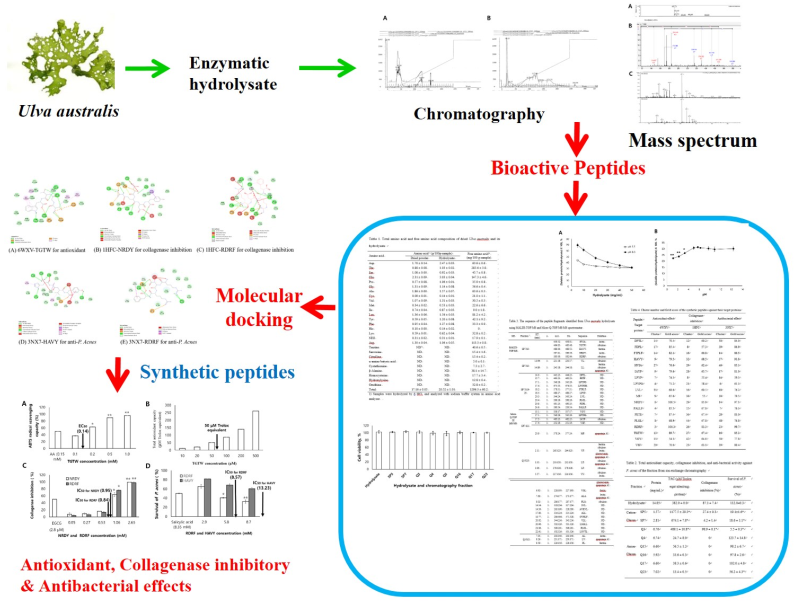
1. Introduction
The genus
Ulva (Ulvophyceae, Chlorophyta), consisting of common green macroalgae that often form algal beds, is found in intertidal zones in bays [
1].
U. australis is commonly known as sea lettuce, an important marine vegetables and feeds, which can be directly collected and eaten.
U. australis has high economic value for aquaculture [
2]. U. australis is widely distributed in the coast of Korea, and particularly, it was appeared in the coast of Jeju island above 10,000 ton per year. Therefore, we need to convert
U. australis to valuable products.
Although red and green algae contain 9-47% protein, most studies focus on a nutritional evaluation of total algae due to the difficulty to extract cytoplasmic compounds from algae [
3,
4,
5]. Recently, the bioactive peptides, including an antioxidant, anti-inflammatory, antimicrobial and antihypertension, from seaweed hydrolysate were widely investigated [
6,
7]. The peptides from the protein of macroalage have a potential application to improve health benefit and to treat disease [
8,
9,
10,
11]. The peptides from Ulva spp. were supposed to have the bioactivity, including the inhibition of angiotensin-I converting enzyme (ACE), dipeptidyl peptidase IV, dipeptidyl peptidase III, renin and α-glucosidase. They also showed antioxidant and anti-inflammatory effects via in silico analysis [
12]. The papain hydrolysate of
U. lactuca showed ACE inhibitory activity [
13]. The peptides form gastro-intestinal digestion of Ulva spp showed anti-inflammatory activity on immune cell [
14].
Due to a great deal of interest in therapeutic peptides, several protein-peptide docking techniques have been developed which lead the study and optimization for drug screening and design [
15]. Protein-peptide interactions serve as structural components in approximately 40% of all macromolecular interactions [
16,
17]. Peptides can be used to prevent diseases involving malfunctioning of proteins due to undesirable protein-protein interactions [
18]. Many databases and algorithms have been developed in the past specifically in the field of peptide-based therapeutics [
19,
20]. There are more than 200 therapeutics peptides, approved by FDA for the treatment of various diseases [
21,
22]. The aim of this research was to screen the multi-functional bioactivities of protein hydrolysate from
U. australis. The protein form
U. australis was hydrolyzed by alcalase and flavourzyme, and the peptides from hydrolysate was purified and identify by column chromatography, MALDT-TOF and Q-TOF mass spectrometry. In vitro ABTS radical scavenging effect, total antioxidant capacity, collagenase inhibitory and anti-bacterial activities of peptides were evaluated after screening peptides through molecular docking.
2. Results
2.1. Properties of Hydrolysate from U. australis
Total amino acids of dried
U. australis and its hydrolysate were 17.17 g/100 g and 20.52 g/100 g, respectively, and were increased to 3.35 g after hydrolysis (
Table 1). Acid amino acid, including aspartic acid and glutamic acid, and proline were increased, while alanine and arginine were decreased after hydrolysis. Total amino acid of dried
U. australis was high in order to glutamic acid, alanine and aspartic acid. Furthermore, free amino acid content from
U. australis hydrolysate was 1296.3 mg/100 g, and high in order to threonine, glutamine and arginine (
Table 1).
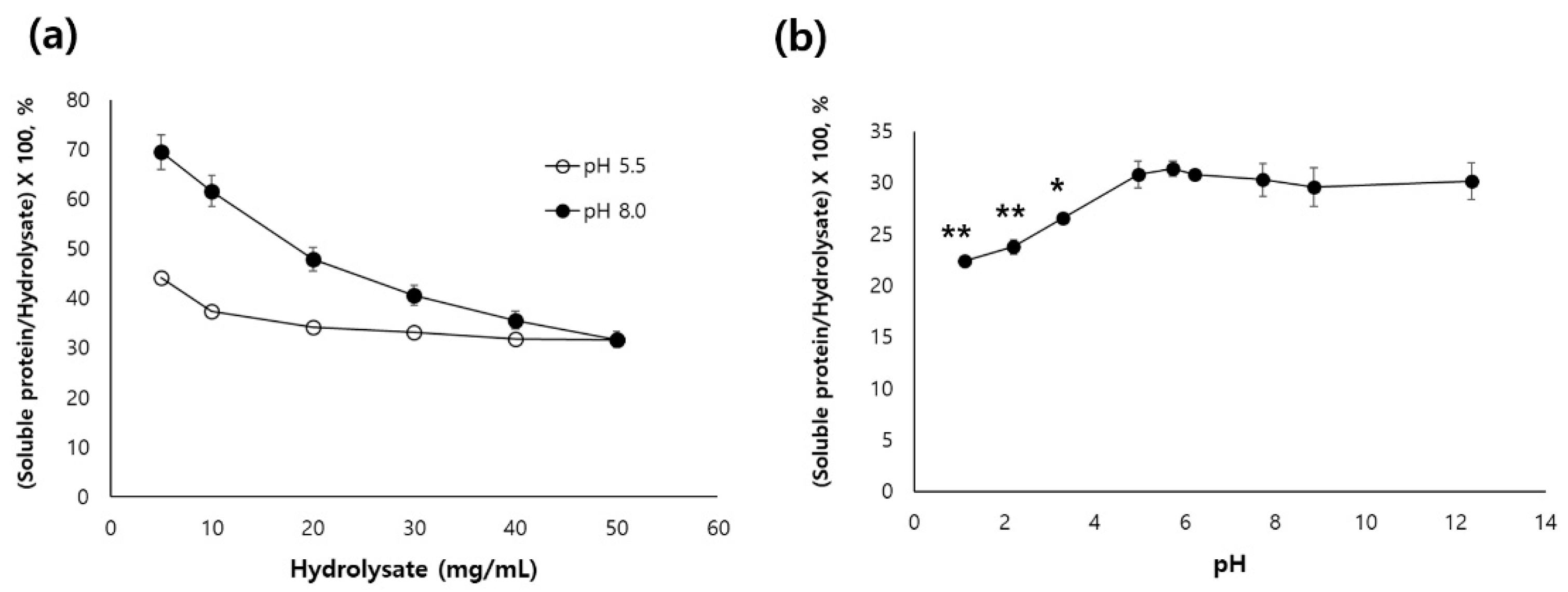
Solubility is important factor to determine the reasonable sample amount for application of chromatography. The solubility on pH of
U. australis hydrolysate didn’t change in the range of pH 5.0~12, significantly, but decreased below pH 4.5 (
Figure 1a). Solubilities of hydrolysate at 25 mM sodium citrate (pH 5.5) and 25 mM Tris-Cl (pH 8.0) were decreased with increase of concentration, and were not difference between pH 5.5 and pH 8.0 at 50 mg/mL. About 30% of hydrolysate was soluble at 50 mg/mL (
Figure 1b).
