1. Introduction
Antibiotic-resistant infections have become a major global health concern, with a staggering prevalence of at least 2.8 million cases and over 35,000 annual deaths reported in the United States alone by the Centers for Disease Control and Prevention (CDC) [
1,
2]. The development of antibiotic resistance is attributed to several factors, including bacterial biofilm formation, enzymatic inactivation of antimicrobials, and alterations in target proteins [
3,
4,
5]. As traditional antibiotics face mounting challenges in combating drug-resistant pathogens, there is a growing interest in exploring alternative therapeutic approaches [
6,
7,
8,
9,
10,
11,
12].
Amid these challenges, antimicrobial peptides (AMPs) have garnered significant interest as promising alternatives to conventional antibiotics for their multifunctional properties [
13]. Their rapid killing mechanism and reduced likelihood of resistance development make them particularly attractive candidates. AMPs possess key features, such as amphipathicity and net cationicity [
12,
14,
15,
16], which enable them to target anionic cell membranes and invade bacterial cytoplasm, leading to membrane permeabilization or intracellular attack, ultimately culminating in cell death [
17,
18,
19,
20,
21,
22,
23]. Beyond their antimicrobial properties, AMPs also exhibit the ability to inhibit biofilm formation and/or eradicate existing biofilms, further adding to their potential efficacy. Moreover, AMPs demonstrate immunomodulatory effects, highlighting their multifunctional nature in the fight against drug-resistant bacteria. These diverse properties render AMPs highly attractive for the development of effective antimicrobial therapeutics capable of circumventing antibiotic resistance.
One major class of AMPs, known as cathelicidins, has been identified in numerous species, including humans, sheep, cows, pigs, rabbits, and other vertebrates [
24,
25]. Typically, cathelicidins comprise a highly conserved N-terminal cathelin portion and an extremely variable cationic domain at the C-terminus, which plays a pivotal role in their antimicrobial function [
18,
26,
27,
28,
29]. Bovine myeloid antimicrobial peptide-27 (BMAP-27), a cathelicidin peptide, adopts an α-helical structure consisting of 27 residues with an amidated C-terminus. Despite its broad-spectrum antimicrobial activity against various pathogens, the application of BMAP-27 has been hampered by its cytotoxic effects on host cells [
18,
24]. Recently, we demonstrated that BMAP-18, a truncated version consisting of the N-terminal 18 residues of BMAP-27, possesses an amphipathic α-helical structure with clustered aromatic residues on the hydrophobic face, resulting in reduced hemolysis and improved cell selectivity compared to the full-length BMAP-27 [
18].
In this study, we synthesized BMAP-18-FL by replacing the aromatic phenylalanine residues at four positions (F3, F6, F10, F14) of BMAP-18 with aliphatic leucine residues and compared the antibacterial, antibiofilm, and anti-inflammatory properties of aromatic BMAP-18 with its aliphatic analogue, BMAP-18-FL. By highlighting the multiple functionalities of both aromatic BMAP-18 and aliphatic BMAP-18-FL, this study aims to provide significant insights into the development of promising peptides as potential alternatives to conventional antibiotics. Through a comprehensive understanding of the multifunctional properties of AMPs, we can pave the way for the development of novel, effective therapies to combat the growing threat of antimicrobial resistance.
2. Results
2.1. Peptide design and characterization
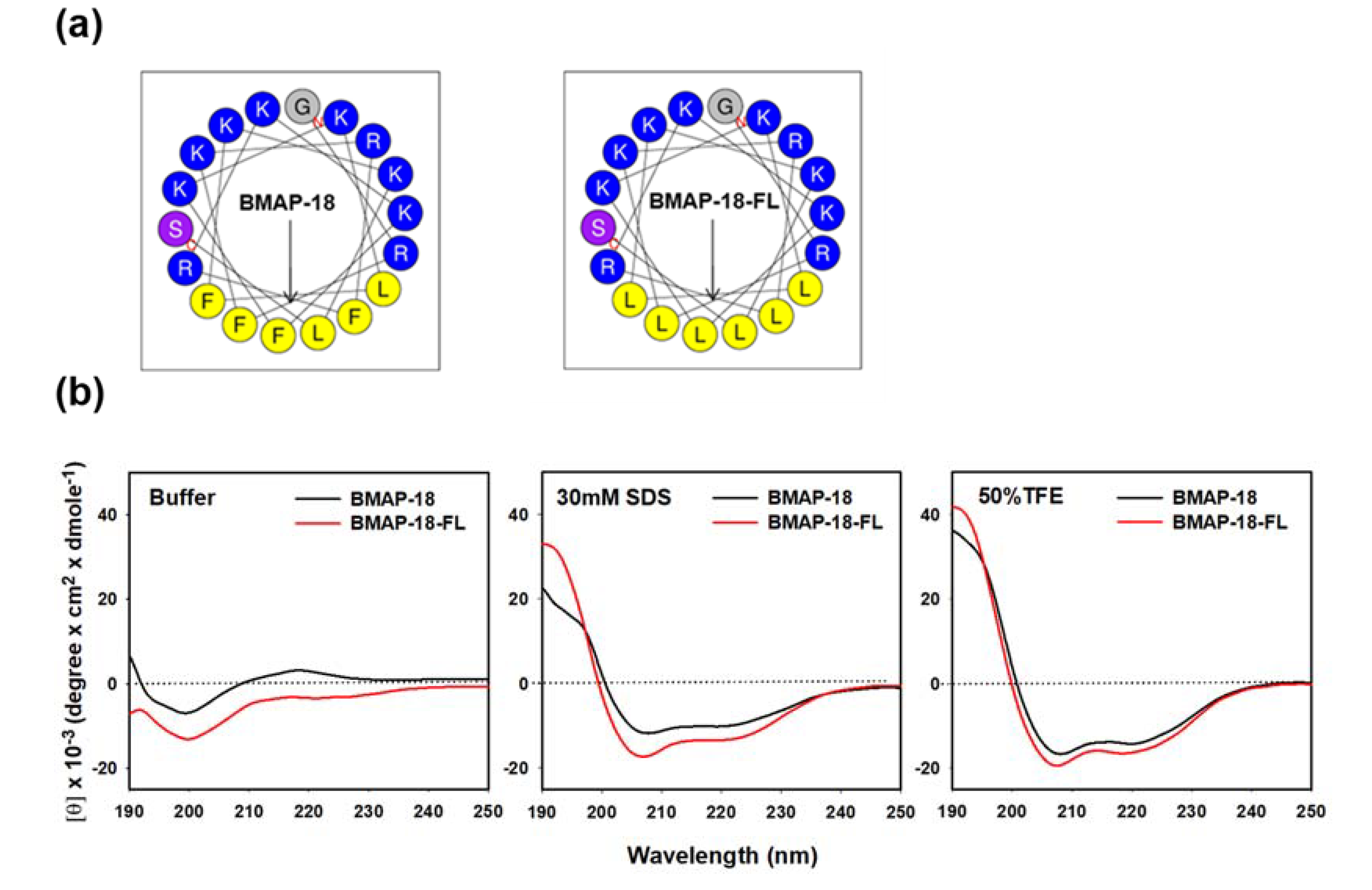
Table 1 presents the amino acid sequences, retention time, molecular mass, ion charge ratio, net charge, and hydrophobic moment of BMAP-18 and its aliphatic analogue, BMAP-18-FL. In BMAP-18-FL, the aromatic amino acid phenylalanine (F) residues at positions F3, F6, F10, and F14 are replaced by the aliphatic amino acid leucine (L) residues. The value found from mass spectroscopy was almost identical to the theoretical value, demonstrating the accuracy of the synthetic peptides (
Figure S1). The analytical RP-HPLC retention time of BMAP-18 and BMAP-18-FL was found 18.1 min and 20.2 min respectively. The α-helical wheel diagram of BMAP-18 and BMAP-18-FL were predicted using the online analysis tool, HeliQuest server (
https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) (
Figure 1a). The results showed that both AMPs carried a net charge of +10 with hydrophobic moment of 0.710 and 0.693, respectively (
Table 1).
