Submitted:
27 July 2023
Posted:
28 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
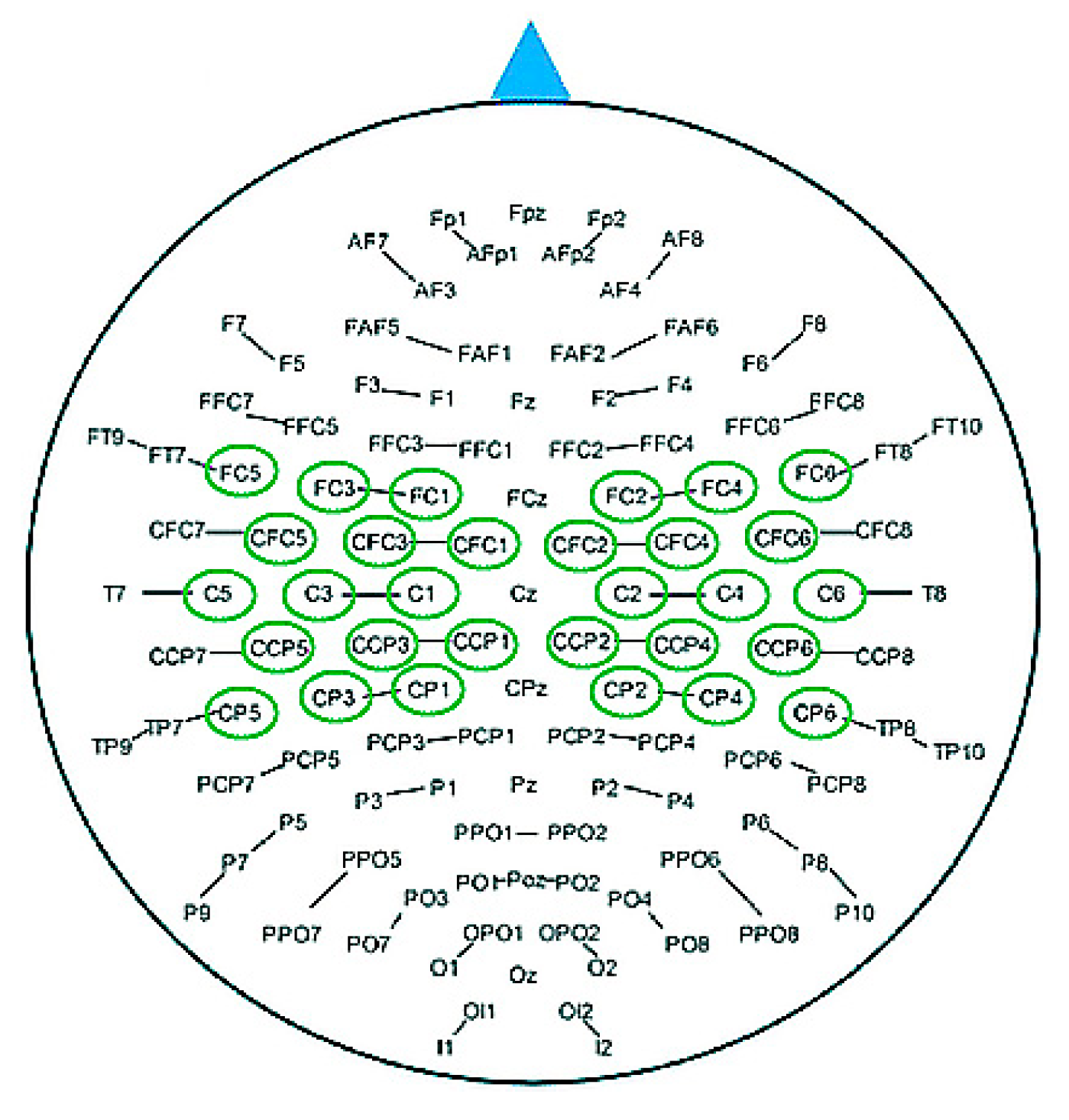
2. Data Description
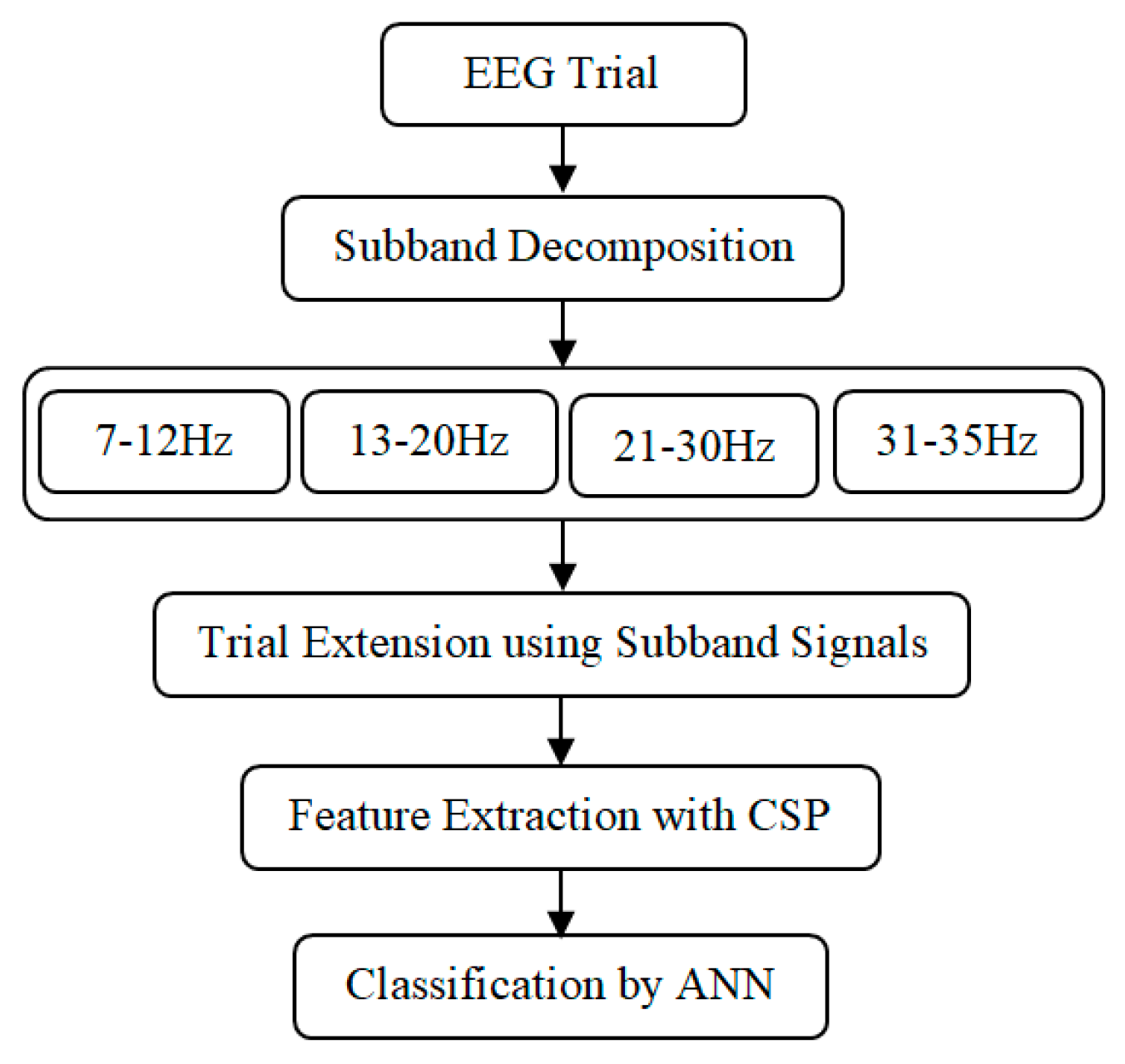
3. Methods
- i.
- The multichannel EEG signal is decomposed into subbands. Each subband represents a rhythmic component.
- i.
- ii. Each trial extended in the spatial dimension by arranging the obtained subband signals.
- i.
- iii. Common spatial pattern (CSP) is applied on the newly generated trials to extract spatial features
- i.
- iv. Separate training and test sets of publicly available datasets are used to train ANN and evaluate the performance of the proposed method respectively.
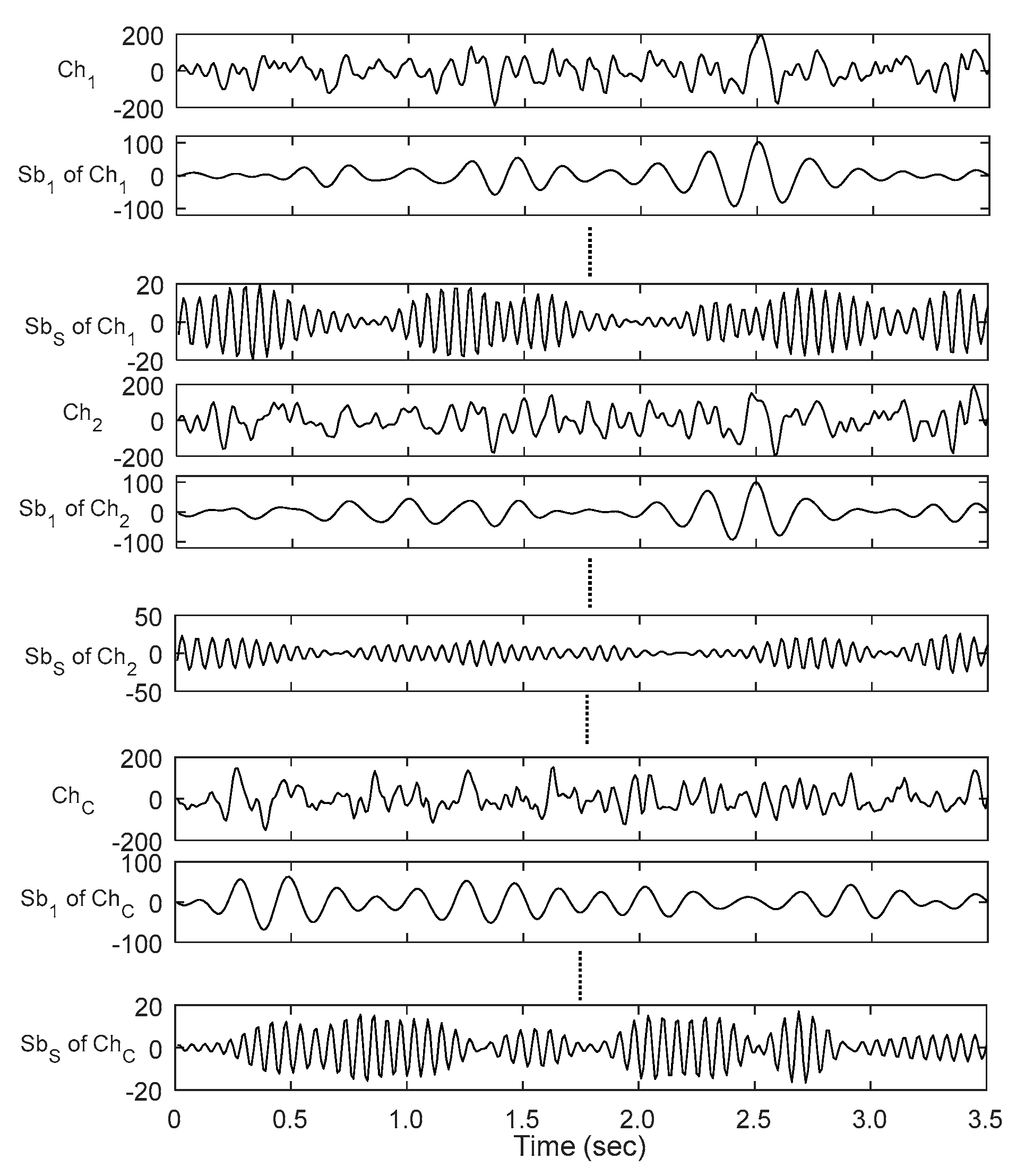
3.1. Subband Decomposition
3.2. Trial Extension
3.3. Feature Extraction
3.4. Classification by ANN
4. Experimental Results
5. Discussion
| Subject | LRFCSP [51] | CTWS [52] | VaS-SVMlk [48] | eTE | Proposed iTE-tP |
| a | 87.40 | 83.00 | 92.50 | 83.45 | 94.14 |