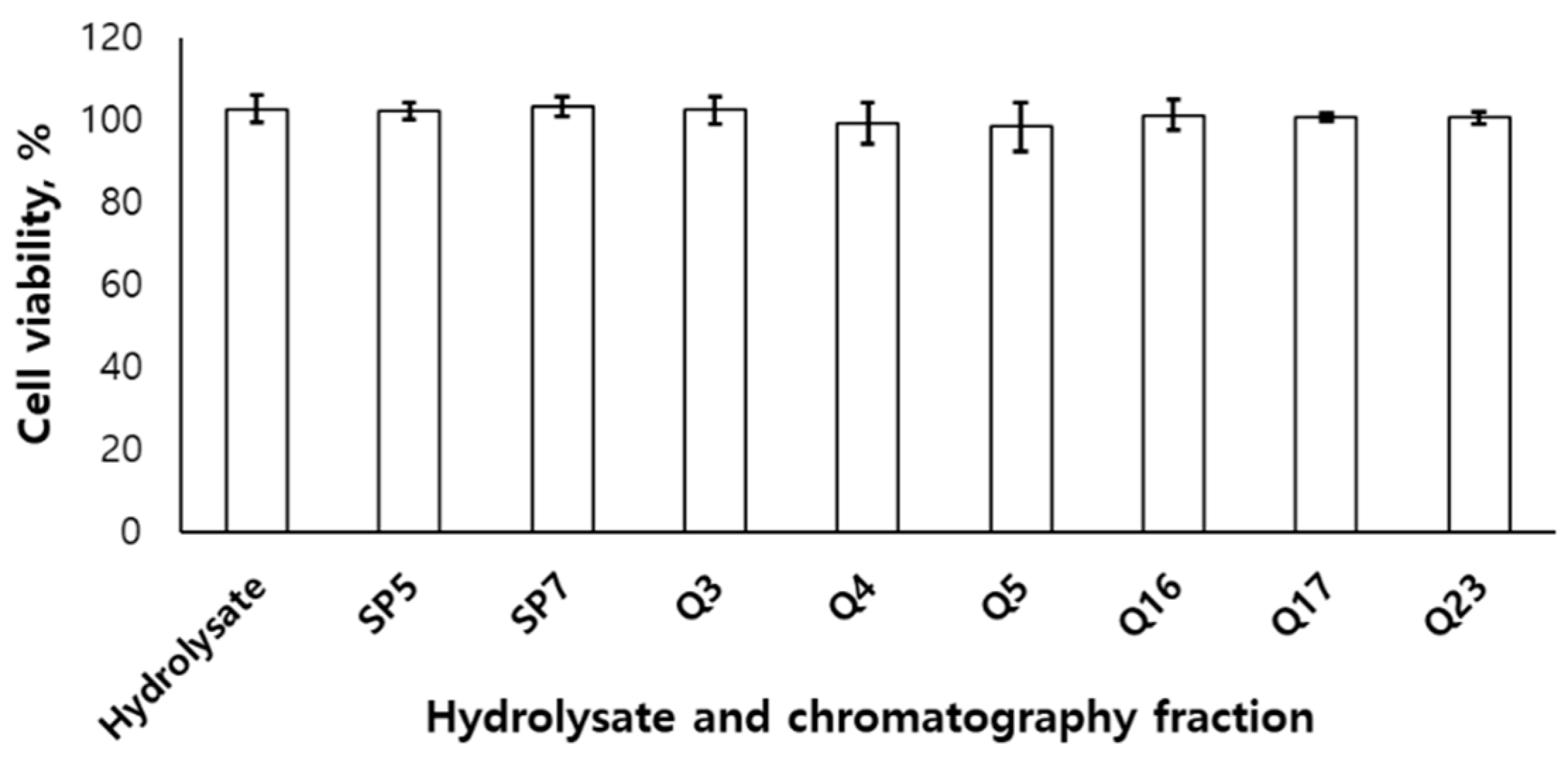
The hydrolysate from
U. australis and the fractions of cation- and anion-exchange chromatography weren’t show the toxicities on RAW 264.7 cell (
Figure 2).
2.2. Purification of Bioactive Peptides
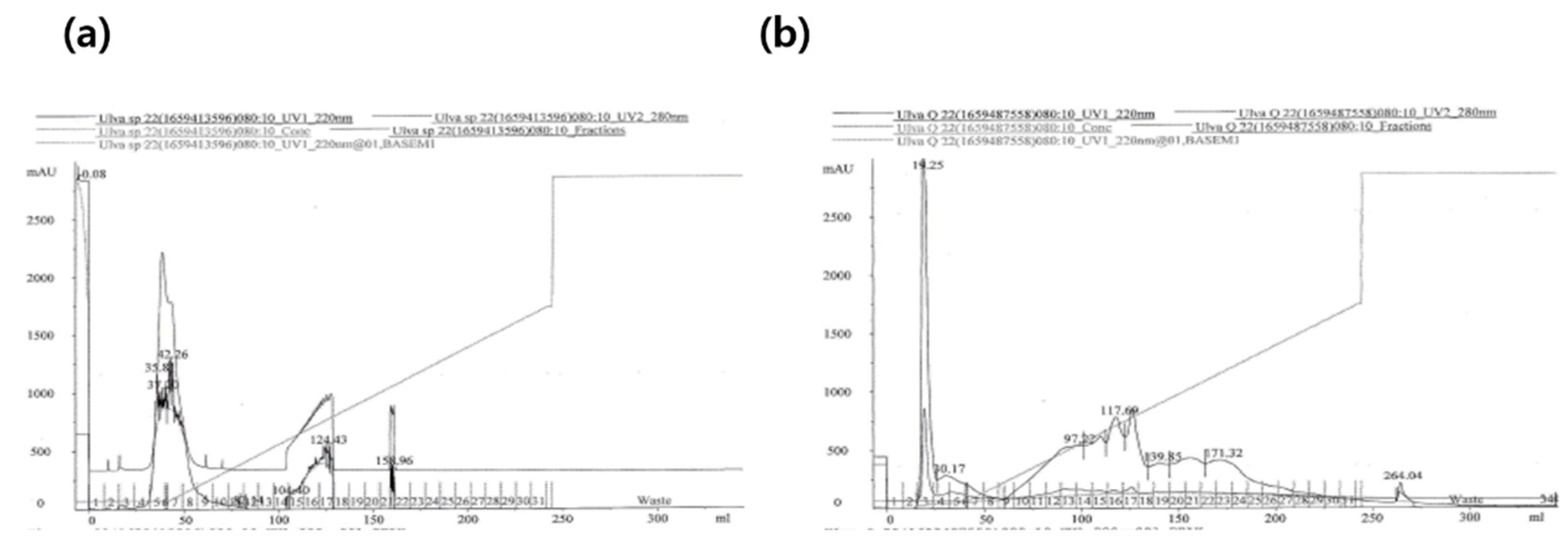
The
U. australis hydrolysate was separated into two fractions using a SP-Sepharose ion-exchange column, while separated into six fractions using a Q-Sepharose ion exchange column (
Figure 3). Relatively high TAC were observed in the three fractions: SP5, SP7, and Q3 (
Table 2). A typical chromatogram of the purification procedure is shown in
Figure 3. Total antioxidant activities of these three fractions were 1177.5, 674.1 and 489.1 μM Trolox equivalent/mg-protein, respectively. Collagenase inhibitory and antibacterial against
P. Acnes were also observed in the three fractions: SP5, SP7 and Q3 (
Table 2). The three active fractions were divided further using size-exclusion chromatography. The SP 5 and SP7 fractions were divided into three fractions (S9, S19, S21), while the Q3 fraction were into two fractions (S20 and S21) (not shown). Bioactivities of fractions from size exclusion chromatography were not assay further due to low protein concentration. The fractions from size exclusion chromatography were used for Q-TOF/MS/MS after completely dryness using speed centrifuge at 40℃. The SP7/S9 fraction was purified on reversed phase (RP) chromatography more for MALDI-TOF analysis and divided into three fractions (SP7/S9/C4; SP7/S9/C5; SP7/S9/C20), while SP5/S9 fraction was not purified on RP chromatography because peptide peaks were not observed in MALDI-TOF analysis (not shown). The specific peptide spectrum of SP5/S9 fraction was also not observed in Q-TOF/MS. The depression of NO production and cox-2 inhibitory activities were not observed in any fractions (not shown).