The secondary structure of the peptides was assessed using CD spectroscopy in various environments: 10mM sodium phosphate buffer (mimicking an aqueous condition), 30mM SDS (mimicking a membrane), and 50% TFE (mimicking a hydrophobic environment) (
Figure 1b). In the aqueous buffer solution, both BMAP-18 and BMAP-18-FL displayed a random distribution of secondary structures. However, upon exposure to SDS or TFE solutions, both peptides exhibited characteristic double-negative maxima at 208 nm and 222 nm, implying that they predominantly adopted an α-helical conformation. These findings suggest significant structural alterations during the transition from the aqueous environment to SDS or TFE conditions. Notably, BMAP-18-FL exhibited a higher degree of helicity compared to its aromatic counterpart, BMAP-18, in both the SDS and TFE environments.
2.2. Antimicrobial activity
For the antimicrobial activity assay, we used two resistant strains: Gram-positive Methicillin-resistant
Staphylococcus aureus (CCARM 3090) and Gram-negative multidrug-resistant
Pseudomonas aeruginosa (CCARM 2095). Using the microdilution method, we assessed the minimum inhibitory concentrations (MICs) of BMAP-18, BMAP-18-FL and some conventional antibiotics (Ciprofloxacin, oxacillin, and tetracycline). As a positive control, we included bee venom melittin for comparison. The MIC values of all tested compounds are listed in
Table 2. Both BMAP-18 and BMAP-18-FL exhibited similar antibacterial activity, with MIC values ranging from 16 to 32 µM, while the conventional antibiotics did not show any activity, even at higher concentrations (up to 1024 µM). These findings highlight the potential of the peptides as promising antimicrobial agents, particularly against the resistant strains tested.
2.3. Mechanism of antibacterial action
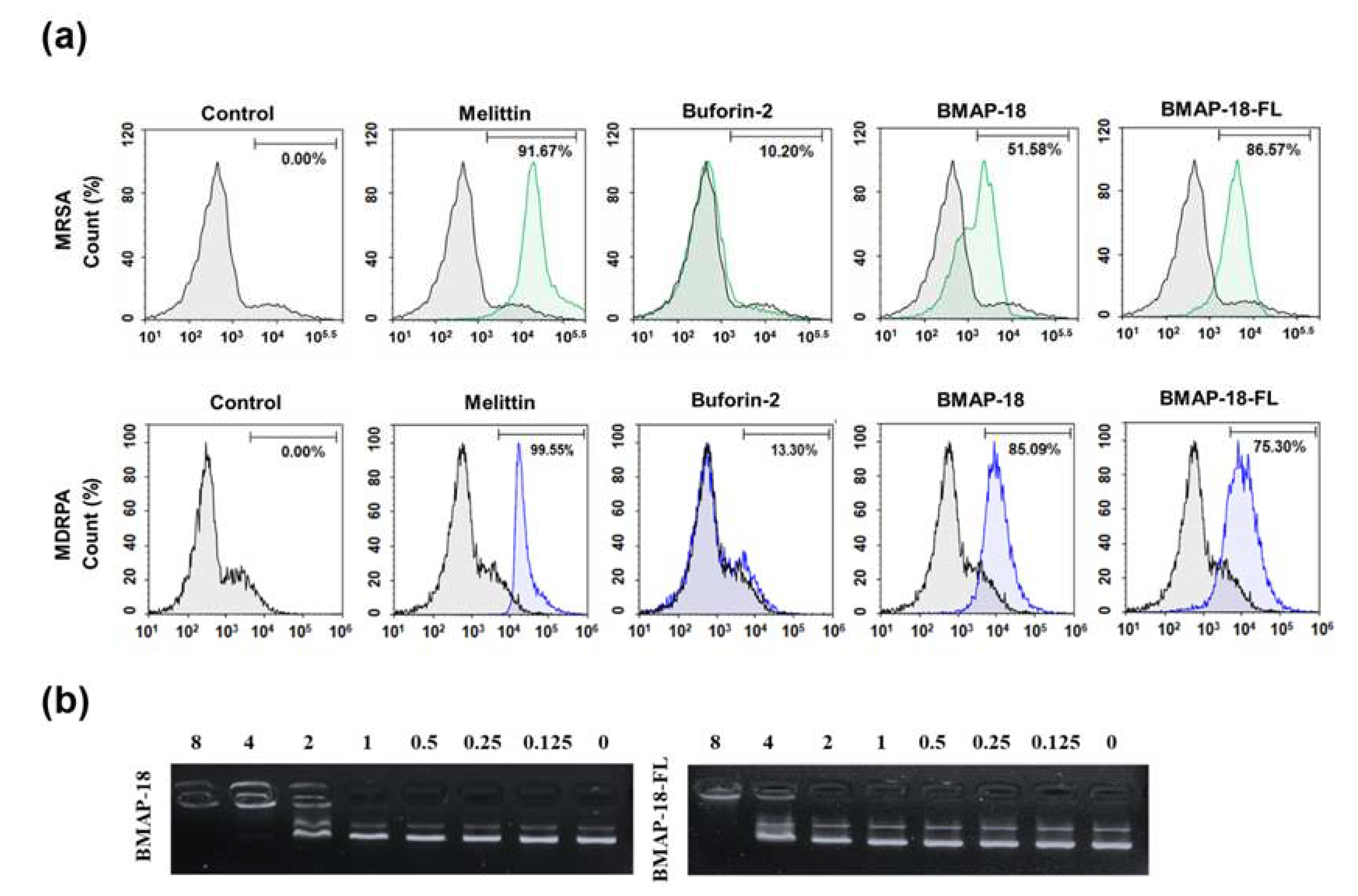
Mechanistic investigations were carried out using the PI uptake assay to assess the membrane disruption caused by the peptides (
Figure 2a). Both resistant bacteria (MRSA and MDRPA) were stained with PI, a fluorescent dye that binds to nucleic acids in cells. For comparison, we included melittin and buforin-2, known as a membrane-lytic peptide and intracellular non-lytic peptide, respectively. The FACScan analysis data indicated that in the absence of any peptide, the bacterial cells remained intact, whereas the membrane were damaged in presence of peptides. Interestingly BMAP-18 exhibited a greater shifting effect against the Gram-negative MDRPA (85%) than the Gram-positive MRSA (51%), indicating its strong membrane-targeting capability against the Gram-negative bacteria. On the other hand, BMAP-18-FL demonstrated a more pronounced membrane-destructive effect against the Gram-positive MRSA (86%) than the Gram-negative MDRPA (75%). These results provide compelling evidence of the membrane-targeting properties of the peptides, enabling them to disrupt the integrity of bacterial cell membranes and potentially contribute to their antimicrobial activity against the resistant strains. The differential effects on Gram-positive and Gram-negative bacteria further highlight the potential of these peptides as promising candidates for combating antibiotic-resistant pathogens.