| b | 70.00 | 67.00 | 77.00 | 60.88 | 78.08 |
| c | 67.40 | 85.50 | 82.70 | 76.44 | 84.89 |
| d | 92.90 | 93.00 | 96.40 | 90.43 | 97.78 |
| e | 93.40 | 99.00 | 97.20 | 92.68 | 100.00 |
| f | 88.80 | 85.50 | 88.80 | 79.54 | 91.04 |
| g | 93.20 | 81.00 | 92.60 | 81.88 | 94.86 |
| Mean | 84.70 | 84.86 | 89.60 | 80.76 | 91.55 |
| SD | 5.78 | 10.4 | 7.41 | 10.48 | 7.11 |
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- S. Gao, Y. Wang, X. Gao, and B. Hong, “Visual and auditory brain– computer interfaces,” IEEE Trans. Biomed. Eng., vol. 61, no. 5, pp. 1436–1447, May 2014.
- van Dokkum, L.; Ward, T.; Laffont, I. Brain computer interfaces for neurorehabilitation – its current status as a rehabilitation strategy post-stroke. Ann. Phys. Rehabilitation Med. 2018, 58, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Nuyujukian, P.; Fan, J.M.; Kao, J.C.; Ryu, S.I.; Shenoy, K.V. A High-Performance Keyboard Neural Prosthesis Enabled by Task Optimization. IEEE Trans. Biomed. Eng. 2015, 62, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain–computer interfaces: a 10 year update. J. Neural Eng. 2018, 15, 031005. [Google Scholar] [CrossRef] [PubMed]
- G. Pfurtscheller, C. Brunner, A. Schlögl, F. Lopes da Silva, "Mu rhythm (de)synchronization and EEG single-trial classification of different motor imagery tasks," NeuroImage, vol. 31, no. 1, pp: 153-159, May 2006.
- Wu, W.; Gao, X.; Hong, B.; Gao, S. Classifying Single-Trial EEG During Motor Imagery by Iterative Spatio-Spectral Patterns Learning (ISSPL). IEEE Trans. Biomed. Eng. 2008, 55, 1733–1743. [Google Scholar] [CrossRef]
- Faust, O.; Acharya, R.; Allen, A.; Lin, C. Analysis of EEG signals during epileptic and alcoholic states using AR modeling techniques. IRBM 2008, 29, 44–52. [Google Scholar] [CrossRef]
- C. Guerrero-Mosquera and A. N. Vazquez, “New approach in features extraction for EEG signal detection,” in Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC '09), pp. 13–16, September 2009.
