1. Introduction
Hepatitis B virus (HBV) infection remains a public health problem for many low and middle-income countries (LMIC). According to the World Health Organization, 296 million people will be living with chronic hepatitis B in 2019, i.e. 3.8% of the world's population [
1,
2]. Despite the availability of an effective HBV vaccine, the number of new infections per year was estimated at 1.5 million [
1]. Africa and the West Pacific are the regions most affected by this infection. In Africa, around 82 million people are chronically infected with HBV [
1]. This infection is associated with the risk of hepatic decompensation, cirrhosis, and hepatocellular carcinoma (HCC) [
3]. It is the leading cause of cirrhosis and hepatocellular carcinoma (HCC) in sub-Saharan Africa [
4].
WHO aims to reduce the number of new cases of hepatitis B by 30% and the number of HBV-related deaths by 10% by 2020, and the number of new cases by 95% and the number of deaths by 65% by 2030, compared with the baseline year 2015 [
5]. Achieving this goal will require an ambitious increase in HBV screening and treatment activities [
6]. Ideally, these chronic carriers should be identified and medical interventions implemented to reduce the risk of premature death [
7]. In the case of chronic HBV infection, viral DNA quantification is the reference test for assessing the stage of infection, and also for treatment decision [
1]. However, access to molecular tests for DNA quantification remains problematic in resource-limited countries. HBeAg, which is a marker of wild-type virus replication and essential for the classification of HBV infection [
6,
8], has been proposed in combination with alanine aminotransferase (ALT) assay for therapeutic decision-making in resource-limited countries [
1,
9]. For HBeAg detection, immunological tests such as RDT, ELISA and ELFA are used. Due to their low cost and ease of use, RDTs are widely used in resource-limited countries. Although offering advantages, the diagnostic performances of RDTs can be influenced by factors such as low detection limits and genetic diversity [
10]. Therefore, it is necessary to evaluate these RDTs with local biological samples before their widespread use. Previous studies evaluating HBeAg RDTs have been conducted in the African setting. In Senegal, diagnostic performance evaluation of three RDTs revealed poor sensitivities ranging from 29.8% to 42.5% [
9]. In Malawi, similar results were reported for three other HBeAg RDTs (28.0% - 72.0%) [
6].
In Burkina Faso, HBV infection is endemic with an estimated prevalence of 9.1% in the general population [
11,
12]. The Ministry of Health (MoH) adopted a strategic plan to combat viral hepatitis in Burkina Faso in 2017. The strategic axes of this plan focused on screening and diagnosis, prevention and standards and protocols for the management of viral hepatitis at different levels of care. At community level and in level 1 and 2 health facilities, screening and diagnosis are based mainly on the use of RDTs. In level 3 health facilities, screening and diagnosis are based on RDTs, enzyme linked-immunosorbent assay (ELISA) and polymerase chain reaction (PCR) for HBV DNA quantification [
13].
In Burkina Faso, despite the widespread use of the SD-Bioline®HBeAg RDT for the clinical management of HBV-infected patients, no studies evaluating the test's diagnostic performance under real-life conditions have been carried out. That is why, this study was carried out to evaluate the diagnostic performance of the SD-Bioline®HBeAg RDT currently used in laboratories in Burkina Faso.
2. Methods
2.1. Study design
This was an evaluation study of a diagnostic tool that was carried out from January 2020 to October 2022. The sample panel used for this evaluation was obtained from HBsAg-positive patients received at the biomedical analysis laboratory of the "Assaut-Hépatites" Center with a duly completed examination form for the detection of HBeAg with the SD-Bioline®HBeAg rapid test. Approximately 8 mL of whole blood from each consenting patient was collected by venipuncture from the elbow. The blood sample was then centrifuged at 4000 rpm for 5 min and the serum obtained was aliquoted in 3 cryotubes. One was used directly for the SD-Bioline®HBeAg test according to the manufacturer's instructions and the second was used for the detection of HBeAg using enzyme-linked fluorescent assay (ELFA) (Gold standard) and the quantification of HBV viral DNA by real-time PCR. Sociodemographic information was collected using a structured questionnaire by the laboratory staff.