To further elucidate the mechanism of action of the peptides we performed the SYTOX green uptake assay of
S. aureus (KCTC 1621). SYTOX Green is known for its high affinity to DNA binding, allowing us to assess the membrane permeability of
S. aureus in the presence of peptides (
Figure S2). In comparison to melittin, both BMAP-18 and BMAP-18-FL showed no significant SYTOX green influx, similar to buforin-2. To confirm the intracellular target mechanism, we performed DNA binding assay of the peptides to
E. coli bacterial plasmid pBR322 by 1% agarose gel electrophoresis (
Figure 2b). Various concentrations of the peptides (ranging from 0 to 8 µM) were employed, and the electrophoretic mobility was detected using a UV illuminator. BMAP-18 exhibited binding up to 2 µM, while BMAP-18-FL showed binding up to 8 µM. This result suggests that the aromatic BMAP-18 had a higher affinity for intracellular DNA than the aliphatic BMAP-18-FL, even though both peptides have intracellular mechanisms of action.
2.4. Antibiofilm activity
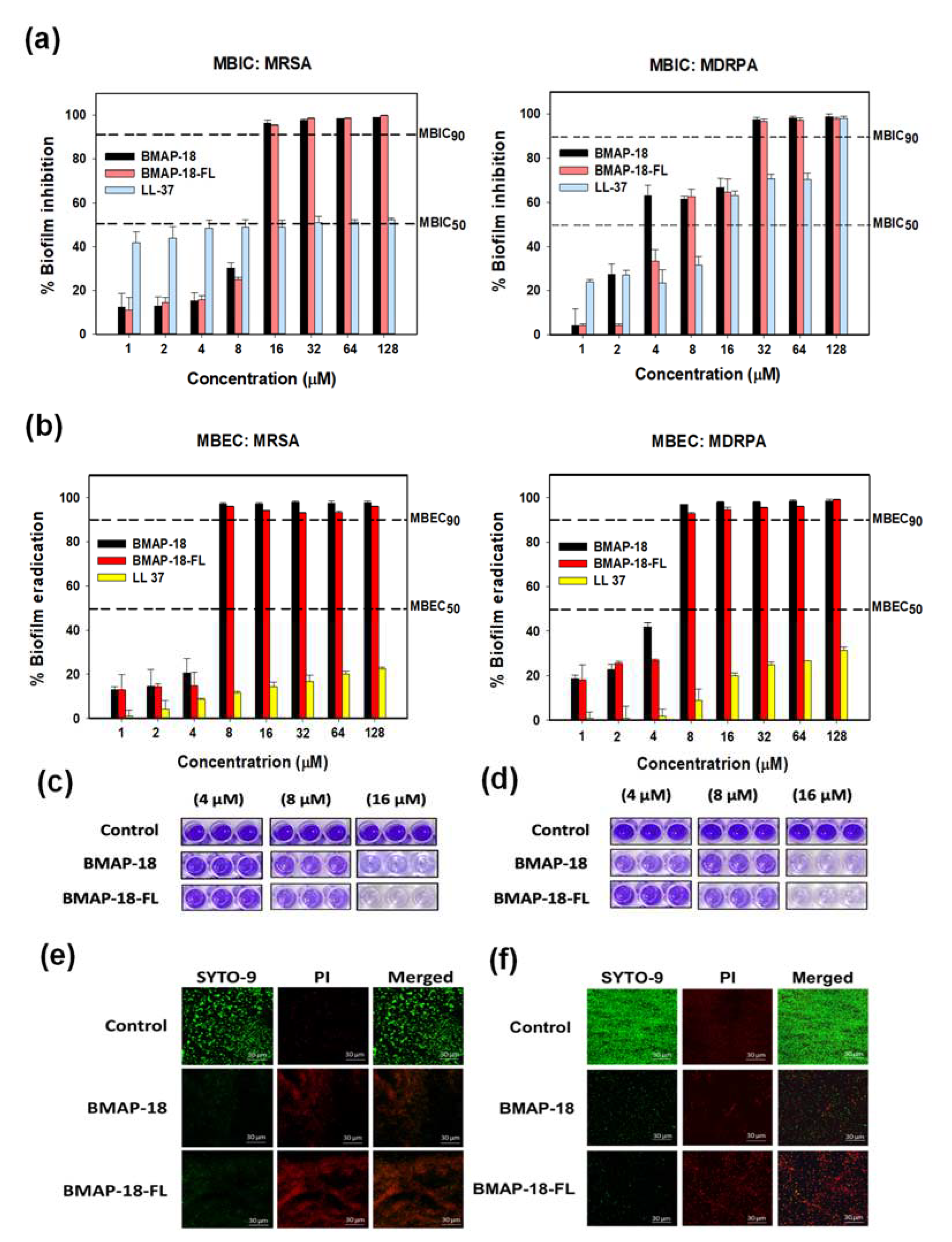
As biofilm formation often leads to antibiotic resistance, we evaluated the antibiofilm activity of BMAP-18 and BMAP-18-FL, comparing LL-37 as a positive control (
Figure 3). We measured the in vitro anti-biofilm action through the minimum biofilm inhibition concentration (
Figure 3a) and minimum biofilm eradication concentration (
Figure 3b). Both BMAP-18 and BMAP-18-FL demonstrated strong inhibition of biofilm formation and eradication of mature biofilms against MRSA and MDRPA strains. Moreover, our peptide analogues effectively inhibited up to 90% of biofilm formation starting at 16 µM (equivalent to 1 MIC). Remarkably, they showed 90% MBEC at a low concentration of 8 µM against both strains. Overall, Both BMAP-18 and BMAP-18-FL exhibited superior biofilm eradication compared to the control peptide LL-37.
Furthermore, we visually confirmed the biofilm eradication by staining with crystal violet (
Figure 3c and 3d) as well as confocal laser-scanning microscopy (CLSM) with a viability staining kit (
Figure 3e and 3f). Biofilm-forming bacterial cells treated with peptides, dried and stained with 0.1% crystal violet. Images showed the concentration-dependent peptide effect visually on mature biofilms of resistant strains. In case of CLSM, mature biofilms were incubated in presence or absence of peptides and then stained with live/dead bacterial staining kit (SYTO-9/PI). Live cells were labeled with a green fluorescence dye (SYTO-9), while dead cells were identified using a red dye (PI). The higher number of dead cells confirmed the eradication activity of our synthesized peptides against the resistant bacteria (MRSA, MDRPA).