- Übeyli, E.D. Analysis of EEG signals by combining eigenvector methods and multiclass support vector machines. Comput. Biol. Med. 2008, 38, 14–22. [Google Scholar] [CrossRef]
- Cvetkovic, D.; Übeyli, E.D.; Cosic, I. Wavelet transform feature extraction from human PPG, ECG, and EEG signal responses to ELF PEMF exposures: A pilot study. Digit. Signal Process. 2008, 18, 861–874. [Google Scholar] [CrossRef]
- Subasi, A.; Kiymik, M.K.; Alkan, A.; Koklukaya, E. Neural Network Classification of EEG Signals by Using AR with MLE Preprocessing for Epileptic Seizure Detection. Math. Comput. Appl. 2005, 10, 57–70. [Google Scholar] [CrossRef]
- Lemm, S.; Blankertz, B.; Curio, G.; Muller, K.-R. Spatio-spectral filters for improving the classification of single trial EEG. IEEE Trans. Biomed. Eng. 2005, 52, 1541–1548. [Google Scholar] [CrossRef]
- Samek, W.; Vidaurre, C.; Müller, K.-R.; Kawanabe, M. Stationary common spatial patterns for brain–computer interfacing. J. Neural Eng. 2012, 9, 026013. [Google Scholar] [CrossRef]
- Ang, K.K.; Chin, Z.Y.; Wang, C.; Guan, C.; Zhang, H. Filter Bank Common Spatial Pattern Algorithm on BCI Competition IV Datasets 2a and 2b. Front. Neurosci. 2012, 6, 39. [Google Scholar] [CrossRef]
- Higashi, H.; Tanaka, T. Simultaneous Design of FIR Filter Banks and Spatial Patterns for EEG Signal Classification. IEEE Trans. Biomed. Eng. 2012, 60, 1100–1110. [Google Scholar] [CrossRef]
- Novi, Q.; Guan, C.; Dat, T.H.; Xue, P. Sub-band common spatial pattern (SBCSP) for brain-computer interface. 3rd Int. IEEE/EMBS Conf. on Neural Engineering 2007, 204–207. [CrossRef]
- Zhang, Y.; Zhou, G.; Jin, J.; Wang, X.; Cichocki, A. Optimizing spatial patterns with sparse filter bands for motor-imagery based brain–computer interface. J. Neurosci. Methods 2015, 255, 85–91. [Google Scholar] [CrossRef]
- Thomas, K.P.; Guan, C.; Lau, C.T.; Vinod, A.P.; Ang, K.K. A New Discriminative Common Spatial Pattern Method for Motor Imagery Brain–Computer Interfaces. IEEE Trans. Biomed. Eng. 2009, 56, 2730–2733. [Google Scholar] [CrossRef]
- Jiao, Y.; Zhang, Y.; Chen, X.; Yin, E.; Jin, J.; Wang, X.Y.; Cichocki, A. Sparse Group Representation Model for Motor Imagery EEG Classification. IEEE J. Biomed. Heal. Informatics 2018, 23, 631–641. [Google Scholar] [CrossRef]
- M. K. I. Molla, M. R. Islam, T. Tanaka, T. Rutkowski, “Artifact suppression from EEG signals using data adaptive time domain filtering,” Neurocomputing, vol. 97, no. 0, pp: 297 – 308, Nov 2012.
- Q. Zhang, A. Benveniste, “Wavelet networks,” IEEE Transactions on Neural Networks, vol. 3, no. 6, pp: 889-898, Nov 1992.
- Oweiss, K.G.; Anderson, D.J. Noise reduction in multichannel neural recordings using a new array wavelet denoising algorithm. Neurocomputing 2001, 38-40, 1687–1693. [Google Scholar] [CrossRef]
- M. R. Islam, T. Tanaka and M. K. I. Molla, “Multiband Tangent Space Mapping and Feature Selection for Classification of EEG during Motor Imagery”, Journal of Neural Engineering, 15(4), May 2018.
- M. K. I. Molla, A. A. Shiam, M. R. Islam and T. Tanaka, "Discriminative Feature Selection Based Motor Imagery Classification Using EEG Signal" IEEE Access, Vol. 8, pp: 98255-98265, May, 2020.
- G. Dornhege, B. Blankertz, G. Curio, and K. R. Müller, Boosting bit rates in non-invasive EEG single-trial classifications by feature combination and multi-class paradigms, IEEE Trans. Biomed. Eng., 51, 993-1002, 2004.
- He, L.; Hu, D.; Wan, M.; Wen, Y.; von Deneen, K.M.; Zhou, M. Common Bayesian Network for Classification of EEG-Based Multiclass Motor Imagery BCI. IEEE Trans. Syst. Man, Cybern. Syst. 2015, 46, 843–854. [Google Scholar] [CrossRef]
- Sreeja, S.R.; Rabha, J.; Samanta, D.; Mitra, P.; Sarma, M. Classification of Motor Imagery Based EEG Signals Using Sparsity Approach. Proc. of International Conference on Intelligent Human Computer Interaction 2017, 47–59. [Google Scholar] [CrossRef]
- J. Rabha, K. Y. Nagarjuna, D. Samanta, P. Mitra and M. Sarma, Motor imagery EEG signal processing and classification using machine learning approach, Proc. of International Conference on New Trends in Computing Sciences (ICTCS), 61-66, 2017.
- Yeh, C.-Y.; Su, W.-P.; Lee, S.-J. An efficient multiple-kernel learning for pattern classification. Expert Syst. Appl. 2013, 40, 3491–3499. [Google Scholar] [CrossRef]
- G. Klem, H. Lüders, H. Jasper HH, C. Elger, “The ten-twenty electrode system of the International Federation,” Electroencephalograph clinical neurophysiology, vol. 52, no. 3, pp: 3-6,1999.