2.2. SD-Bioline®HBeAg RDT
The SD-Bioline®HBeAg rapid diagnostic test is a one-step in vitro immunochromatographic test designed for the detection of HBeAg in human serum or plasma. It is a single-step, easy-to-use test that can be stored at 2°C to 30°C. The test was performed according to the manufacturer's instructions and the Good Laboratory Practices (GLP). After removing the test device from the foil pouch, 100 µL of the collected sample was added to the sample well, and the result was interpreted after 5-20 minutes. Interpretation of the test results was based on the appearance of lines visible to the naked eye. A test was negative if the control zone line appeared in the absence of the test zone line. A positive test was characterized by the appearance of two lines, one in the test area and one in the control area. The test was invalid if the control zone line was absent. The performance characteristics of the test provided by the manufacturer were as follows: sensitivity = 95.5% (95% CI: 88.9 - 98.2%); specificity = 98.6% (95% CI: 96.6 - 99.5%).
3.3. Gold standard: VIDAS HBe/Anti-HBe (ELFA)
All samples were retested for HBeAg by the ELFA technique using the VIDAS HBe/Anti-HBe kit. The principle of the HBe test combines the enzyme immunoassay with a final detection by fluorescence. To perform the test, the MINI VIDAS was first calibrated using the standards provided in the kit: S1 (HBeAg), C1 (positive control), C2 (negative control). Then, 150 µL of vortexed serum was added to each sample well of the corresponding cartridge and the analysis was started. Results were obtained after 90 minutes. Two fluorescence measurements in the reading cuvette are performed for each assay. The first one takes into account the background due to the substrate cuvette before the substrate is brought into contact with the cone (SPR). The second reading is taken after the substrate is incubated with the enzyme present in the cone (SPR). The calculation of the relative fluorescence value (RFV) is the result of the difference of the two measurements. The index of the test is calculated by dividing the RFV of the sample or control by the RFV of the standard: index (i) = RFV of the sample / RFV of the standard S1. The result is negative if i < 0.1 and positive if i ≥ 0.1. All tests were performed according to the manufacturer's instructions.
2.4. Quantification of HBV viral DNA
Viral DNA extraction was performed on the GenoXtract® automated instrument using GXT NA extraction kits (Hain Lifescience, Nerhen, Germany). During extraction, an internal control supplied by the manufacturer was associated with each sample to validate the extraction and amplification process. To detect and quantify HBV DNA, quantitative real time polymerase chain reaction (qPCR) was performed with GENERIC HBV CHARGE VIRALE (GHBV-CV) kit (BIOCENTRIC, Bandol, France) using FluoroCycler®XT (Hain Lifescience, Nerhen, Germany) platform in a reaction volume of 20 µL. The detection limit of the technique is 95 IU/mL (1.98 log10 UI/mL). All steps were performed according to the manufacturer’s instructions and the GLP.
2.5. Statistical analysis
Data were collected using Excel and statistical analyses were performed with R software version 4.0.1. The comparison of the results of the SD-Bioline®HBeAg RDT with those of the ELFA allowed the calculation of the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) by the Maskill method with their 95% confidence intervals (CI). Agreement between the two tests was determined using Cohen’s Kappa test. These performance characteristics were also determined based on different viral DNA quantity thresholds. The significance level of the analyses was set at p ≤ 0.05.
2.6. Ethical approval
This study was approved by the institutional ethics committee of the “Institut de Recherche en Sciences de la Santé” (IRSS) (A01-2020/CEIRES du 23 janvier 2020). The participant consent was obtained for the use of the collected samples for this research.