2.5. Cytotoxic activity of peptides
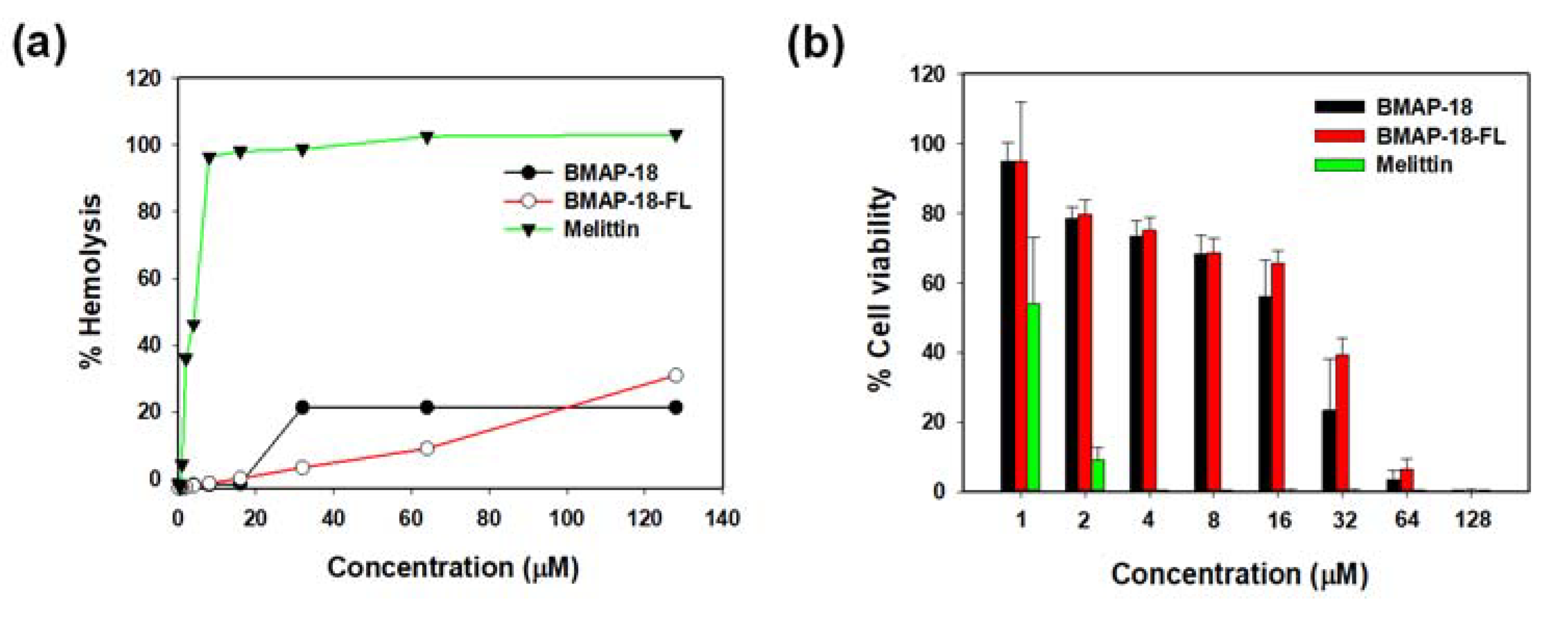
To evaluate the toxicity of BMAP-18 and BMAP-18-FL, we conducted hemolysis assays using sheep red blood cells (
Figure 4a) and examined their cytotoxic effects on mouse macrophage RAW 264.7 cells using the MTT dye reduction assay (
Figure 4b). Both peptides did not induce hemolytic activity below 16 µM. At a concentration of 64 µM, BMAP-18-FL caused less than 10% hemolysis, while BMAP-18 induced about 20% hemolysis (
Figure 4a). In the RAW 264.7 cells, both peptides showed a similar pattern, with over 70% cell survival (
Figure 4b). Compared to melittin, both peptides demonstrated significantly lower hemolysis of sheep red blood cells and lower cytotoxicity against RAW 264.7 cells, suggesting their potential as safer candidates for further exploration as antimicrobial agents.
2.6. Anti-inflammatory activity
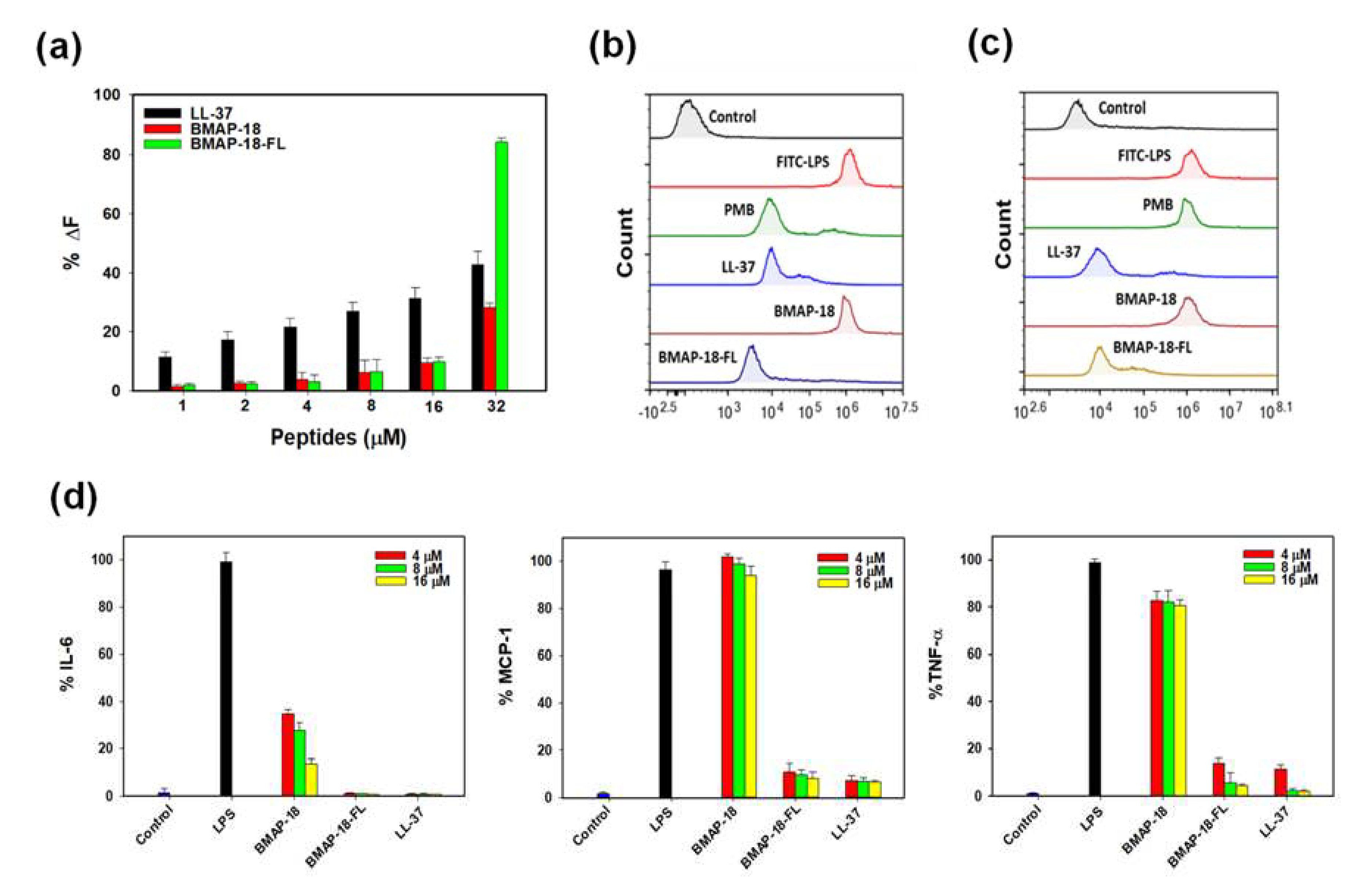
To explore the anti-inflammatory activity of BMAP-18 and BMAP-18-FL, we conducted the LPS binding assay using the BODIPY-TR cadaverine (BC) displacement assay to investigate whether their inhibitory mechanism against cytokines in LPS-stimulated RAW 264.7 cells is due to direct neutralization of LPS. In this assay, BC fluorescence is quenched upon binding to free LPS. However, the introduction of AMPs displaces BC, resulting in alleviation of quenching, and thus an increase in fluorescence, indicating successful binding of the compounds with LPS. As illustrated in
Figure 5a, all peptides exhibited binding in a dose-dependent manner, with BMAP-18-FL demonstrating stronger LPS-binding affinity compared to BMAP-18 or the control peptide (LL-37).