- Juri, D. Kropotov, "Functional Neuromarkers for Psychiatry–Applications for Diagnosis and Treatment", Science Direct Publication, 2016.
- Hsu, K.-C.; Yu, S.-N. Detection of seizures in EEG using subband nonlinear parameters and genetic algorithm. Comput. Biol. Med. 2010, 40, 823–830. [Google Scholar] [CrossRef]
- Molla, K.I.; Tanaka, T.; Osa, T.; Islam, M.R. EEG signal enhancement using multivariate wavelet transform Application to single-trial classification of event-related potentials. IEEE Int. Conference on Digital Signal Processing 2015, 804–808. [Google Scholar] [CrossRef]
- Shiam, A. A.; Islam, M.R.; Tanaka, T.; Molla, M.K.I. Electroencephalography Based Motor Imagery Classification Using Unsupervised Feature Selection,” Proc. of Cyberworld, 2019.
- Zhang, Y.; Wang, Y.; Zhou, G.; Jin, J.; Wang, B.; Wang, X.; Cichocki, A. Multi-kernel extreme learning machine for EEG classification in brain-computer interfaces. Expert Syst. Appl. 2018, 96, 302–310. [Google Scholar] [CrossRef]
- Suefusa, K.; Tanaka, T. Asynchronous Brain–Computer Interfacing Based on Mixed-Coded Visual Stimuli. IEEE Trans. Biomed. Eng. 2017, 65, 2119–2129. [Google Scholar] [CrossRef]
- G. Bebis, M. Georgiopoulos, “Feed-forward neural networks.” IEEE Potentials 1994, 13, 27–31.
- M. K. I. Molla, N. Morikawa, M. R. Islam and T. Tanaka, “Data-adaptive Spatiotemporal ERP Cleaning for Single-trial BCI Implementation,” IEEE Transactions on Neural Systems and Rehabilitation Engineering, Vol. 26, Issue 7, pp:1334 – 1344, June 2018.
- S. Selim, M. M. Tantawi, A. S. Howida, and A. Badr, “A CSP\AM-BA-SVM Approach for Motor Imagery BCI System,” IEEE Access, 6, pp. 49192–49208, 2018.