3. Results
A total of 340 sera obtained from HBsAg positive patients were included in this evaluation. Compared to the VIDAS HBe/Anti-HBe ELFA test, the sensitivity of the SD-Bioline®HBeAg RDT was 33.3% (95% CI: 18.5 - 50.9), the specificity was 98.6% (95% CI: 96.6 - 99.6), the PPV was 75.0% (47.6 - 92.7), the NPV was 92.5% (89.1 - 95.1). The kappa value of agreement with reference test was 0.42 (95 CI: 0.25 - 0.59) (Table 2).
Table 1.
Cross-tabulation of the SD-Bioline®HBeAg RDT results with the reference test results.
Table 1.
Cross-tabulation of the SD-Bioline®HBeAg RDT results with the reference test results.
| |
Results |
VIDAS HBe/Anti-HBe |
SD-Bioline®HBeAg RDT |
|
Positive (%) |
Negative (%) |
Total |
| Positive |
12 |
4 |
16 |
| Negative |
24 |
300 |
324 |
| Total |
36 |
304 |
340 |
Table 2.
Diagnostic performances of the SD-Bioline®HBeAg RDT compared to the reference test.
Table 2.
Diagnostic performances of the SD-Bioline®HBeAg RDT compared to the reference test.
| Performance parameters |
SD-Bioline®HBeAg RDT |
| |
Estimate (%) |
95% CI |
| Se |
33.3 |
18.5 – 50.9 |
| Sp |
98.6 |
96.6 -99.6 |
| VPP |
75.0 |
47.6 – 92.7 |
| VPN |
92.5 |
89.1 – 95.1 |
| Kappa |
0.42 |
0.25 – 0.59 |
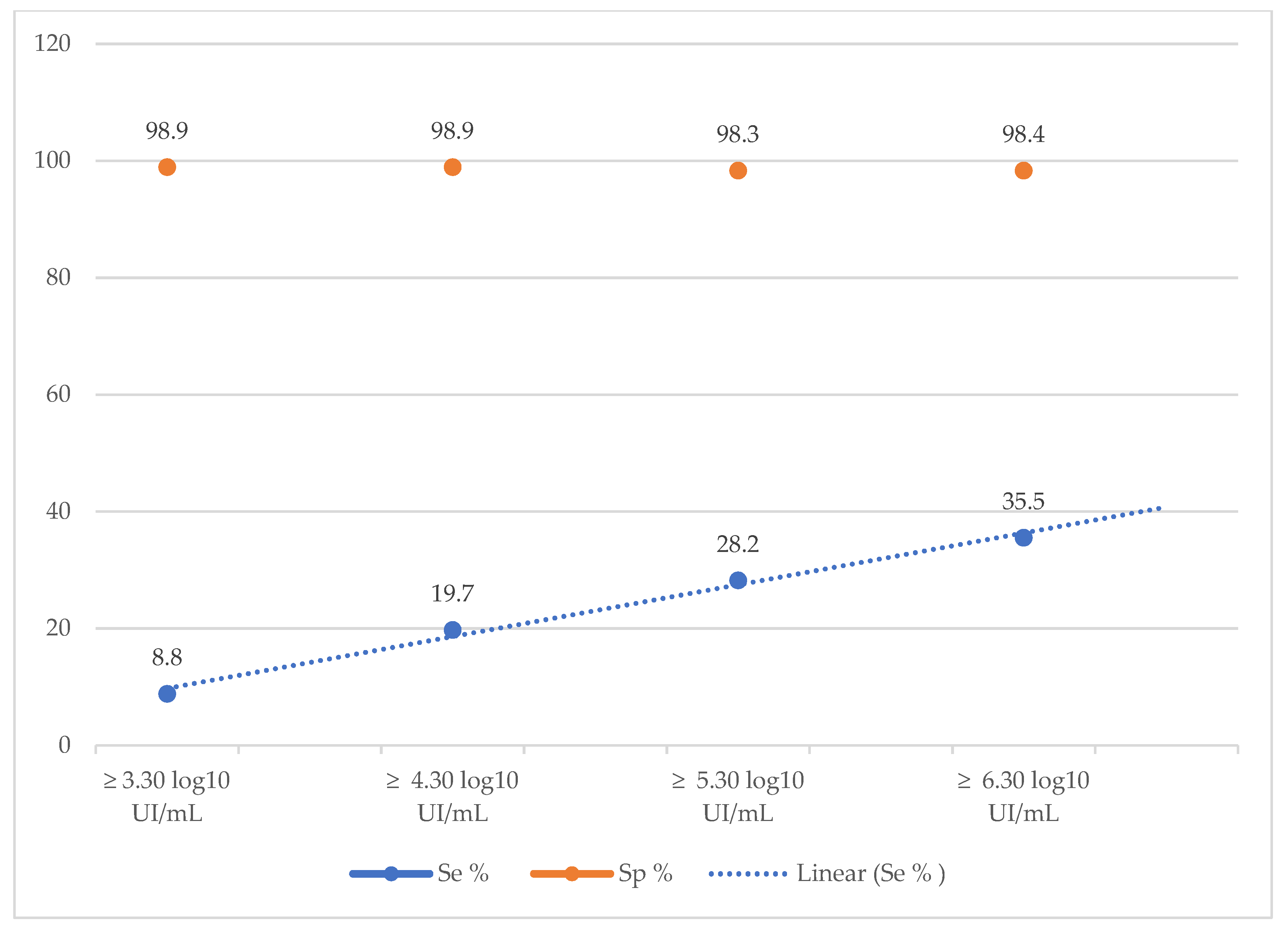