To gain insights into the mechanism of action of the peptides in macrophage cells, we pursued two approaches. First, we examined the effects of peptides on free LPS by pre-incubating the FITC-LPS/peptide mixture and then adding it to RAW 264.7 cells. As depicted in
Figure 5b, BMAP-18-FL significantly suppressed the binding of FITC-LPS to macrophage cells, indicating successful peptide-LPS binding compared to its parent analogue. Furthermore, we evaluated the neutralizing ability of peptides on receptor-bound LPS in RAW 264.7 cells. As shown in
Figure 5c, similar to LL-37, BAMP-18-FL effectively bound to receptor-bound LPS, suggesting antagonistic activity towards LPS-binding receptors (CD14 or TLR4) by neutralizing LPS. These findings were validated by Polymyxin B (PMB) as it cannot neutralize receptor-bound LPS, while LL-37 showed the opposite pattern.
We also assessed the effect of BAMP-18 peptides on the expression of proinflammatory cytokines (IL-6, MCP-1, and TNF-α) in LPS-stimulated RAW 264.7 cells using ELISA. The transcriptional expression levels of these cytokines decreased upon peptide treatment in a dose-dependent manner (
Figure 5d). However, BMAP-18 could not inhibit MCP-1 expression in LPS-stimulated RAW264.7 cells, whereas BMAP-18-FL showed excellent inhibitory activities against all these cytokines. Even at the lowest concentration of 4 µM, BMAP-18-FL demonstrated significantly lower expression (≤10%) of cytokines, similar to the anti-inflammatory peptide LL-37.
3. Discussion
Antimicrobial peptides (AMPs) are an important part of the immune system and have shown promise as a first line of defense against invading pathogens [
30,
31]. With the increasing resistance of antibiotics, AMPs have emerged as promising candidates due to their ability to target a wide range of microorganisms, while also having lower susceptibility to developing resistance [
17,
30,
32]. To enhance their antimicrobial activity, AMPs require an adequate positive charge for electrostatic interactions with the bacterial membrane [
30]. Additionally, a hydrophobic domain is crucial for membrane permeabilization, allowing the peptides to disrupt and penetrate the bacterial membrane effectively [
33,
34,
35]. In this study, we synthesized the aromatic BMAP-18 and its aliphatic analogue, BMAP-18-FL to compare their antimicrobial, antibiofilm, and anti-inflammatory activities.
Previous studies have demonstrated the significance of aromatic residues, such as phenylalanine, in AMPs, as they play a crucial role in membrane disruption and antimicrobial activity owing to their hydrophobic nature and their ability to interact with bacterial membranes [
36]. Conversely, aliphatic residues, such as leucine, may exhibit different interactions with the membrane. Considering the escalating threat of antibiotic-resistant infections caused by MRSA and MDRPA, we assessed the MIC values of our synthesized BMAP-18 and BMAP-18-FL against these two resistant pathogens. Despite the variation in amino acid composition, our findings reveal that both BMAP-18 and BMAP-18-FL exhibited comparable MIC results, ranging from 16 to 32 µM (
Table 2). The outcomes from flow cytometry analysis (
Figure 2a), SYTOX green uptake assay (
Figure S2), and DNA binding assay (
Figure 2b) suggest that both peptides not only disrupt the bacterial membrane but also engage in an intracellular antimicrobial mechanism. However, BMAP-18 demonstrated a stronger membrane-targeting capability against the Gram-negative MDRPA (85%) compared to the Gram-positive MRSA (51%), indicating its preference for Gram-negative bacteria. Moreover, BMAP-18 exhibited a higher affinity for intracellular DNA compared to BMAP-18-FL. Conversely, BMAP-18-FL showed a more pronounced membrane-destructive effect against the Gram-positive MRSA (86%) than the Gram-negative MDRPA (75%), suggesting its preference for Gram-positive bacteria. Although there were some distinctions between BMAP-18 and BMAP-18-FL in terms of membrane permeabilization and DNA binding affinity, both peptides displayed similar antimicrobial activity, primarily due to the shared mechanism involving membrane permeabilization combined with intracellular targeting. Interestingly, both peptides also exhibited strong antibiofilm activity against pre-formed biofilms of MRSA and MDRPA, further emphasizing their potential as alternative therapeutic agents against antibiotic-resistant infections. The ability to disrupt and prevent biofilm formation is crucial in combating bacterial resistance, as biofilms provide a protective environment that enables bacteria to evade conventional antibiotics. While the antimicrobial and antibiofilm activities of aromatic BMAP-18 and aliphatic BMAP-18-FL were found to be comparable, a notable difference emerged in their anti-inflammatory properties. BMAP-18-FL exhibited superior anti-inflammatory activity compared to BMAP-18, as demonstrated by its significant reduction in the expression levels of pro-inflammatory cytokines.
Several studies have demonstrated that Lipopolysaccharide (LPS), crucial component of Gram-negative bacteria, actively controls the membrane insertion and antibacterial properties of numerous AMPs [
37,
38,
39]. As the binding capacity of AMPs to LPS is considered a potential therapeutic target, we looked at different anti-inflammatory effects associated with LPS against our target peptides to assess its potential as a treatment for septic shock. The aliphatic BMAP-18-FL exhibited direct binding to LPS and demonstrated a strong neutralizing effect on LPS-conjugated macrophages. Initially, we evaluated the direct binding to LPS with BODIPY-TR cadaverine (BC) displacement assay and found the dislocation of dye commenced at 1 µM (
Figure 5a). Notably, BMAP-18-FL showed over two-fold (>80%) stronger binding to LPS than BMAP-18 at higher concentrations. To investigate the mechanism of LPS-binding, we studied the effects of our peptides with FITC-labeled LPS on macrophage cells through flow cytometry analysis. As depicted in
Figure 5b and 5c, the aliphatic BMAP-18-FL significantly bound to both free LPS and receptor (CD-14) bound LPS. Our data fully complied with former studies on LL-37 that binds only with receptor bound FITC-LPS [
37,
40,
41,
42,
43]. Previous reports indicate that Toll-like receptor-4 (TLR-4), a transmembrane protein, interacts with the LPS-CD14 complex to initiate intracellular signaling, subsequently leads to the production and release of pro-inflammatory cytokines, ultimately contributing to septic shock [
37,
38]. Based on the receptor binding evidence obtained from flow cytometry, we sought to evaluate the inhibitory effect of our peptides on pro-inflammatory cytokines: Interleukin-6 (IL-6), monocyte chemoattractant protein-1 (MCP-1), and Tumor necrosis factor-alpha (TNF-α). Our findings revealed a remarkable reduction in the levels of these cytokines with BMAP-18-FL treatment, even at lower concentrations such as 4 µM (
Figure 5d). Collectively, our results suggest that BMAP-18-FL works on LPS by competitively binding to the CD-14 receptor initially and successively binds to free LPS, resulting in a significant inhibition of pro-inflammatory cytokines. These findings hold promise for the potential use of BMAP-18-FL as an effective anti-inflammatory agent in addition to its antimicrobial properties.