- Arvaneh, M.; Guan, C.; Ang, K.K.; Quek, H.C. Spatially sparsed common spatial pattern to improve BCI performance,” in Proc. of IEEE International Conference on Acoustics, Speech and Signal Processing 2011, 2412–2415. [CrossRef]
- Lotte, F.; Guan, C. Spatially regularized common spatial patterns for EEG classification,” in Proc. of 20th International Conference on Pattern Recognition 2010, 3712–3715. [Google Scholar] [CrossRef]
- Dai, M.; Zheng, D.; Liu, S.; Zhang, P. Transfer Kernel Common Spatial Patterns for Motor Imagery Brain-Computer Interface Classification. Comput. Math. Methods Med. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Lal, S.; Guesgen, H.W. Small Sample Motor Imagery Classification Using Regularized Riemannian Features. IEEE Access 2019, 7, 46858–46869. [Google Scholar] [CrossRef]
- Park, C.; Looney, D.; Rehman, N.U.; Ahrabian, A.; Mandic, D.P. Classification of Motor Imagery BCI Using Multivariate Empirical Mode Decomposition. IEEE Trans. Neural Syst. Rehabilitation Eng. 2012, 21, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Miao, Y.; Daly, I.; Zuo, C.; Hu, D.; Cichocki, A. Correlation-based channel selection and regularized feature optimization for MI-based BCI. Neural Networks 2019, 118, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Chung, W. Optimal Channel Selection Using Correlation Coefficient for CSP Based EEG Classification. IEEE Access 2020, 8, 111514–111521. [Google Scholar] [CrossRef]
- Park, Y.; Chung, W. Selective Feature Generation Method Based on Time Domain Parameters and Correlation Coefficients for Filter-Bank-CSP BCI Systems. Sensors 2019, 19. [Google Scholar] [CrossRef]