The diagnostic performance of the SD-Bioline®HBeAg RDT was also calculated according to different viral load thresholds. The sensitivity of the rapid test increased as the viral load increased, ranging from 8.8% for a VL ≥ 3.30 log10 IU/mL to 35.5% a VL ≥ 6.30 log10 IU/mL (Table 4, Figure 1). In contrast, specificity remained stable (approximately 98%) for all viral load thresholds (Table 4, Figure 1).
Figure 1.
Evolution of the sensitivity and specificity of the TDR SD Bioline®HBeAg according to the viral load.
Figure 1.
Evolution of the sensitivity and specificity of the TDR SD Bioline®HBeAg according to the viral load.
Table 3.
Cross-tabulation of the SD-Bioline®HBeAg RDT results with the viral load of HBV.
Table 3.
Cross-tabulation of the SD-Bioline®HBeAg RDT results with the viral load of HBV.
Viral load
|
|
SD-Bioline®HbeAg RDT |
| Positive |
Negative |
Total |
VL ≥ 3.3 log10 UI/mL
(VL ≥ 2 000 UI/mL)
|
Detectable |
14 |
145 |
159 |
| Undetectable |
2 |
179 |
181 |
| Total |
16 |
324 |
340 |
VL ≥ 4.3 log10 UI/mL
(VL ≥ 20 000 UI/mL) |
Detectable |
13 |
53 |
66 |
| Undetectable |
3 |
271 |
274 |
| Total |
16 |
324 |
340 |
VL ≥ 5.3 log10 UI/mL
(VL ≥ 200 000 UI/mL)
|
Detectable |
11 |
28 |
39 |
| Undetectable |
5 |
296 |
301 |
| Total |
16 |
324 |
340 |
VL ≥ 6.3 log10 UI/mL
(VL ≥ 2 000 000 UI/mL)
|
Detectable |
11 |
20 |
31 |
| Undetectable |
5 |
304 |
309 |
| Total |
16 |
324 |
340 |
Table 4.
Diagnostic performances of the SD-Bioline®HBeAg RDT compared to viral load of HBV.
Table 4.