In conclusion, our study investigated the antimicrobial, antibiofilm, and anti-inflammatory properties of aromatic BMAP-18 and aliphatic BMAP-18-FL. Both peptides demonstrated potent antimicrobial activity against Gram-positive MRSA and Gram-negative MDRPA. Importantly, these peptides exhibited relatively low cytotoxicity, making them promising candidates for combating antibiotic-resistant infections. Mechanistic analysis provided valuable insights into the mode of action of both BMAP-18 and BMAP-18-FL, inducing membrane permeability and leading to intracellular targeting, which contributed to their potent antimicrobial activity. Additionally, both peptides effectively prevented biofilm formation and eradicated existing biofilms of MRSA and MDRPA. Notably, our study uncovered a distinct advantage of BMAP-18-FL over BMAP-18, as the aliphatic analogue exhibited superior anti-inflammatory activity. It significantly reduced the expression levels of pro-inflammatory cytokines in LPS-stimulated macrophages, suggesting its potential as a valuable anti-inflammatory agent alongside its antimicrobial properties. Overall, this study underscores the significance of understanding the similarities and differences between aromatic BMAP-18 and aliphatic BMAP-18-FL in their antimicrobial, antibiofilm, and anti-inflammatory properties. These multifunctional attributes of both peptides offer promising prospects for the development of novel antimicrobial therapies capable of effectively combating drug-resistant bacteria.
4. Materials and Methods
4.1. Materials
The solid phase peptide synthesis (SPPS) materials, including the rink amide-methylbenzhydrylamine (MBHA) resin and the 9-fluorenyl-methoxycarbonyl (Fmoc) protected amino acids, were bought from Novabiochem (California, USA). The following substances were provided by Sigma Aldrich (St. Louis, MO, USA): Lipopolysaccharide (LPS) purified from Escherichia coli O111:B4, FITC-labeled LPS (E. coli O111:B4), 2,2,2-triflluoroethanol (TFE), sodium dodecyl sulphate (SDS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT), propidium iodide (PI). From R&D Systems (Minneapolis, MN, USA), ELISA kits for TNF-α, IL-6, and MCP-1 were purchased. Invitrogen live/dead bacterial viability kits, SYTOX green and E. coli bacterial plasmid pBR322 were purchased from Thermo Fisher Scientific, South Korea. Mouse macrophage RAW 264.7 cells were bought from the American Type Culture Collection (Manassas, VA). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were acquired from Lonza (USA) and Atlas biologicals (Korea) respectively. Two resistant bacteria of this study: Methicillin-resistant Staphylococcus aureus strains (CCARM 3090) and multidrug-resistant Pseudomonas aeruginosa strains (CCARM 2095) were obtained from the Culture Collection of Antibiotic-Resistant Microbes (CCARM) of Seoul Women's University in Korea. Other strains were obtained from the Korean Collection for Type Cultures (KCTC)
4.2. Peptide synthesis
BMAP-18 (GRFKRFRKKFKKLFKKLS) and BMAP-18-FL (GRLKRLRKKLKKLLKKLS) were synthesized by solid-phase synthesis method using Fmoc (9-fluorenyl-methoxycarbonyl) chemistry (0.06 mmol scale) on Rink amide 4-methyl benzyldrylamin (MBHA) resin. Both the peptides were purified with RP-HPLC (Shimadzu, Japan) on an analytical Vydac C18 column (length: 250mm, internal diameter: 20 mm, particle size: 15 μm, pore size: 300 Å) which was monitored by UV detector at 224 nm. The purified products were determined by LC-MS analysis (API2000, AB SIEX, USA).
4.3. Circular dichroism spectroscopy
The secondary structure of peptides was detected in different solutions in a J-715 CD spectropolarimeter (Jasco, Japan) equipped with a 1 mm path length cell. CD spectra was monitored in 10 mM sodium phosphate buffer (pH 7.4), 30mM SDS micelles and 50% TFE between 190 nm to 250 nm wavelength at 25°C. The results from three scans were collected and calculated for each peptide. The CD data were expressed as the mean residue ellipticity [θ] in degree square centimeter per decimole (deg.cm2/dmol).
4.4. Antimicrobial activity
According to the guidelines of clinical and laboratory standard institute (CLSI), antimicrobial activity of the peptides against resistant bacteria was determined [
44]. Minimum inhibitory concentrations (MICs) were measured by using microtiter broth dilution method. All strains were cultured overnight to stationary phase at 37°C. The overnight cultures were 10-fold diluted in Muller-Hinton (MH) broth (Difco,USA) and incubated for a few hours to attain mid-log phase growth. The mid-log phase cultures were diluted with MHB media and added to sterile 96-well plates having two-fold serially diluted peptides and antibiotics. The plate was incubated for 18-24 hours at 37°C and MIC was detected as the lowest concentration of peptide that prevented visible turbidity. The MIC was determined by optical density (OD) at 600 nm using a microplate ELISA (Bio-Tek Instruments EL800, USA) reader.
4.5. Antibiofilm activity
Antibiofilm properties of the peptides were evaluated by measuring minimum biofilm inhibition concentration (MBIC) and minimum biofilm eradication concentration (MBEC) against MRSA (CCARM 3090) and MDRPA (CCARM 2095). Both experiments followed previously described methods [
45,
46]. For biofilm eradication, these bacteria (~10
6 CFU/mL) were incubated at 37 °C for 24 h, in 96-well Calgary microtiter plates containing PEG, in absence or presence of serially diluted peptides. After the incubation, lids were washed with PBS to remove free bacteria and then air-dried. Later fixation was done by methanol and dried wells were stained with 100 μl of 0.1% crystal violet for 5 min. After discarding the stain, 95% ethanol was added in wells and kept in a shaker for 30 min. Finally, the absorbance was measured at 600 nm.
4.6. Confocal laser-scanning microscopy
MRSA (CCARM 3090) and MDRPA (CCARM 2095) were cultured (~ 106 cells/well) in 6-well plates with coverslips in MHB-glucose media to form biofilms for 48 h followed by peptide treatment for another 24 h. After that, cells were washed with PBS and a staining probe mixture (LIVE/DEAD™ BacLight™ Bacterial Viability kit; Invitrogen) was used according to the manufacturer’s instructions. The two nucleic acid stains of the kit, SYTO-9 and PI were added to the cells and incubated in the dark for 30 min. The cells were then washed to remove the stain and fixation was done by methanol. Later, the coverslips were dried and prepared in glass slide. Biofilm mass in coverslips were visualized using a Confocal laser-scanning microscopy (Zeiss Microscopy, Jena, Germany) and analyzed by ZEN 2009 Light edition software (Zeiss Microscopy, Jena, Germany).