2.3. Amino Acid Sequence of Bioactive Peptides from Hydrolysate
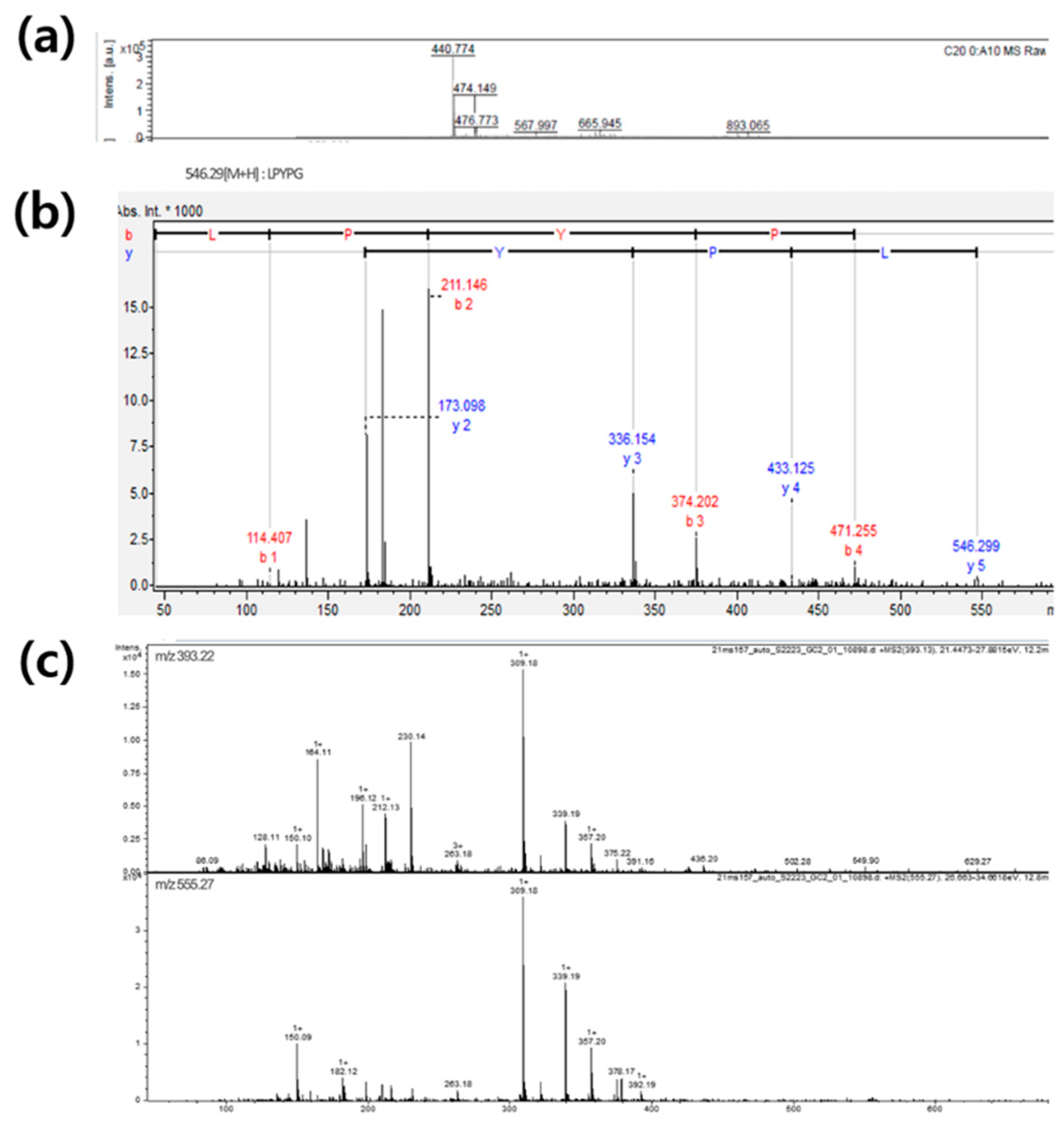
To identify the sequence of peptides, the fractions (SP7/S9, SP7/S9, SP7/S19 and SP7/S21) were subjected to MALDI-TOF/MS and Q-TOF/MS/MS. Five peptides was identified in MALDI-TOF/MS, while 35 peptides was identified in Q-TOF/MS/MS. (
Table 3). In MALDI-TOF/MS, the amino acid sequences from protein database of Ulva australis (
www.uniprot.org) were cleaved according to the substrate specificity of alcalase proposed by Doucet et al. (2003) [
23]. Database suggested the five protein sequences, including ferritin, ribulose biphosphate carboxylase-large chain, lectin, plastocyanin and chlorophyll apoprotein A1, derived from Ulva asutralis. The peptide sequence was identified from the molecular weight of cleaved fragments of protein sequences matched with m/z value of spectrum obtained from MALDI-TOF/MS. The sequences of 14 peptide sequences obtained from Q-TOF/MS/MS were identified from database, but 21 peptide sequences not identified. A typical mass spectrum of MALDI-TOF and Q-TOF/MS/MS is shown in
Figure 4. The spectrum of SP7/S21 fraction using Q-TOF/MS/MS proposed the presence of glycan, and suggested the possibility of N-acetylhexsosamine attached to serine and threonine residues of protein. Since the protein from database have 9-60 serine residues and 4-41 threonine residues, glycoprotein is a possible in U. australis hydrolysate.