Diagnostic performances of the SD-Bioline®HBeAg RDT compared to viral load of HBV.
| Viral load |
SD-Bioline®HBeAg RDT |
| Se |
Sp |
VPP |
VPN |
| |
Estimate (%) |
95% CI |
Estimate (%) |
95% CI |
Estimate (%) |
95% CI |
Estimate (%) |
95% CI |
VL ≥ 3.3 log10 UI/mL
(VL ≥ 2 000 UI/mL) |
8.8 |
4.9 – 14.3 |
98.9 |
96.1 – 99.9 |
87.5 |
61.6 – 98.4 |
55.2 |
49.6 – 60.7 |
VL ≥ 4.3 log10 UI/mL
(VL ≥ 20 000 UI/mL) |
19.7 |
10.9 – 31.3 |
98.9 |
96.7 – 99.7 |
81.3 |
54.3 – 95.9 |
83.6 |
79.1 – 87.7 |
VL ≥ 5.3 log10 UI/mL
(VL ≥ 200 000 UI/mL) |
28.2 |
15.0 – 44.9 |
98.3 |
96.2 – 99.4 |
68.8 |
41.3 – 89.0 |
91.4 |
87.7 – 94.2 |
VL ≥ 6.3 log10 UI/mL
(VL ≥ 2 000 000 UI/mL) |
35.5 |
19.2 – 54.6 |
98.4 |
96.3 – 99.5 |
68.8 |
41.3 – 89.0 |
93.8 |
90.2 – 95.6 |
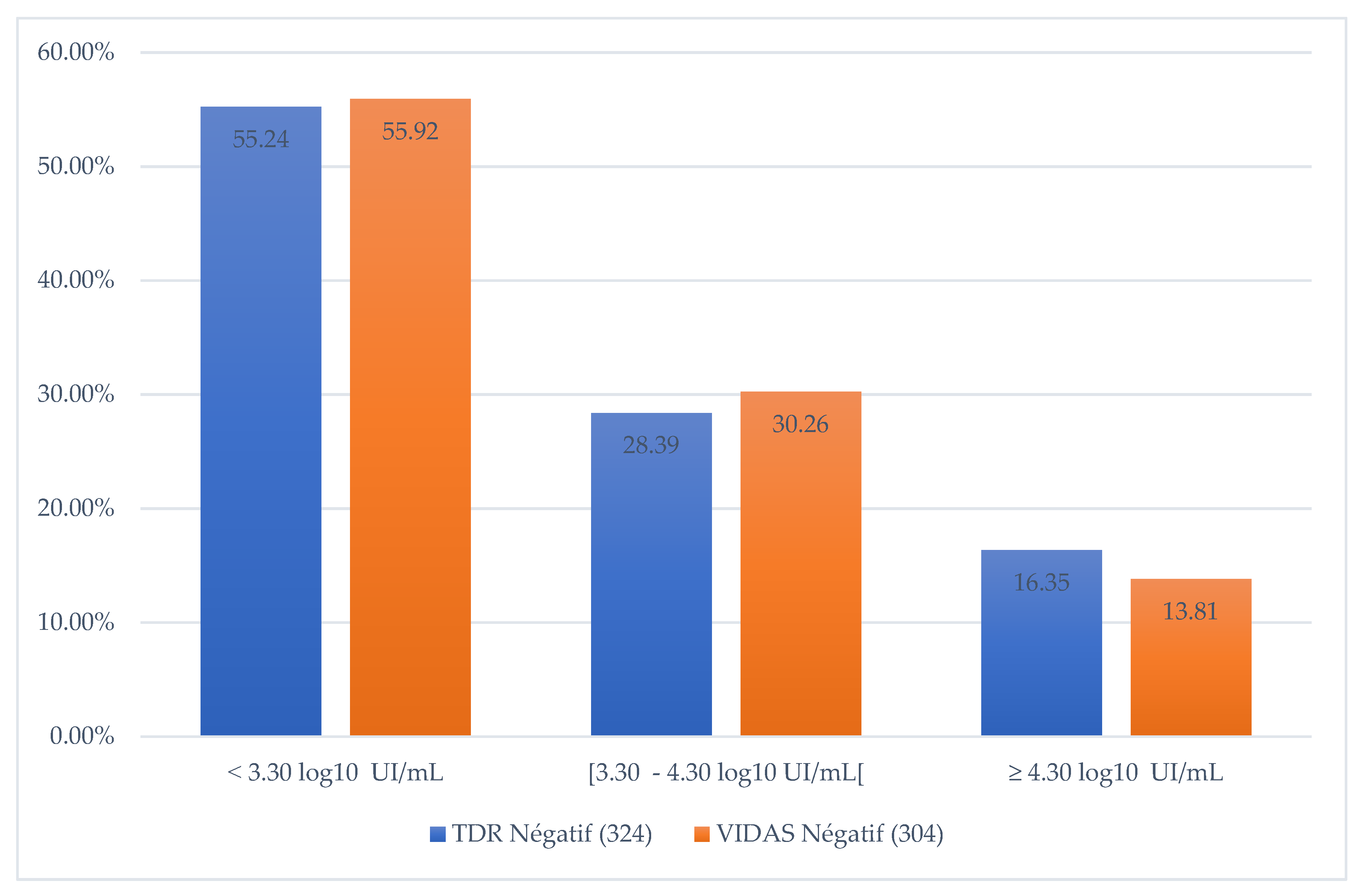
In addition, when comparing the VIDAS HBe/Anti-HBe ELFA test results with VL, we noted that 55.92% of VIDAS test results were negative when VL < 3.30 log10 IU/mL, 30.3% when VL [ 3.3 - 4.3 log10 IU/mL] and 13.8% for VL > 4.3 log10 IU/mL (Figure 2).
Figure 2.
Proportion of negative RDTs according to the quantity of DNA detected.
Figure 2.
Proportion of negative RDTs according to the quantity of DNA detected.
4. Discussion
This study evaluated the diagnostic performance of the SD-Bioline®HBeAg RDT compared to VIDAS HBe/Anti-HBe ELFA test as gold standard. We found that the Bioline®HBeAg SD RDT had a low sensitivity (33.3%) and a high specificity (98.6%). The sensitivity does not reach those provided by the manufacturer (95.5%) indicating that this RDT should be used with caution for the clinical management of HBV infected patients. Similar sensitivities of 28.0 % and 29.8% for Bioline®HBeAg SD RDT has been reported respectively by studies conducted in Malawi [
6] and Senegal [