- Wang, H.; Xu, T.; Tang, C.; Yue, H.; Chen, C.; Xu, L.; Pei, Z.; Dong, J.; Bezerianos, A.; Li, J. Diverse Feature Blend Based on Filter-Bank Common Spatial Pattern and Brain Functional Connectivity for Multiple Motor Imagery Detection. IEEE Access 2020, 8, 155590–155601. [Google Scholar] [CrossRef]
- Jin, J.; Liu, C.; Daly, I.; Miao, Y.; Li, S.; Wang, X.; Cichocki, A. Bispectrum-Based Channel Selection for Motor Imagery Based Brain-Computer Interfacing. IEEE Trans. Neural Syst. Rehabilitation Eng. 2020, 28, 2153–2163. [Google Scholar] [CrossRef]
- Park, Y.; Chung, W. Frequency-Optimized Local Region Common Spatial Pattern Approach for Motor Imagery Classification. IEEE Trans. Neural Syst. Rehabilitation Eng. 2019, 27, 1378–1388. [Google Scholar] [CrossRef]
- Feng, J.; Yin, E.; Jin, J.; Saab, R.; Daly, I.; Wang, X.; Hu, D.; Cichocki, A. Towards correlation-based time window selection method for motor imagery BCIs. Neural Networks 2018, 102, 87–95. [Google Scholar] [CrossRef]
- Molla, K.I.; Saha, S.K.; Yasmin, S.; Islam, R.; Shin, J. Trial Regeneration With Subband Signals for Motor Imagery Classification in BCI Paradigm. IEEE Access 2021, 9, 7632–7642. [Google Scholar] [CrossRef]
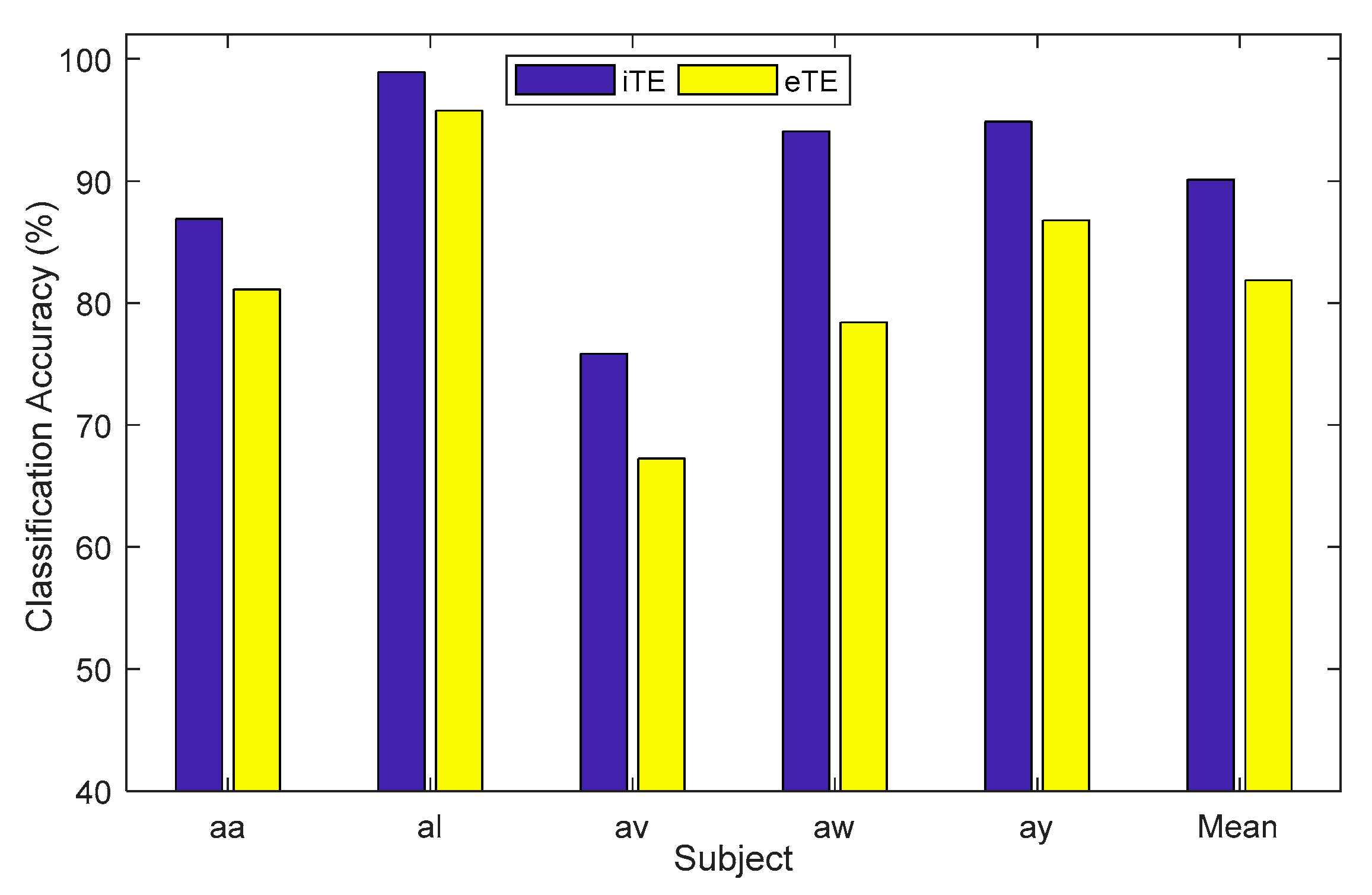
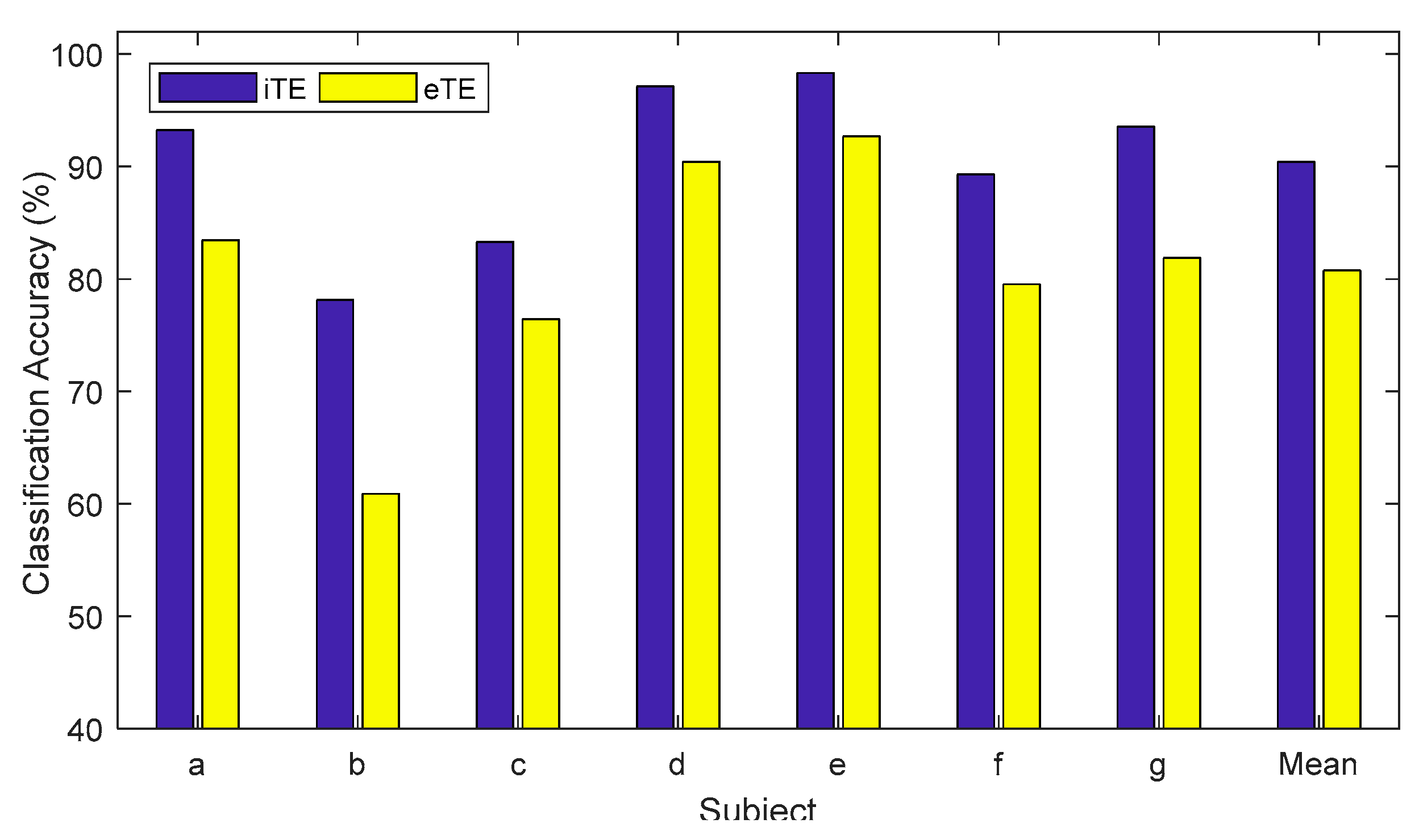
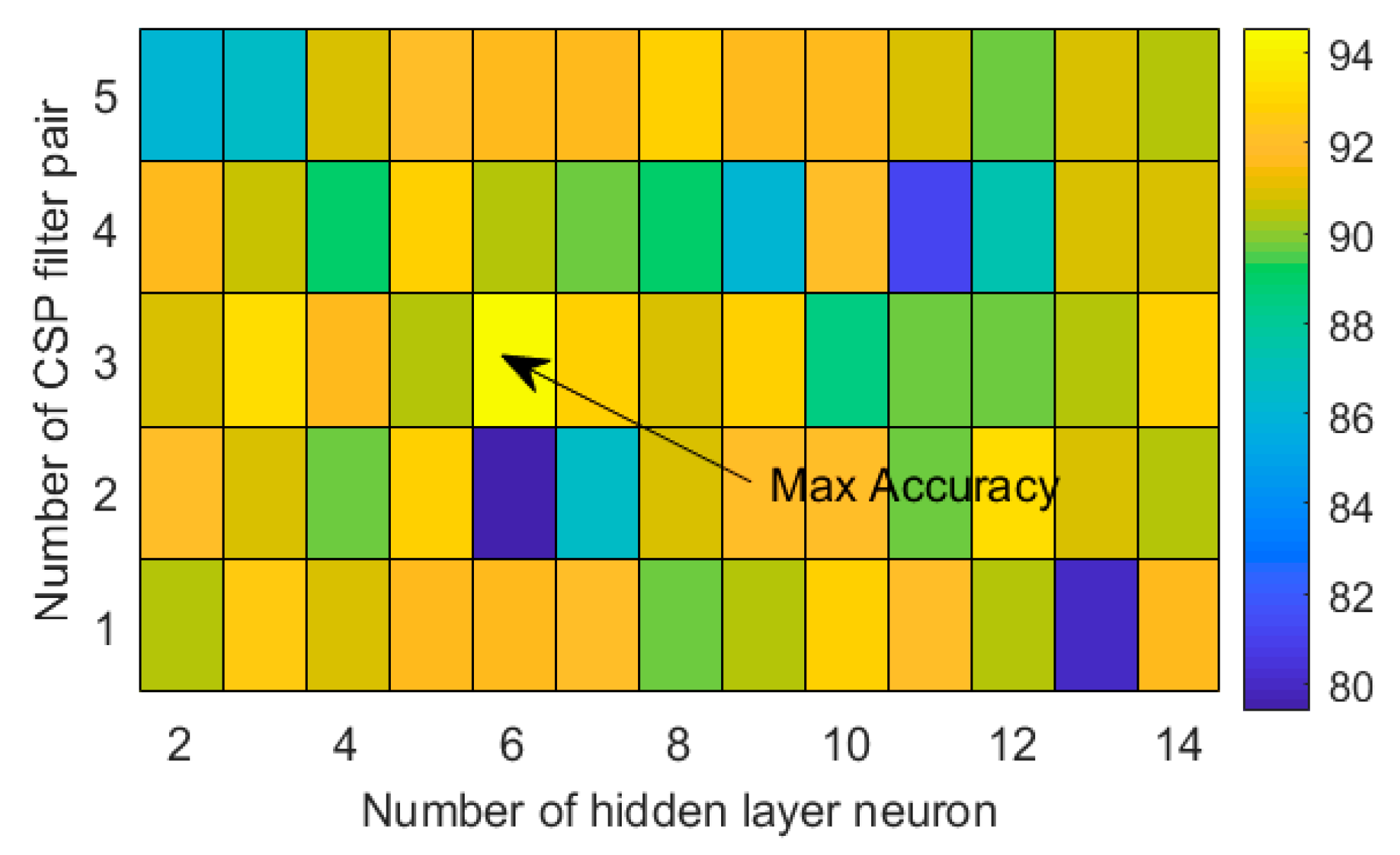
| Subject | iTE | iTE-tP | eTE |
| aa | 86.90 | 94.94 | 81.12 |
| al | 98.92 | 98.18 | 95.76 |
| av | 75.84 | 76.25 | 67.24 |
| aw | 94.06 | 100.00 | 78.42 |
| ay | 94.86 | 100.00 | 86.78 |
| Mean | 90.12 | 93.88 | 81.87 |
| SD | 9.08 | 9.00 | 10.53 |
| Subject | iTE | iTE-tP | eTE |
| a | 93.24 | 94.14 | 83.45 |