2.4. Molecular Docking for Screening Bioactive Peptides
In order to study the interaction mechanism between peptides and target proteins, eighteen peptides were docked into the active sites of target proteins, including 6WXV (pdb code) for antioxidant, iHFC (pdb code) for collagenase inhibition, and 3NX7 (pdb code), by CDOCKER ligand docking module in BIOVIA Discovery Studio (Dassault Systems, Walsham, MA, USA). Protein, 6 WXV, catalyzes production of hydrogen peroxide as FAD oxidase family, FAD and FDAH is important coenzyme for the mechanism of antioxidant. Protein, 1HFC, is collagenase in human fibroblast cell. Protein, 3NX7, is human matrix metallopeptidase-12 (MMP-12) [
24]. The mechanisms of non-membrane targeting for antibacterial effect are divide into an inhibition of protein and nucleic acid synthesis, an inhibition of serine protease and a cell division [
25]. Especially, eNAP-2 and indolcidine as antimicrobial peptides inhibited an elastase and chymotrypsin of microorganisms [
26]. We supposed that inhibition of the selected protein for molecular docking lead to screen the bioactive peptides.
The reasonable docked conformations were selected based on cluster analysis and the Gold fitness score. FDPL, NRDY and TGTW for antioxidant, NRDY and RDRF for collagenase inhibitor, and FDPL, HAVY, NRDY and RDRF for antibacterial effect were screened for in vitro validation based on molecular docking. The cluster number from 50 trials was 13 for TGTW as antioxidant, 29 and 26 for NRDY and RDRF as collagenase inhibitor, and 37 and 23 for HAVY and RDRF as antibacterial effect, respectively. The Gold scores of peptides were in the range of 83.3- 100.5 (
Table 4).
Non-covalent interactions play an important role in bio-macromolecues, not only maintaining the three dimensional structure of large molecules but also are responsible for the molecular recognition process [
27]. The interaction models between the target proteins and peptides were analyzed by Discovery Studio (
Figure 5). In 6WXV/TGTW complex, TGTW has a hydrogen bond with Arg 1106, Met 1104, Tyr 1232, Arg 1214, FAD 1601 within 6WXV, and a hydrophobic interaction with Ile 1100, Met 1205, His 1319, Trp 1222. The electrostatic interactions were formed with FAD 1601, Glu 1317 and Tyr 1228. In 1HFC/peptides, NRDY didn’t show an electrostatic interaction with Zn-ion in collagenase, while showed Glu 219, His 218 and His 222. In most case of collagenase inhibition, inhibitory peptides formed an electrostatic interaction with Zn-ion. Although NRDY didn’t have an electrostatic interaction, the reason to have the collagenase inhibitory comparable to RDRF is ambiguous. HAVY as antibacterial agent had a lot of hydrogen bonds compared to RDRF, and showed higher antibacterial effect than RDRF in vitro validation.
2.5. In Vitro Validation of Bioactive Peptides
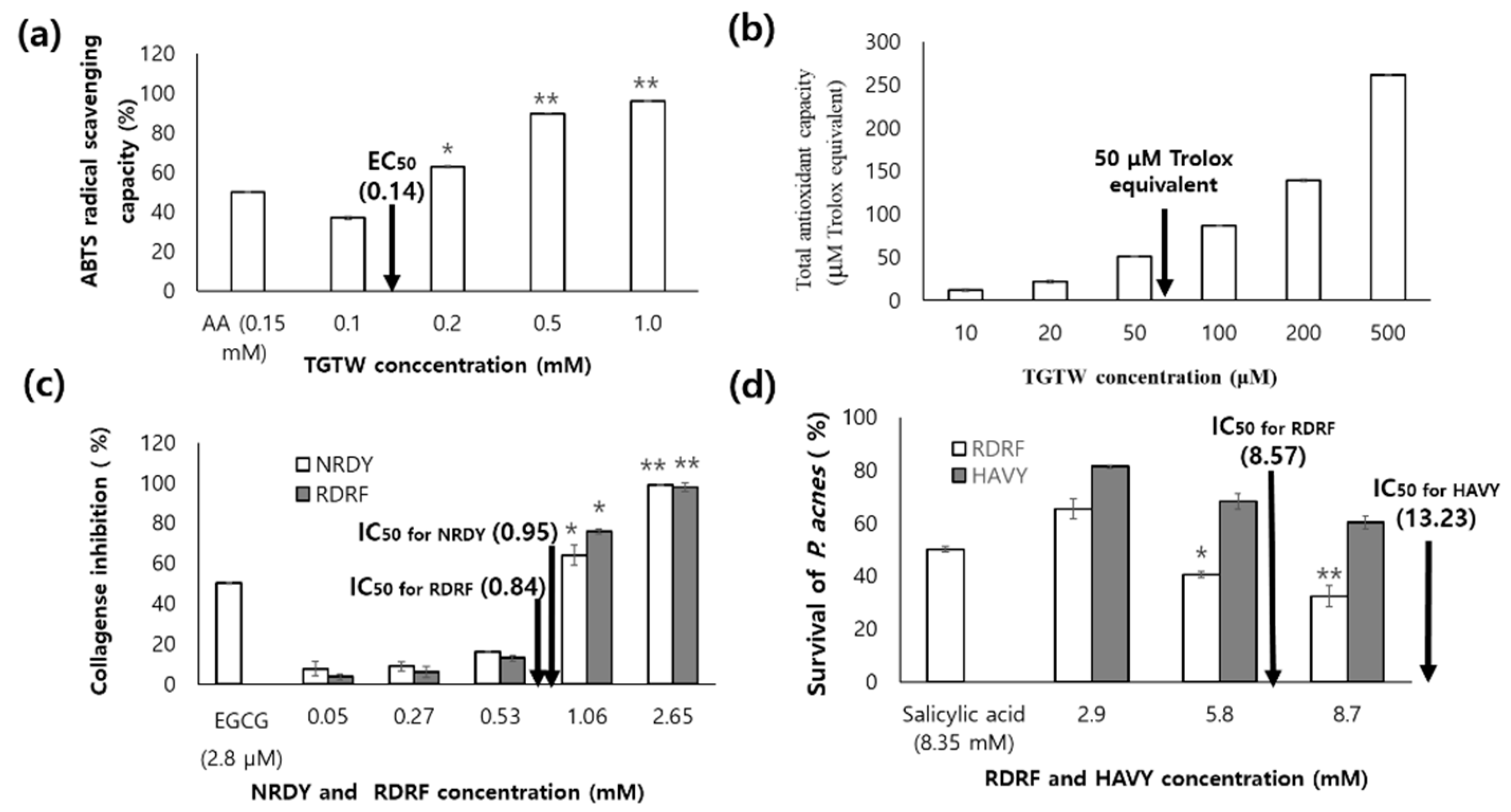
ABTS [2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid] scavenging and total antioxidant capacity of TGTW were dependent on the concentration. EC
50 value of ABTS scavenging capacity was 0.14 mM comparable to 0.15 mM of ascorbic acid (
Figure 6a). Total antioxidant capacity was 50 μM Trolox equivalent at the concentration of 53.9 μM (
Figure 6b). The results suggest the U. australis containing TGTW is available antioxidant agents for food application. NRDY and RDRF inhibited the collagenase with concentration dependent manner, the IC
50 values of them were 0.95 mM and 0.84 mM, respectively (
Figure 6c). Peptide, VICE, from the peptic hydrolysate of flounder skin inhibited collagenase up to 83.25% at 2.16 mM [
28]. The collagenase inhibitory activity of RDRF was about 1.5 times higher than one of VICE. The survival of P. Acnes was greatly decreased with increasing of the concentration of peptide, RDRF and HAVY, and the IC
50 values of them were 8.57 mM and 13.23 mM, respectively (
Figure 6d). The inhibition effect of RDRF was comparable with salicylic acid as positive control (8.35 mM).