9]. Low sensitivity of HBeAg RDTs appears to be a widespread problem. Indeed, evaluations of the diagnostic performance of other RDTs used for HBeAg detection other than SD-Bioline have reported low sensitivities. These include the HBeAg Rapid Test (Biopanda Reagents) and the HBeAg serum rapid test (Creative Diagnostics) for which sensitivities of 53.2% and 72.3% were observed respectively [
6]. The same is true for the Insight HBeAg (Tulip Diagnostics Ltd., Goa, India) and OneStep HBeAg (AMSUK Ltd., Antrim, UK) tests for which sensitivities of 31.1% and 42.5% were found [
9].
The coefficient kappa observed in our study was 0.42. According to the criteria of Landis and Koch [
14], the agreement between the SD-Bioline®HBeAg RDT and the VIDAS HBe/Anti-HBe ELFA test was moderate. Indeed, the Landis and Koch criteria stipulate that: Kappa < 0, no agreement; 0 < kappa ≤ 0.2; slight agreement; 0.2 < kappa < 0.4, fair agreement; 0.4 < kappa < 0.6, moderate agreement; 0.6 < kappa < 0.8, substantial agreement; 0.8 < kappa < 1, near perfect agreement.
Moreover, the diagnostic performance of the SD-Bioline®HBeAg RDT was also calculated according to different viral load thresholds. We noted an increase of the sensitivity as the viral load increased, ranging from 8.8% for a VL ≥ 3.30 log
10 IU/mL to 35.5% a VL ≥ 6.30 log
10 IU/mL. Similar finding was reported by a Cambodian study where they noted an increase of sensitivities from 76.5% to 89.3% for VL > 5.30 log
10 IU/mL and > 7.30 log
10 IU/mL respectively [
15]. This observation highlights the low analytical sensitivity of HBeAg RDT, reflecting its poor ability to be correlated with HBV replication. This low sensitivity could therefore prevent patients with chronic hepatitis B who are eligible for treatment from accessing it. The main consequence would be to prevent the infection from progressing to the complications of cirrhosis and cancer.