4.7. FACScan analysis
The integrity of bacterial cell membrane was evaluated by measuring the propidium iodide (PI) uptake by flow cytometry. In short, mid-log phase MRSA and MDPRA bacterial cells were diluted to until the OD600 absorbance was approximately 0.5. The mixture was prepared by adding same volume of cell suspension and PBS (1x) and centrifuged for 5mins at 8000 rpm. After discarding PBS, cell pellets were resuspended in PBS. Then 10 µl PI was added and incubated for 15 mins. 1 MIC of peptides were added to the mixture followed by incubation for another 15 min. Finally, PI fluorescence was quantified using a FACScan instrument (Agilent, USA).
4.8. DNA binding assay
An agarose gel-retardation assay was carried out to determine the binding of peptides with DNA as described previously [
40]. Different concentrations of peptides were mixed with 100 ng of bacterial plasmid pBR322 in a binding buffer (10 mM Tris-HCl, 5% glucose, 50 mg/mL BSA, 1 mM EDTA, and 20 mM KCl). Then the mixture of DNA and peptide was incubated at 37°C for 1 h followed by 1% agarose gel electrophoresis in 0.5% TBE buffer. Bands were detected using UV illuminator (Bio-Rad, USA).
4.9. Hemolytic activity
The hemolytic activity of the peptides was determined by measuring free hemoglobin of sheep red blood cells (sRBCs). Fresh sRBCs were washed with PBS, centrifuged at 4°C and resuspended in PBS to make a dilution of erythrocytes (4% v/v). 100 μl of sRBCs were suspended to sterilized 96 well plate with 100 μl serially diluted peptides then incubated for 1 h at 37°C. Centrifugation was carried out for 5min at 1000 RCF and the supernatant was transferred to a new 96 well clean plate. Using a microplate ELISA reader (Bio-Tek Instruments EL800, USA) hemoglobin release was recorded by measuring absorbance at 405 nm. Melittin was used as a reference peptide whereas 0.1% Triton X-100 was used for inducing 100% hemolysis. Zero hemolysis was calculated using PBS only. The percentage of hemolytic activity was calculated by the following formula: % Hemolysis = [(Abs in peptide solution- Abs in PBS]/ (Abs of 0.1% Triton X-100 - Abs of PBS)] x 100.
4.10. Cytotoxicity
MTT dye reduction assay was carried out against RAW 264.7 cells to determine cytotoxicity of the peptides. Briefly, the cells were seeded in 96 well plates and incubated for 18-24 h in the presence 5% CO2 at 37°C. Peptides were added in varied concentrations to the cells and incubated for 24 h followed by the addition of 20 µl MTT. After 3 h of incubation, formed formazan crystals were dissolved in 200 µl DMSO and absorbance was measured at 550 nm for cell viability calculation.
4.11. LPS binding assay
By using BODIPY-TR cadaverine (BC) (Sigma, USA) displacement assay, the ability of peptides to bind with LPS was analyzed as in previous studies [
47]. LPS from
E. coli 0111: B4 (25 µg/mL) was incubated with BC (2.5 µg/mL) in a quartz cuvette with 50 mM Tris buffer (pH 7.4). Peptides were added by dose dependent manner up to 32 µM within 60 s of time interval and the fluorescence was recorded at an excitation wavelength of 580 nm and an emission wavelength of 620 nm with a Spectrofluorophotometer (Shimadzu, Japan). The values were converted to %∆F using the following equation: %∆F = [(F
obs – F
0) / (F
100 -F
0)] x 100, where F
obs is the observed fluorescence at a given peptide concentration, F
0 is the initial fluorescence of BC with LPS in absence of peptide, and F
100 is the fluorescence of BC with LPS in presence of polymyxin B which is used as positive control.
4.12. Effect of peptides on LPS binding
The ability of peptides to bind with FITC-LPS was evaluated as a previously described method [
37]. First, FITC-LPS (1 µg/mL) from
E. coli 0111: B4 was incubated with peptides at 4°C for 1 h, then RAW264.7 macrophage cells (3 x10
5 cells/mL) were treated with FITC-LPS/peptide mixture and incubated for 30 min at 37°C. After that cells were washed with PBS to remove unbound LPS and the binding affinity was measured by fluorescence intensity from a FACScan machine (Agilent, USA). Whether the peptides affect receptor bound LPS, we assessed another flow cytometry analysis. In brief, RAW264.7 cells were pre-incubated with FITC-LPS for 1 h in absence of peptides. After washing with PBS, the cells were incubated with peptides for 1 h followed by further washing. Finally, the binding of FITC-LPS to RAW264.7 cells was analyzed by flow cytometry. In both experiments, LPS binding inhibitor PMB (Polymyxin B) and LL-37 were used as control. Background fluorescence was assessed by using RAW 264.7 cells incubated without FITC-LPS or peptides.
4.13. Evaluation of pro-inflammatory cytokines and chemokines
To examine whether the tested peptides were able to block the inflammatory responses induced by LPS, mouse macrophage RAW 264.7 cells were stimulated with LPS (20 ng/ml) with or without peptides. Untreated cells and only LPS stimulated cells were considered as controls. After incubation, the inhibitory effect of pro-inflammatory cytokines (IL-6, MCP-1 and TNF-α) was measured by ELISA KIT according to manufacturer’s (R&D Systems) guidelines. All samples were quantified in triplets.
4.14. Statistical analysis
All the graphs and statistical analysis was carried out using one-way analysis of variance (ANOVA) with SPSS 16.0 software and Sigma plot v12.0. The data in this study was expressed as the means ± standard deviation (SD) from three independent experiments.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, Figure S1: ESI-MS spectra of BMAP-18 and BMAP-18-FL; Figure S2: SYTOX green uptake caused by peptides.
Author Contributions
Conceptualization, S.-H.S. and S.Y.; methodology, I.J. and S.D.K; validation, I.J., S.Y.S, C.W.L and S.Y.; investigation, I.J. and S.Y.; writing—original draft preparation, I.J.; writing—review and editing, S.-H.S. and S.Y.; supervision, S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2023-00210292 and No. 2022R1F1A1068595) and the Ministry of Education (No. 2018R1D1A1B07042620).
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable.
Data Availability Statement
We encourage all authors of articles published in MDPI journals to share their research data. In this section, please provide details regarding where data supporting reported results can be found, including links to publicly archived datasets analyzed or generated during the study. Where no new data were created, or where data is unavailable due to privacy or ethical restrictions, a statement is still required. Suggested Data Availability Statements are available in section “MDPI Research Data Policies” at
https://www.mdpi.com/ethics.