Antimicrobial peptides are mainly composed of 12–100 amino acid residues, and total charge of them is in the range of 2
+–10
+ as amphoteric peptide [
29]. However, a long chain peptide is not transported into the cell without transporter. Therefore, we limited amino acid residue of peptide below 5 amino acids in this study. Huan et al., (2020) classified the antimicrobial peptides based on amino acid-rich species as following; proline-rich, tryptophan and arginine-rich, histidine-rich and glycine-rich peptide [
30]. The stage of the action mechanism of small cationic peptides with antimicrobial activity is ruled by electrostatic interaction between the peptide and the pathogen cell membrane. An increase in its activity could be expected with an increase in the positive charge on the peptide [
31]. Most cationic antimicrobial peptide act by accumulating on the surface of bacterial membrane and causing the formation of defects when a threshold is reached [
32]. RDRF and HAVY had two positive charge, and the number of total positive charge was not much compared to other antimicrobial peptides. The peptide, SFIQRFTT, had two positive charge, and suppressed the growth of Listeria innocus and E. coli [
33].
3. Discussion
Many research efforts have focused on identifying effective peptide possessing biofunctional properties that might have preventive or therapeutic value to combat aging or disease. Protein hydrolysates are of interest as materials for medicinal purposes because of their pharmaceutical activity as inhibitor to reduce other related diseases and aging [
34]. Some peptides having a potential angiotensin I-converting enzyme-inhibitory activity are inactive within the protein sequence, but they can reveal a biological activity when they are released by hydrolysis [
35]. Moreover, among the various activities of protein hydrolysates, the inhibition of tyrosinase and elastase attracted attention due to their pharmaceutical activities in the field of agriculture, food and cosmetics as well as medicine [
36].
In our study, total and free amino acid composition of extracts and enzymatic hydrolysate analyzed. As the results, acid amino acid, including aspartic acid and glutamic acid, and proline were increased, while alanine and arginine were decreased after hydrolysis. Total amino acid of dried
U. australis was high in order to glutamic acid, alanine and aspartic acid. Glutamic acid, proline, glycine, alanine and amino sulfonic acid is abundant in green algae [
37]. Glutamic acid, aspartic acid and leucine is abundant in
Enteromprpha prolifera [
38]. Our results were comparable to the amino acid composition of the previous reports. Furthermore, free amino acid content from
U. australis hydrolysate was 1296.3 mg/100 g, and high in order to threonine, glutamine and arginine. The nitrogen compounds of extract were composed of amino acid, peptide, ammonia in the range of 40-70% [
39]. Free amino acid from
Enteromorpha prolifera depended on a harvested month, and arginine and glutamic acid were higher than other amino acids [
40]. Green algae accumulate taurine and citrulline to regulate a osmotic pressure of seawater. Arginylglutamine as dipeptide is in
U. australis and
U. linza, and a major compound of free amino acid [
41]. A lot of taurine and sarcosine were detected, while a little homocysteine and hydrolxylysine were also detected [
40].
Solubility is important factor to determine the reasonable sample amount for application of chromatography. A previous study reported that the oyster hydrolysate didn’t show the pH-dependence [
42]. In this study, the solubility on pH of
U. australis hydrolysate didn’t change in the range of pH 5.0~12, significantly, and Solubilities of hydrolysate at 25 mM sodium citrate (pH 5.5) and 25 mM Tris-Cl (pH 8.0) were decreased with increase of concentration, and were not difference between pH 5.5 and pH 8.0 at 50 mg/mL.
Numerous docking methods have been developed in the past for structural determination of protein-peptide complexes and these methods can be classified broadly into the following 3 categories; i) protein-peptide docking, ii) protein-protein docking and iii) protein-small molecule docking [
43]. Protein-peptide docking methods have been specifically developed to dock peptide on protein [
44]. Though protein-protein docking methods have been developed for docking two proteins, some of the software developed for docking small-molecules on a protein can also be used to dock peptide on a protein [
45]. Therefore, a wide range of docking methods can be used directly or indirectly for docking peptide on a protein. In this study, ABTS radical scavenging effect, TAC, collagenase inhibitory and anti-bacterial activities from enzymetic hydrolysate of
U. pertusa were determined by protein-peptide docking techniques for the development of value-added products such as ingredients in foods or cosmetics. Eighteen peptides were docked into the active sites of target proteins. FDPL, NRDY and TGTW for antioxidant, NRDY and RDRF for collagenase inhibitor, and FDPL, HAVY, NRDY and RDRF for antibacterial effect were selected based on cluster analysis and the Gold fitness score. Finally, TGTW, NRDY, NRDY and HAVY confirmed by the interaction models of protein/peptide complex after molecular dynamics simulation by Discovery Studio. The final selected peptides showed ABTS radical scavenging effect (TGTW), total antioxidant capacity (TGTW), collagenase inhibitory (NRDY and RDRF) and antibacterial effect against P. Acnes (RDRF and HAVY). Overall,
U. australis hydrolsate showed multi-function including ABTS radical scavenging effect, TAC, collagenase inhibition and antibacterial effect against
P. Acnes and it can be used in food or cosmetic industries as a bioactive ingredient.
4. Materials and Methods
4.1. Materials
U. australis was collected from the coast of Jeju island, South Korea in December 2021 and stored it at freezer of −20 °C after pulverizing dried U. ustralis under sunlight for 1 week. Alcalase 2.4 L (2.4 AU/g, endopeptidase, Bacillus licheniformis) and Flavourzyme 500 MG (500 LAPG/g, endoprotease and exopeptidase, Aspergillus oryzae) were purchased from Biosis company (Busan, South Korea). Water and acetonitrile (AH365-4, Burdick & Jackson Co. Europe) were high performance liquid chromatography (HPLC) grade. The mouse macrophage cell line RAW 264.7 was obtained from Korean Cell Line Bank (Seoul, Korea). Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin were supplied by Gibco BRL (Grand Island, NY, USA). Assay kits for total antioxidant capacity (ab65329) and collagenase inhibitory activity (EnzChek gelatinase/collagenase) were purchased from Abcam Inc. (Cambridge, England) and Invitrogen Co. (Waltham, MA, USA), respectively. HiLoad SP- and Q-Sepharose, and HiLoad 16/60 Superdex 30 prep grade columns were purchased from GE Healthcare (Parsoppany, NJ, USA). Eight peptides (FDPL, TGTW, HAVY, HVIA, NRDY, LPYPG, PETF and RDRF) were synthesized at A&PEP Co. (Cheongju, Korea).