The HBV genome is a partially circularized DNA, composed of four overlapping and shifting reading frames (ORFs): Pol (P), Core (C), Surface (S) and X. These ORFs encode the seven HBV proteins and also code for four promoter regions that initiate transcription and two enhancers that promote gene transcription [
16]. Given its genomic organization, any mutation occurring in a specific region of the genome can potentially have an impact on other regions, thus affecting the viral cycle. HBeAg is a protein encoded from the pre-C transcription of the open reading frame of the HBV nucleus, so a mutation in this area could affect HBeAg synthesis [
17]. Indeed, pre-C/C mutations are most commonly found in HBeAg-negative patients with detectable viral loads [
18]. Among these mutations, we have the G1896A substitution, which leads to the appearance of a premature stop codon and the arrest of HBeAg translation, without impact on viral replication [
18]. For a wild-type virus, the absence of HBeAg in the serum of an HBV-infected individual would correlate with a lack of viral replication. However, we found that 13.8% of HBeAg-negative sera with VIDAS HBe/Anti-HBe had a VL ≥ 4.30 log10 IU/mL (VL ≥ 20 000 IU/mL). This observation could be explained by the presence of pre-C/C mutations in the HBV genome that impact HBeAg production [
19].
Our study has some limitations. The reference test used (VIDAS HBe/Anti-HBe ELFA) did not allow the quantification of HBeAg. This could allow a better appreciation of the analytical sensitivity of the RDT by determining the detection threshold. Furthermore, the sequencing of the Pre-Core region was not performed. This would give an idea of the prevalence of mutations, the type of mutations, and their impact on the performance of the diagnostic tests.
5. Conclusion
To our knowledge, this is the first study to evaluate the diagnostic performance of an RDT for the detection of HBeAg in Burkina Faso. The results showed a low Se of the SD-Bioline®HBeAg test indicating that its use is inappropriate for the clinical management of HBV-infected patients. These results and other evaluation studies highlight the urgent need to develop HBeAg rapid tests with better sensitivities. This would facilitate access to quality diagnostic tools for low-income populations and, consequently, better control of HBV infection.
Author’s Contributions
AMS, AD and MKG conceived and designed the study. AD, MNGO and AMS performed laboratory investigations, acquired and curated the data. AKI undertook analysis and interpretation of data. AD and AMS wrote the original draft of the manuscript. AMS, AD, MNGO, AKI, DNZ, BDL, DI and MKG reviewed and edited the final manuscript. All authors approved the final manuscript.
Funding Information
This work was supported by the Non-Governmental Organization “Assaut-Hépatites”.
Data Availibility Statement
Data generated during this study are available from the corresponding author on reasonable request.
Aknowlegments
The authors thank the study participants. We also thank the medical staff for their active participation in the patient enrollment and sample collection phases. Finally, we thank our colleagues for their critical review of this article.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- OMS. Interim Guidance for Country Validation of Viral Hepatitis Elimination. TECHNICAL REPORT. 2021;96.
- Word Health Organization. Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021. 2021. https://www.who.int/publications-detail-redirect/9789240027077.
- Chang ML, Liaw YF. Hepatitis B Flare in Hepatitis B e Antigen-Negative Patients: A Complicated Cascade of Innate and Adaptive Immune Responses. IJMS. 2022 Jan 28;23(3):1552. https://www.mdpi.com/1422-0067/23/3/1552.
