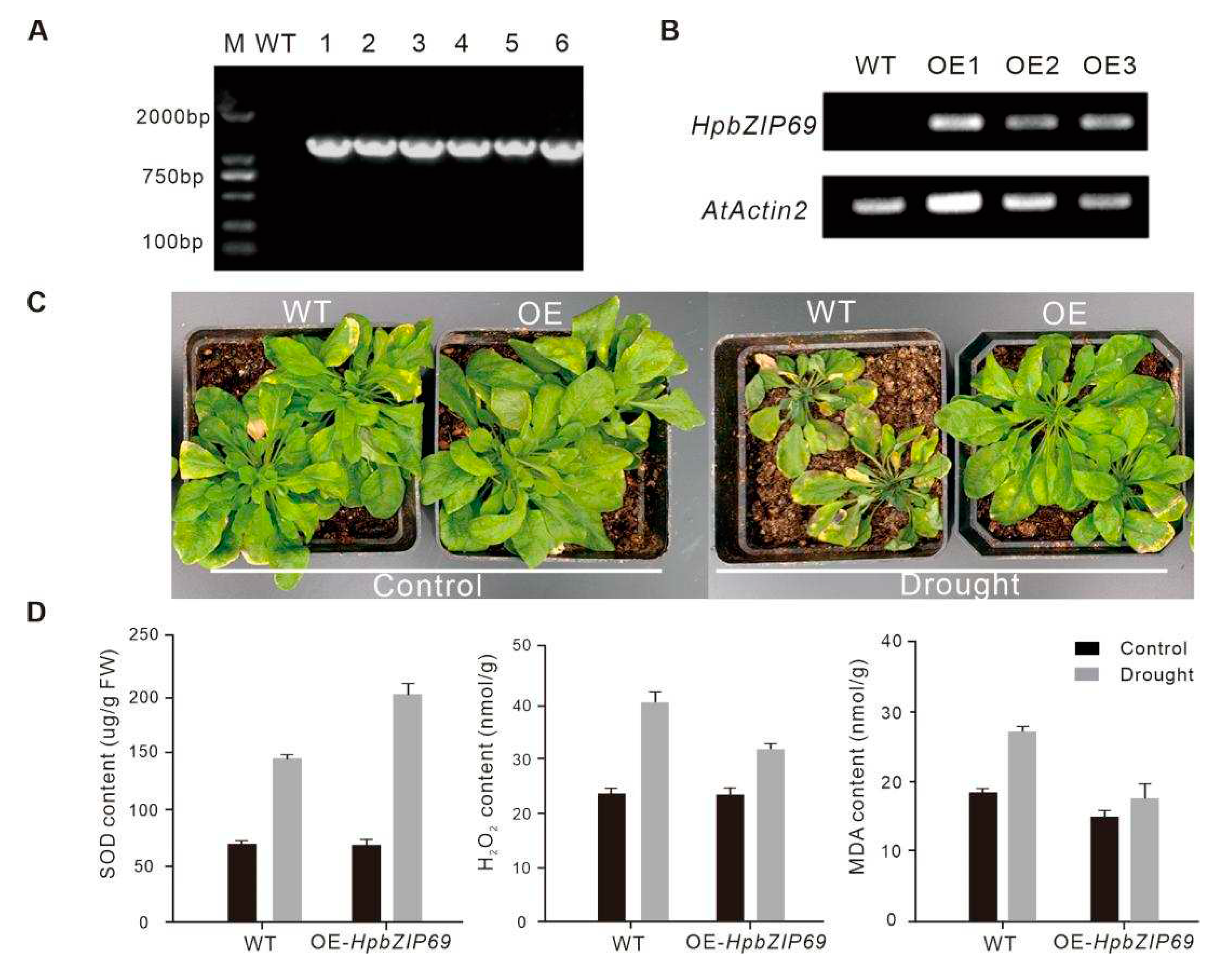
2.7. Overexpression of HpbZIP69 in Arabidopsis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Identification and sequence analysis
5.2. Phylogenetic analysis, cis-acting element predictions, and Gene Ontology annotations
5.3. Plant materials, stress treatments, and expression analysis
5.4. Subcellular localization of the HpbZIP69
5.5. Transcriptional Activation and Y1H Assay
5.6. Overexpressing HpbZIP69 in Arabidopsis
5.7. Physiological index measurement
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hurst, H.C. Transcription factors 1: bZIP proteins. Protein Profile 1995, 2, 101–168. [Google Scholar]
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant physiology 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends in plant science 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Foster, R.; Chua, N.H. Plant bZIP protein DNA binding specificity. Journal of molecular biology 1993, 230, 1131–1144. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, T.E.; Brandl, C.J.; Struhl, K.; Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 1992, 71, 1223–1237. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Srivastava, R.; Howell, S.H. Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ 2008, 31, 1735–1743. [Google Scholar] [CrossRef]
- Weltmeier, F.; Ehlert, A.; Mayer, C.S.; Dietrich, K.; Wang, X.; Schütze, K.; Alonso, R.; Harter, K.; Vicente-Carbajosa, J.; Dröge-Laser, W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. Embo j 2006, 25, 3133–3143. [Google Scholar] [CrossRef]
- Hartmann, L.; Pedrotti, L.; Weiste, C.; Fekete, A.; Schierstaedt, J.; Göttler, J.; Kempa, S.; Krischke, M.; Dietrich, K.; Mueller, M.J.; Vicente-Carbajosa, J.; Hanson, J.; Dröge-Laser, W. Crosstalk between Two bZIP Signaling Pathways Orchestrates Salt-Induced Metabolic Reprogramming in Arabidopsis Roots. The Plant cell 2015, 27, 2244–2260. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC genomics 2015, 16, 227. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant molecular biology 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Cai, W.; Yang, Y.; Wang, W.; Guo, G.; Liu, W.; Bi, C. Overexpression of a wheat (Triticum aestivum L.) bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs. Plant Physiol Biochem 2018, 124, 100–111. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.; Qian, W.; Yao, L.; Hao, X.; Li, N.; Yang, Y.; Wang, X. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Annals of botany 2017, 119, 1195–1209. [Google Scholar] [CrossRef]
- Yao, T.; Zhang, J.; Xie, M.; Yuan, G.; Tschaplinski, T.J.; Muchero, W.; Chen, J.G. Transcriptional Regulation of Drought Response in Arabidopsis and Woody Plants. Front Plant Sci 2020, 11, 572137. [Google Scholar] [CrossRef]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. The Plant cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhou, J.; Jie, Y.; Xing, H.; Zhong, Y.; Yu, W.; She, W.; Ma, Y.; Liu, Z.; Zhang, Y. A Ramie bZIP Transcription Factor BnbZIP2 Is Involved in Drought, Salt, and Heavy Metal Stress Response. DNA and cell biology 2016, 35, 776–786. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Gao, L.; Hao, C.; Zhao, G.; Jia, J.; Kong, X. A Novel Wheat C-bZIP Gene, TabZIP14-B, Participates in Salt and Freezing Tolerance in Transgenic Plants. Front Plant Sci 2017, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol 2019, 19, 260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, J.; Yuan, W.; Wang, Y.; Hu, P.; Jiao, C.; Xia, H.; Wang, D.; Cai, Q.; Li, J.; Wang, C.; Zhang, X.; Chen, Y.; Wang, Z.; Ou, Z.; Xu, Z.; Shi, J.; Chen, J. Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int J Biol Macromol 2021, 181, 1207–1223. [Google Scholar] [CrossRef]
- Wang, B.; Li, L.; Peng, D.; Liu, M.; Wei, A.; Li, X. TaFDL2-1A interacts with TabZIP8-7A protein to cope with drought stress via the abscisic acid signaling pathway. Plant science : an international journal of experimental plant biology 2021, 311, 111022. [Google Scholar] [CrossRef]
- Rolly, N.K.; Imran, Q.M.; Shahid, M.; Imran, M.; Khan, M.; Lee, S.U.; Hussain, A.; Lee, I.J.; Yun, B.W. Drought-induced AtbZIP62 transcription factor regulates drought stress response in Arabidopsis. Plant Physiol Biochem 2020, 156, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Zavaliev, R.; Wu, Q.; Zhou, Y.; Cheng, J.; Dillard, L.; Powers, J.; Withers, J.; Zhao, J.; Guan, Z.; Borgnia, M.J.; Bartesaghi, A.; Dong, X.; Zhou, P. Structural basis of NPR1 in activating plant immunity. Nature 2022, 605, 561–566. [Google Scholar] [CrossRef]
- Fan, W.; Dong, X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. The Plant cell 2002, 14, 1377–1389. [Google Scholar] [CrossRef]
- Zhang, Y.; Tessaro, M.J.; Lassner, M.; Li, X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. The Plant cell 2003, 15, 2647–2653. [Google Scholar] [CrossRef]
- Zhang, R.; Ji, Y.; Zhang, X.; Kennelly, E.J.; Long, C. Ethnopharmacology of Hypericum species in China: A comprehensive review on ethnobotany, phytochemistry and pharmacology. J Ethnopharmacol 2020, 254, 112686. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Lakra, N.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice (New York, N.Y.) 2019, 12, 58. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr Opin Plant Biol 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 2015, 6, 895. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, Z.; Ren, W.; Yan, S.; Xing, N.; Zhang, Z.; Li, H.; Ma, W. Identification of the bZIP gene family and regulation of metabolites under salt stress in isatis indigotica. Front Plant Sci 2022, 13, 1011616. [Google Scholar] [CrossRef]
- Que, F.; Wang, G.L.; Huang, Y.; Xu, Z.S.; Wang, F.; Xiong, A.S. Genomic identification of group A bZIP transcription factors and their responses to abiotic stress in carrot. Genetics and molecular research : GMR 2015, 14, 13274–13288. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Xu, C.; Wang, X.; Wang, P.; Huang, S. ABA collaborates with lignin and flavonoid to improve pre-silking drought tolerance by tuning stem elongation and ear development in maize (Zea mays L.). The Plant journal : for cell and molecular biology 2023.