4.2. Preparation of Hydrolysate from U. australis
Extract of protein from
U. australis was conducted by alkaline method [
45]. Dried sample (100 g) mixed with 15 volumes of 1N NaOH for 4 h and alkali soluble protein as supernatants obtained by centrifuge (Supra R22, Hanil Sci. CO. Ltd., Dajeon, Korea) at 8,000 rpm for 10 min. After adjustment into pH 4.2 using 1N HCL, the precipitated proteins collected by centrifuge at 8,000 rpm for 10 min. After suspend with 3 times of distilled water, adjusted them into pH 7.0 and proteins solubilized. To hydrolyze the solubilized proteins, one percentage (w/w) of alcalase 2.4 L and flavourzyme 500 MG were added, the mixture was then incubated at 60 °C for 4 h with stirring at 60 rpm. After inactivating the protease in a 95–100 °C water bath for 1h, the hydrolysate was centrifuged at 8,000 rpm for 15 min, and the supernatant was lyophilized and stored at −20 °C.
4.3. Solubility of Hydrolysate
To analyze solubility according to pH, distilled water added into enzymatic hydrolysate (0.2 g) and adjusted at 20 mL. Each 2 mL from them adjusted into pH 1–13 ranges with 1N HCL or 1N NaOH. The proteins concentration of supernatant by centrifuging (Labogen 1248, Gyrozen CO. Ltd., Gimpo, Korea; 2,500 × g and 10 min) measured with Biuret method [
22]. The relative solubility was expressed as percentage of soluble proteins/sample proteins.
4.4. Analysis of Total and Free Amino Acid Composition
The dried power of U. australis (10 mg) and enzymatic hydrolysate (10 mg) were exactly weighted in test tube. After adding 1.5 mL of 6 N HCL, the test tube was filled with N2 gas and sealed. Acid hydrolysis was conducted using heating block (HB-96D, Daihan Scientific Co, Ltd, Seoul, South Korea) for 24 h. The hydrolysate was filtered with 3G-4 glass filter, and the hydrochloride of filtrate was completely evaporated with vacuum rotatory evaporator (N-1110, Eyela, Tokyo, Japan) below 50 °C. The acid hydrolysate was dissolved in 0.02 N HCl solution, filtered with 0.02 μm cylinder-type filter. After injecting 40 μL of filtrate, amino acid composition was analyzed by the Amino Acid Analyzer (Biochrom 30, Biochrom, Cambridge, UK). Total amino acid was analyzed with sodium buffer system, while free amino acid composition was analyzed with lithium buffer system.
4.5. Purification of Bioactive Peptides from Hydrolysate
U. australis hydrolysates was dissolved in 25 mM citrate buffer (pH 5.5) for cation exchanger chromatography and 25 mM Tris-Cl buffer (pH 7.5) for anion exchanger chromatography, respectively, and the bioactive peptide fractions were eluted using a HiLoad SP-Sepharose column (16 × 100 mm) and HiLoad Q-Sepharose column (16 × 100 mm). Elution was performed using a linear gradient system from solvent A (25 mM citrate, pH 5.5) to solvent B (25 mM citrate, pH 5.5 containing 0.6 M NaCl) for cation-exchanger chromatography, and from solvent A (25 mM Tris-Cl, Ph 7.5) to solvent B (25 mM Tris-Cl, pH 7.5 containing 0.6 M NaCl) for anion-exchange chromatography with 10 column volumes at a flow rate of 4 mL/min, and detected at 220 nm and 280 nm. Antioxidant effect, collagenase inhibition, NO production and antibacterial effect against P. Acnes were assayed for each peak. The bioactive fractions were pooled, and concentrated with vacuum rotary evaporator. The concentrated fractions were loaded on a HiLoad Superdex 30 prep grade column (16 × 600 mm) and eluted with HPLC grade water at flow 1 mL/min. Detection was then carried out at 220 nm and 280 nm. Each fractions were lyophilized, and used for Q-TOF/MS/MS analysis. For MALDI-TOF analysis, each fraction from size exclusion chromatography was dissolved in 0.1% TFA/water and fractionated using the UHPLC system (Ultimate 3000, Thermo Fisher Scientific, Walsham, MA, USA) with a Biobasic C18 column (4.6 × 250 mm, 5 μm; Thermo Fisher Scientific, Walsham, MA, USA). Elution was performed using a linear gradient system from solvent A (0.1% TFA/water) to solvent B (0.1% TFA/50% ACN) over 10 min at a flow rate of 1 mL/min at 40 °C and detected at 220 nm. The column was equilibrated with solvent A, after which 50 μL of sample was applied to the column and elution was carried as follows; 10 min of solvent A, 2 min of A-B gradient, 10 min of solvent B, and 10 min of solvent A for equilibration. Each fraction was dried in speed vacuum concentrator, and determines the m/z value in MALDI-TOF spectrometry.
4.6. Cell Toxicity of the Hydrolysate
The cytotoxicity of hydrolysate and each fractions were measured at murine macrophage cell line RAW 264.7 (KCLB, Seoul, South Korea) by MTS assay described previously [
41]. Cell viability calculated by the following equation: Cell viability (%) = (1-At/Ac) ×100, where At is the sample treated absorbance and Ac is the absorbance of the non-treated control.
4.7. ABTS Radical Scavenging Activity and Total Antioxidant Capacity (TAC)
ABTS radical scavenging activity was determined by modification of a previously reported method [
25]. ABTS was calculated by the following equation: ABTS (%) = (Ac-At/Ac) ×100, where At is the sample treated absorbance and Ac is the absorbance of the non-treated control. TAC (%) was expressed as Trolox equivalent capacity (μM).
4.8. Collagenase Inhibitory Activity
Collagenase inhibitory activity was determined using EnzChek® Gelatinase/Collagenase Assay Kit (Invitrogen, Grand Island, NY) [
32]. Collagenase inhibitory activity (%) was calculated by the following equation: Collagenase inhibitory activity (%) =[100-(B-Bc/A-Ac)] × 100, where A is non-sample and collagenase treated absorbance, B is sample and collagenase treated absorbance, Ac is non-sample and non-collagenase treated absorbance, and Bc is non-sample and collagenase treated absorbance.
4.9. Antibacterial Effect on P. Acnes
The P. Acnes were cultivated in Reinforced Clostridial medium broth. The concentration of P. Acnes was adjusted to 0.6 OD 600 nm. The diluted microbial suspension (100 µL) and samples (40 µL) were inoculated into a 96-well microplate. After incubation for 1 day for 37 °C, the absorbance was measured at 600 nm with a microplate reader (Molecular Devices, VersaMax ELISA Microplate Reader, USA). The results were transformed to a percentage of the controls.
4.10. MALDI-TOF Mass and Q-TOF Mass/Mass Spectrometry
The fractions from reversed-phase chromatography were completely dried in vacuum centrifuge. The dried samples were dissolved in TA30 (30:70 (v/v) acetonitril:0.1% trifluoroacetic acid in water) and mixed with a saturated solution of α-cyano-4-hydroxycinnamic acid. The mixture was spotted onto a polished steel targets (Bruker Daltonics, Bremen, Germany). Mass spectra were acquired using an autoflex maX MALDI-TOF/TOF (Bruker Daltonics; High-Tech Materials Analysis Core Facility at Gyeongsang National University, Jinju, Korea), equipped with Smartbeam II laser working at 355 nm. The instrument was operated in the positive ion reflection mode. An external calibration was performed using a Peptide Calibration Standard (Bruker Daltonics), which includes Angiotensin II (MH+ = 1046.5418), Angiotensin I (MH+ = 1296.6848), Substance P (MH+ = 1347.7354), Bombesin (MH+ = 1619.8223), ACTH clip 1-17 (MH+ = 2093.0862), ACTH clip 18-39 (MH+ = 2465.1983), Somatostatin 28 (MH+ = 3147.4710). Mass spectra in the range 300-4,000 Da were obtained by averaging 500 laser shots and peptide peaks were generated using FlexAnalysis software ver 3.4 (Bruker Daltonics). The MS/MS spectra were obtained in the LIFT mode. De novo peptide sequencing of the major peaks was performed by manual interpretation of the spectra compared with the sequence obtained from Ulva protein database (
www.uniprot.org).