- Lemoine M, Thursz MR. Battlefield against hepatitis B infection and HCC in Africa. Journal of Hepatology. 2017 Mar;66(3):645–54. [CrossRef]
- Sheena BS, Hiebert L, Han H, Ippolito H, Abbasi-Kangevari M, Abbasi-Kangevari Z, et al. Global, regional, and national burden of hepatitis B, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. The Lancet Gastroenterology & Hepatology . 2022 Sep;7(9):796–829. [CrossRef]
- Stockdale AJ, Silungwe NM, Shawa IT, Kreuels B, Gordon MA, Geretti AM. Diagnostic performance evaluation of hepatitis B e antigen rapid diagnostic tests in Malawi. BMC Infect Dis. 2021 Dec;21(1):487. [CrossRef]
- Clement F, Dewint P, Leroux-Roels G. Evaluation of a New Rapid Test for the Combined Detection of Hepatitis B Virus Surface Antigen and Hepatitis B Virus e Antigen. J Clin Microbiol. 2002 Dec;40(12):4603–6. [CrossRef]
- Stoeckl L, Funk A, Kopitzki A, Brandenburg B, Oess S, Will H, et al. Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proceedings of the National Academy of Sciences. 2006 Apr 25;103(17):6730–4. [CrossRef]
- Maylin S, Akbar SMF, Funk AL, Bercion R, Mishiro S, Ndiaye B, et al. Poor Sensitivity of Commercial Rapid Diagnostic Tests for Hepatitis B e Antigen in Senegal, West Africa. The American Journal of Tropical Medicine and Hygiene. 2018 Aug 2;99(2):428–34. [CrossRef]
- Sanou AM, Toyé R, Kagoné T, Nikiéma A, Testa J, Sakandé J, et al. Analytical performance of eight rapid point-of-care tests routinely used for the detection of HBsAg in Burkina Faso: A cross-sectional study. Journal of Clinical Virology. 2020 Aug;129:104546. [CrossRef]
- Meda N, Tuaillon E, Kania D, Tiendrebeogo A, Pisoni A, Zida S, et al. Hepatitis B and C virus seroprevalence, Burkina Faso: a cross-sectional study. Bull World Health Organ. 2018 Nov 1;96(11):750–9. [CrossRef]
- Lingani M, Akita T, Ouoba S, Sanou AM, Sugiyama A, Tarnagda Z, et al. High prevalence of hepatitis B infections in Burkina Faso (1996–2017): a systematic review with meta-analysis of epidemiological studies. BMC Public Health. 2018 Dec;18(1):551. [CrossRef]
- Ministère de la santé BF. Normes et protocoles de prise en charge des hépatites virales au BURKINA FASO. 2019. http://www.scgecm.com/download/Normes%20et%20protocoles%20de%20Prise%20en%20charge%20HV%20au%20BF%20Vf%202019.pdf.
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977 Mar;33(1):159–74.
- Ségéral O, S. N’Diaye D, Prak S, Nouhin J, Chhun S, Khamduang W, et al. Usefulness of a serial algorithm of HBsAg and HBeAg rapid diagnosis tests to detect pregnant women at risk of HBV mother-to-child transmission in Cambodia, the ANRS 12328 pilot study. Journal of Clinical Virology. 2018 Dec;109:29–34. [CrossRef]
- Valaydon ZS, Locarnini SA. The virological aspects of hepatitis B. Best Practice & Research Clinical Gastroenterology. 2017 Jun;31(3):257–64. [CrossRef]
- Kramvis A, Arakawa K, Yu MC, Nogueira R, Stram DO, Kew MC. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J Med Virol. 2008 Jan;80(1):27–46. [CrossRef]
- Besharat S, Poustchi H, Mohamadkhani A, Katoonizadeh A, Moradi A, Roshandel G, et al. Association of Mutations in the Basal Core Promoter and Pre-core Regions of the Hepatitis B Viral Genome and Longitudinal Changes in HBV Level in HBeAg Negative Individuals: Results From a Cohort Study in Northern Iran. Hepat Mon. 2015 Feb 21;15(2). [CrossRef]
- Pivert A, Servant-Delmas A, Lunel-Fabiani F, Le Guillou-Guillemette H, Laperche S, Ducancelle A. Correlation between the promoter basal core and precore mutations and HBsAg quantification in French blood donors infected with hepatitis B virus: HBV in French Blood Donors. J Med Virol. 2015 Mar;87(3):529–35. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).