- Zhou, Q.; Li, Y.; Wang, X.; Yan, C.; Ma, C.; Liu, J.; Dong, S. Effects of Different Drought Degrees on Physiological Characteristics and Endogenous Hormones of Soybean. Plants (Basel) 2022, 11, 2282. [Google Scholar] [CrossRef]
- Li, L.; Yi, H. Enhancement of drought tolerance in Arabidopsis plants induced by sulfur dioxide. Ecotoxicology (London, England) 2022, 31, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Saed-Moucheshi, A.; Sohrabi, F.; Fasihfar, E.; Baniasadi, F.; Riasat, M.; Mozafari, A.A. Superoxide dismutase (SOD) as a selection criterion for triticale grain yield under drought stress: a comprehensive study on genomics and expression profiling, bioinformatics, heritability, and phenotypic variability. BMC Plant Biol 2021, 21, 148. [Google Scholar] [CrossRef] [PubMed]
- Reiser, L.; Subramaniam, S.; Zhang, P.; Berardini, T. Using the Arabidopsis Information Resource (TAIR) to Find Information About Arabidopsis Genes. Current protocols 2022, 2, e574. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic acids research 2018, 46, W200–w204. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Li, B.; Petijová, L.; Hu, S.; Zhang, Q.; Niu, J.; Wang, D.; Wang, S.; Dong, Y.J.J. o. P. R. Whole-genome sequence data of Hypericum perforatum and functional characterization of melatonin biosynthesis by N-acetylserotonin O-methyltransferase. 2021, 70, e12709.
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; Finn, R.D.; Bateman, A. Pfam: The protein families database in 2021. Nucleic acids research 2021, 49, D412–d419. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; Yang, M.; Zhang, D.; Zheng, C.; Lanczycki, C.J.; Marchler-Bauer, A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic acids research 2020, 48, D265–d268. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic acids research 2021, 49, W216–w227. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic acids research 2015, 43, W39–49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic acids research 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. International journal of plant genomics 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; Zhang, X.; Bolund, L.; Chen, Q.; Wang, J.; Yang, H.; Fang, L.; Shi, C. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic acids research 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Q.; Sun, Y.; Yang, L.; Wang, Z. Genome-wide identification and characterization of R2R3-MYB family in Hypericum perforatum under diverse abiotic stresses. Int J Biol Macromol 2020, 145, 341–354. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, S.; Yang, L.; Xiao, R.; Chen, S.; Wang, D.; Wang, S.; Wang, Z.J.I.J. o. M. S. Genome-Wide Identification of the Hypericum perforatum WRKY Gene Family Implicates HpWRKY85 in Drought Resistance. 2023, 24, 352.
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostatistics, bioinformatics and biomathematics 2013, 3, 71–85. [Google Scholar]
- Zhou, W.; Wang, S.; Yang, L.; Sun, Y.; Zhang, Q.; Li, B.; Wang, B.; Li, L.; Wang, D.; Wang, Z.J.P. Reference genes for qRT-PCR normalisation in different tissues, developmental stages, and stress conditions of Hypericum perforatum. 2019, 7, e7133.
- Reece-Hoyes, J.S.; Walhout, A.J.M. Gateway Recombinational Cloning. Cold Spring Harbor protocols 2018, 2018, pdb–top094912. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, S.; Zhang, Q.; Xiao, R.; Li, B.; Wang, D.; Niu, J.; Wang, S.; Wang, Z. Functional Characterization of Serotonin N-Acetyltransferase Genes (SNAT1/2) in Melatonin Biosynthesis of Hypericum perforatum. Front Plant Sci 2021, 12, 781717. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature protocols 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