UHPLC-Q-TOF/MS/MS analyses were performed using a Vanquish UHPLC (Thermo Fisher Scientific, Walsham, MA, USA) coupled to a Q Extractive plus (Thermo Fisher Scientific, Walsham, MA, USA) mass spectrum spectrometer equipped with a electrospray ionization (ESI) source. Each lyophilized fraction from size-exclusion chromatography was dissolved in 200 μL of 0.1% formic acid and filtered with 0.20 μm membrane filter after centrifuge at 13,000 rpm for 10 min.
Chromatographic separation was carried out on Zobax eclipse plus C18 column (3.0 × 100 mm, 1.8 μm, Agilent). The mobile phase consisted of solvent A (0.1% formic acid/water, v/v) and solvent B (0.1% formic acid/acetonitrile, v/v) and elution conditions were as follows: 0.0-2.0, 2% B; 2.0-30.0 min, 30% B; 30.0-31.0, 98% B; 31.0-36.0, 98% B; 36.0–37.0, 2.0% B, 37–45.0 min, 2% B. The flow rate was set at 0.3 mL/min. The column and autosampler were maintained at 30 °C and 4 °C, respectively. The injection volume was 5 μL. For Mass spectrometry was set to acquire over the m/z range 100–1500 with full MS scan type. For electrospray ion source, spray voltage and capillary temperature were set to 3.5 kV and 250 °C, respectively. MS/MS parameter was set to Dd-MS2(HCD) as scan type, 17,500 as resolution, 100 ms as maximum ion time and 100 m/z as fixed first mass. Both the peptide sequencing module of the software and manual calculations were used to process the MS/MS data and to perform peptide sequencing.
4.11. Molecular Docking
The calculation of target protein-peptide docking was performed with genetic optimization (GOLD version 5.2.2, Cambridge Crystallographic Data Centre, Cambridge, UK) according a previous method [
46]. Synthesized peptides (HVIA, TGTW, HAVY, NRDY, RDRF, PETF, RDRF and FDPL) were used as the ligand, and the 3D structures of them were obtained from Discovery Studio (DS) program (BIOVIA, San Diego, CA, USA). Geometry was optimized by energy minimizing using minimized ligand tool existing DS. Ligand converted to SD file for use of GOLD and contained all active ligands within 10 Å radius of center for calculation. The consistency of 3D structure in each peptide was identified by clustering of 50 times. The mouse DUOX1 (pdb, 6WXV), human fibroblast collagenase (pdb, 1HFC) and human MMP-12 (pdb, 3NX7) as target for protein-peptide docking were downloaded from
www.rcsb.org.
4.12. Statistical Analysis
Data were expressed as the mean with standard deviation of triplicate determinations. Analysis of variance was carried out by the Tukey HSD test using the JMP (version 12, SAS Institute, NC, USA). Significance was indicated at a p < 0.05 and p < 0.01.
5. Conclusions
The fractions from U. australis hydrolysate using cation-exchange chromatography showed high ABTS scavenging and TAC, the inhibition of collagenase, and antibacterial effect against P. Acnes. U. australis hydrolysate and the fractions from ion-exchange chromatography were not show toxicities on RAW 264.7 cell. In TGTW, the ABTS radical scavenging capacity was comparable to ascorbic acid, and total antioxidant capacity 50 uM Trolox equivalent at the concentration of 53.9 μM. The IC50 values of NRDY and RDRF for collagenase inhibition were 0.95 mM and 0.84 mM, respectively. Antibacterial effect of RDRF against P. Acnes showed the IC50 values of 8.57 mM comparing with salicylic acid. Since the U. australis hydrolsate showed multi-function including antioxidant, collagenase inhibition and antibacterial effect against P. Acnes, it has a potential for the cosmetic application.
Author Contributions
The in vitro experiments, Y.J. and S.K.; writing—original draft preparation, H.J. and Y.A.; study concept, writing—review and editing, Y.A., S.K. and H.J.; funding acquisition, Y.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Regional Innovation Cluster Upbringing (R&D), Ministry of Trade, Industry and Energy, South Korea (grant no. 1415171432-P0015257).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All the data generated or analyzed during this study were included in this article.
Conflicts of Interest
There are no conflicts of interest.
References
- Mitsuhashi, C.; Teramura, H.; Shimada, H. Construction of genomic marker sets based on the chloroplast genome of a green alga, Ulva pertusa (syn. Ulva australis), leads to simple detection of Ulva species. Genes Genet. Syst. 2020, 95, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Tabarsa, M.; Rezaei, M.; Ramezanpour, Z.; Waaland, J.R. Chemical compositions of the marine algae Gracilaria salicornia (Rhodophyta) and Ulva lactuca (Chlorophyta) as a potential food source. J. Sci. Food Agric. 2012, 92, 2500–2506. [Google Scholar] [CrossRef] [PubMed]
- Harnedy, P.A.; FitzGerald, R.J. Extraction and enrichment of protein from red and green macroalgae. In Natural products from marine algae. 1nd ed.; Stengel, D.B. Connan, S., EDs.; Humana press, Springer Science, N.Y., 2015, pp. 103–108.