| b | 78.15 | 78.08 | 60.88 |
| c | 83.30 | 84.89 | 76.44 |
| d | 97.12 | 97.78 | 90.43 |
| e | 98.33 | 100.00 | 92.68 |
| f | 89.28 | 91.04 | 79.54 |
| g | 93.55 | 94.86 | 81.88 |
| Mean | 90.43 | 91.55 | 80.76 |
| SD | 7.39 | 7.11 | 10.48 |
| Methods | Subjects | Mean±STD | ||||
| aa | al | av | aw | ay | ||
| RRF [43] | 81.25 | 100.00 | 76.53 | 87.05 | 91.26 | 87.22±9.08 |
| SGRM [19] | 73.90 | 94.50 | 59.50 | 80.70 | 79.90 | 77.70±12.67 |
| SSCSP [40] | 72.32 | 96.42 | 54.10 | 70.54 | 73.41 | 73.36±13.50 |
| SRCSP [41] | 69.64 | 96.43 | 59.18 | 70.09 | 86.51 | 76.37±13.31 |
| TKCSP [42] | 68.10 | 93.88 | 68.47 | 88.40 | 74.93 | 78.76±10.54 |
| AM-SVM [39] | 86.61 | 100.00 | 66.84 | 90.63 | 80.95 | 85.00±11.00 |
| UDFS [34] | 86.98 | 97.45 | 76.04 | 93.93 | 94.94 | 89.86±8.65 |
| NCFS [24] | 90.00 | 98.93 | 76.71 | 98.21 | 97.14 | 92.20±9.36 |
| eTE | 81.12 | 95.76 | 67.24 | 78.42 | 86.78 | 81.87±10.53 |
| Proposed iTE-tP | 94.94 | 98.18 | 76.25 | 100.00 | 100.00 | 93.88±9.00 |
| Method | Subject | Mean±STD | |||
| a | b | f | g | ||
| VaS-SVMlk [48] | 92.50 | 77.00 | 88.80 | 92.60 | 87.72±7.36 |
| NA-MEMD [44] | 85.90 | 77.60 | 78.80 | 90.90 | 83.30±6.25 |
| CCS-RCSP [45] | 85.50 | 67.00 | 79.50 | 94.50 | 81.60±11.50 |
| CC-FBCSP [46] | 86.50 | 69.50 | 87.50 | 94.00 | 84.40±8.20 |
| TDP-CC [47] | 86.50 | 57.25 | 92.50 | 90.50 | 81.69±16.80 |
| NS-CSP [49] | 82.00 | 67.50 | 65.00 | 87.50 | 75.50±11.00 |
| BCS-CSP [50] | 79.00 | 79.00 | 92.00 | 88.00 | 84.50±5.69 |
| eTE | 83.45 | 60.88 | 79.54 | 81.88 | 76.44±10.50 |
| Proposed iTE-tP | 94.14 | 78.08 | 91.04 | 94.86 | 89.53±6.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