- Fleurence, J. Seaweed proteins, biochemical, nutritional aspects and potential use. Trends in Food Sci. Technol. 1999, 10, 25–28. [Google Scholar] [CrossRef]
- McDermid, K.L.; Stuercke, B. Nutritioanl composition of edible Hawaiian seaweeds. J. Appl. Phycol. 2003, 15, 513–524. [Google Scholar] [CrossRef]
- Rengasamy, K.R.R.; Mahomoodally, M.F.; Aumeeeruddy, M.Z.; Zengin, G.; Xiao, J.; Kim, D.H. Bioactive compounds in seweeds: An overview of their biological properties and safety. Food Chem. Toxicol. 2020, 135, 111013. [Google Scholar] [CrossRef]
- Echave, J.; Fraga-Cocrral, M.; Garcia-Perez, P.; Popović-Djordjević, J.; Avdović, E.H.; Radulović, M.; Xiao, J.; Prieto, M.P.; Simal-Gandara, J. Seaweed protein hydrolysates and bioactive peptides: Extraction, purification, and applications. Mar. Drugs 2021, 19, 500. [Google Scholar] [CrossRef]
- Aneiros, A.; Garateix, A. Bioactive peptides from marine sources: pharmacological properties and isolation procedures. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 803, 41–53. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Gallagher, E.; Tasdemir, D.; Hayes, M. Heart health peptides from macroalgae and their potential use in functional foods. J. Agric. Food Chem. 2011, 59, 6869–6836. [Google Scholar] [CrossRef]
- Gupt, S.; Sabu-Ghannam, N. Bioactive potential and possible health effects of edible brown seaweeds. Trend. Food Sci. Technol. 2011, 315–326. [Google Scholar] [CrossRef]
- Fan, X.; Bai, L.; Zhu, L.; Yang, L.; Zhang, X. Marine algae-derived bioactive peptides for human nutrition and health. J. Agric. Food Chem. 2014, 62, 9211–9222. [Google Scholar] [CrossRef]
- Amin, M.A.; Chondra, U.; Mostafa, E.; Alam, M.M. Green seaweed Ulva lactuca, a potential source of bioactive peptides revealed by in silico analysis. Informatics Med. Unloacked 2022, 101099. [Google Scholar] [CrossRef]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. In vitro and in silico approaches to generating and identifying angiotensin-converting enzyme I inhibitory peptides from green Macroalga Ulva lactuca. Mar. Drugs 2019, 17, 204. [Google Scholar] [CrossRef]
- Cian, R.E.; Hernadez-Chirlaque, C.; Gamez-Belmonte, R.; Drago, S.R.; de Medina, F.S.; Martínez-Augustin, O. Green alga Ulva spp. hydrolysates and their peptide fractions regulate cytokine production in splenic macrophages and lymphocytes involving the TLR-NFkB/MAPK oathways. Mar. Drugs 2018, 16, 235. [Google Scholar] [CrossRef]
- Masomian, M.; Lalani, S.; Poh, C.L. Molecular Docking of SP40 Peptide towards cellular receptors for enterovirus 71 (EV-A71). Molecules 2021, 26, 6576. [Google Scholar] [CrossRef]
- Petsalaki, E.; Russell, R.B. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr. Opin. Biotechnol. 2008, 19, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Neduva, V.; Russell, R.B. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006, 17, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Saint-Maur, P.D. Is malignant mesenchymoma a mildly aggressive tumor? Ann. Pathol. 1992, 12, 146. [Google Scholar]
- Nevola, L.; Giralt, E. Modulating protein–protein interactions: the potential of peptides. Chem. Commun. 2015, 16, 3302–3315. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Agrawal, P.; Kumar, R.; Bhalla, S.; Usmani, S.S.; Varshney, G.C.; Raghava, G.P.S. Prediction of cell-penetrating potential of modified peptides containing natural and chemically modified residues. Front. Microbiol. 2018, 9, 725. [Google Scholar] [CrossRef]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: database of FDA-approved peptide and protein therapeutics. PLoS One 2017, 12, e0181748. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Doucet, D.; Otter, D.E.; Gauthier, S.F.; Fodgeding, E.A. Enzyme-induced gelation of extensively hydrolyzed whey proteins by Alcalase: Peptide identification and determination of enzyme specificity. J. Agric. Food Chem. 2003, 51, 6300–6308. [Google Scholar] [CrossRef] [PubMed]
- Sun, J. Structure of mouse DUOX1-DOUXA1 provide mechanistic insights into enzyme activation and regulation. Nat. Struct. Mol. Biol. 2020, 27, 1086–1093. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracelluar targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, e02340–16. [Google Scholar] [CrossRef]
- Cerny, J.; Hobza, P. Non-covalent interactions in biomacromolecules. Phys. Chem. Chem. Phys. 2007, 9, 5291–5303. [Google Scholar] [CrossRef]
- Dawnay, A.B.; Hirst, A.D.; Perry, D.E.; Chambers, R.E. A critical assessment of current analytical methods for the routine assay of serum total protein and recommendations for their improvement. Ann. Clin. Biochem. 1991, 28, 556–567. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 49, 491–511. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Cascales, J.J.L.; Zenak, S.; de la Torre, J.G.; Lezama, O.G; Garro, A.; Daniel Enriz, R.D. Small cationic peptides: Influence of charge on their antimicrobial activity. ACS Omega 2018, 3, 5390–5398. [Google Scholar] [CrossRef]
- Bortolotii, A.; Troiano, C.; Bobone, S.; Konai, M.M.; Ghosh, C.; Bocchinfuso, G.; Acharya, Y.; Santucci, V.; Bonacorsi, S.; Di Stefano, C.; Haldar, J.; Stella, L. Mechanism of lipid bilayer perturbation by bactericidal membrane active small molecules. Biochim. Biophys. Acta Biomembr. 2023, 1865, 184079. [Google Scholar] [CrossRef] [PubMed]
- Ennaas, N.; Hammami, R.; Beaulieu, L.; Fliss, I. Purification a characterization of four antibacterial peptides from protamex hydrolysate of Atlantic mackerel (Scomber scombrus) by-products. Biochem. Biophys. Res. Commun. 2015, 462, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kim, K.T.; Kim, S.M. Biofunctional properties of enzymatic squid meat hydrolysate. Prev. Nutr. Food Sci. 2015, 20, 67–72. [Google Scholar]
- Jun, S.Y.; Park, P.J.; Jung, W.K.; Kim, S.K. Purification and characterization of an antioxidative peptide from enzymatic hydrolysate of yellowfin sole (Limanda aspera) frame protein. European Food Res. Technol. 2004, 219, 20–26. [Google Scholar]
- Ames, B.N.; Shigenaga, M.K.; Hagen, T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA 1993, 90, 7915–7922. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Botta, J.R. Seafoods: Chemistry, Processing Technology and Quality; Springer: New York, NY, 1994; pp. 331–356. [Google Scholar]
- Ganesan, K.; Kumar, K.S.; Rao, P.V.S.; Tsukui, Y.; Bhaskar, N.; Hosokawa, M.; Miyashita, K. Studies on chemical composition of three species of Enteromorpha. Biomed. Preventive Nutr. 2014, 365–369. [Google Scholar] [CrossRef]
- Thiviya, P.; Gamage, A.; Gama-Arachchige, N.S.; Merah, O.; Madhujith, T. Seaweeds as a source of functional proteins. Phycol. 2022, 216–243. [Google Scholar] [CrossRef]
- Oliveira, A.P.; Pereira, D.M.; Andrade, P.B.; Valentão, P.; Sousa, C.; Pereira, J.A.; Bento, A.; Rodrigues, M.A.; Seabra, R.M.; Silva, B.M. Free amino acids of tronchuda cabbage (Brassica oleracea L. Var. costata DC): influence of leaf position (internal or external) and collection time. J. Agric. Food Chem. 2008, 56, 5216–5221. [Google Scholar] [CrossRef]
- Lim, A.S.; Jeong, H.J.; Kim, S.J.; Ok, J.H. Amino acids profiles of six dinoflagellate species belonging to diverse families: possible use as animal feeds in aquaculture. Algae 2018, 33, 279–290. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Xing, R.; Liu, S.; Chen, X.; Li, P. Optimization of oyster (Crassostrea talienwhanensis) protein hydrolysates using response surface methodology. Molecules 2020, 25, 2844. [Google Scholar] [CrossRef]
- Agrawal, P.; Singh, H.; Srivastava, H.K.; Singh, S.; Kishore, G.; Raghava, G.P.S. Benchmarking of different molecular docking methods for protein-peptide docking. BMC Bioinformatics 2019, 19, 426. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.Y. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450.45. Lyskov S.; Gray, J.J. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008, 36, W233–W238. [Google Scholar]
- Vanvi, A.; Tsopmo, A. Pepsin digested oat bran proteins: separation, antioxidant activity, and identification of new peptides. J. Chem. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracelluar targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, e02340–e20516. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).