Acknowledgments
We thank Eun Young Kim, Chosun University, South Korea, for assistance with the evaluation of pro-inflammatory cytokines.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hunter, P. A war of attrition against antibiotic resistance: current strategies try to keep antibiotic resistance at bay and further encourage research to produce genuinely novel antibacterials. EMBO reports 2020, 21, e50807. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Nagel, J.L.; Kaye, K.S.; LaPlante, K.L.; Albin, O.R.; Pogue, J.M. Antimicrobial stewardship and the infection control practitioner: A natural alliance. Infectious Disease Clinics 2021, 35, 771–787. [Google Scholar] [PubMed]
- Zhong, C.; Zhu, N.; Zhu, Y.; Liu, T.; Gou, S.; Xie, J.; Yao, J.; Ni, J. Antimicrobial peptides conjugated with fatty acids on the side chain of D-amino acid promises antimicrobial potency against multidrug-resistant bacteria. European Journal of Pharmaceutical Sciences 2020, 141, 105123. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, J.; De La Fuente-Núñez, C.; Wang, Z.; Hancock, R.E.; Roberts, C.R.; Ma, J.; Li, J.; Haapasalo, M.; Wang, Q. Experimental and theoretical investigation of multispecies oral biofilm resistance to chlorhexidine treatment. Scientific reports 2016, 6, 27537. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed research international 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Lau, Q.Y.; Ng, F.M.; Cheong, J.W.D.; Yap, Y.Y.A.; Tan, Y.Y.F.; Jureen, R.; Hill, J.; Chia, C.S.B. Discovery of an ultra-short linear antibacterial tetrapeptide with anti-MRSA activity from a structure–activity relationship study. European journal of medicinal chemistry 2015, 105, 138–144. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nature biotechnology 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Current opinion in pharmacology 2006, 6, 468–472. [Google Scholar] [CrossRef]
- Zhang, L.; Falla, T.J. Antimicrobial peptides: therapeutic potential. Expert opinion on pharmacotherapy 2006, 7, 653–663. [Google Scholar] [CrossRef]
- Brogden, N.K.; Brogden, K.A. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? International journal of antimicrobial agents 2011, 38, 217–225. [Google Scholar] [CrossRef]
- Yeung, A.T.; Gellatly, S.L.; Hancock, R.E. Multifunctional cationic host defence peptides and their clinical applications. Cellular and Molecular Life Sciences 2011, 68, 2161–2176. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: form follows function. Nature reviews Drug discovery 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Chen, H.; Yuan, X.; Tian, J.; Dong, N.; Feng, X.; Shan, A. Designing double-site lipidated peptide amphiphiles as potent antimicrobial biomaterials to combat multidrug-resistant bacteria. Frontiers in Microbiology 2022, 13, 1074359. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, C.; Ren, B.; Yin, X.; Shim, S.H.; Gao, Y.; Zhu, J.; Zhao, P.; Liu, C.; Yu, R. Two optimized antimicrobial peptides with therapeutic potential for clinical antibiotic-resistant Staphylococcus aureus. European Journal of Medicinal Chemistry 2019, 183, 111686. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Chen, L.; Guang, H.; Li, Z.; Yang, H.; Li, J.; You, D.; Yu, H.; Lai, R. Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PLoS One 2011, 6, e22120. [Google Scholar] [CrossRef]
- Chen, W.; Yang, B.; Zhou, H.; Sun, L.; Dou, J.; Qian, H.; Huang, W.; Mei, Y.; Han, J. Structure–activity relationships of a snake cathelicidin-related peptide, BF-15. Peptides 2011, 32, 2497–2503. [Google Scholar] [CrossRef]
- Seefeldt, A.C.; Graf, M.; Pérébaskine, N.; Nguyen, F.; Arenz, S.; Mardirossian, M.; Scocchi, M.; Wilson, D.N.; Innis, C.A. Structure of the mammalian antimicrobial peptide Bac7 (1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Research 2016, 44, 2429–2438. [Google Scholar] [CrossRef]
- Yang, S.; Lee, C.W.; Kim, H.J.; Jung, H.-H.; Kim, J.I.; Shin, S.Y.; Shin, S.-H. Structural analysis and mode of action of BMAP-27, a cathelicidin-derived antimicrobial peptide. Peptides 2019, 118, 170106. [Google Scholar] [CrossRef]
- Melo, M.N.; Ferre, R.; Castanho, M.A. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nature Reviews Microbiology 2009, 7, 245–250. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends in biotechnology 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacological reviews 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Peptide Science: Original Research on Biomolecules 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochimica et Biophysica Acta (BBA)-Biomembranes 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Kim, Y.-C.; Nan, Y.H.; Shin, S.Y. Cell selectivity, mechanism of action and LPS-neutralizing activity of bovine myeloid antimicrobial peptide-18 (BMAP-18) and its analogs. Peptides 2011, 32, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Frontiers in microbiology 2020, 11, 2559. [Google Scholar] [CrossRef]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: family of antimicrobial peptides. A review. Molecular biology reports 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M. The role of cathelicidins in the innate host defenses of mammals. Current issues in molecular biology 2005, 7, 179–196. [Google Scholar]
- Nizet, V.; Ohtake, T.; Lauth, X.; Trowbridge, J.; Rudisill, J.; Dorschner, R.A.; Pestonjamasp, V.; Piraino, J.; Huttner, K.; Gallo, R.L. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 2001, 414, 454–457. [Google Scholar] [CrossRef]
- Zhang, L.-j.; Guerrero-Juarez, C.F.; Hata, T.; Bapat, S.P.; Ramos, R.; Plikus, M.V.; Gallo, R.L. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 2015, 347, 67–71. [Google Scholar] [CrossRef]
- Zhang, L.; Rozek, A.; Hancock, R.E. Interaction of cationic antimicrobial peptides with model membranes. Journal of Biological Chemistry 2001, 276, 35714–35722. [Google Scholar] [CrossRef]
- Juba, M.L.; Porter, D.K.; Williams, E.H.; Rodriguez, C.A.; Barksdale, S.M.; Bishop, B.M. Helical cationic antimicrobial peptide length and its impact on membrane disruption. Biochimica et Biophysica Acta (BBA)-Biomembranes 2015, 1848, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Thompson, Z.; Yu, Z.; Cowan, J. Cu-ATCUN derivatives of Sub5 exhibit enhanced antimicrobial activity via multiple modes of action. ACS Chemical Biology 2019, 14, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lim, S.I.; Shin, S.H.; Lim, Y.; Koh, J.W.; Yang, S. Conjugation of cell-penetrating peptides to antimicrobial peptides enhances antibacterial activity. ACS omega 2019, 4, 15694–15701. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.T.; Shin, S.Y.; Shin, S.H. The central PXXP motif is crucial for PMAP-23 translocation across the lipid bilayer. International Journal of Molecular Sciences 2021, 22, 9752. [Google Scholar] [CrossRef]
- Lee, H.; Shin, S.H.; Yang, S. Rationally designed PMAP-23 derivatives with enhanced bactericidal and anticancer activity based on the molecular mechanism of peptide–membrane interactions. Amino Acids 2023, 1–10. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, M.; Lai, R.; Zhang, Z. Chemical modifications to increase the therapeutic potential of antimicrobial peptides. Peptides 2021, 146, 170666. [Google Scholar] [CrossRef]
- Rosenfeld, Y.; Papo, N.; Shai, Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides: peptide properties and plausible modes of action. Journal of Biological Chemistry 2006, 281, 1636–1643. [Google Scholar] [CrossRef]
- Shang, D.; Zhang, Q.; Dong, W.; Liang, H.; Bi, X. The effects of LPS on the activity of Trp-containing antimicrobial peptides against Gram-negative bacteria and endotoxin neutralization. Acta Biomaterialia 2016, 33, 153–165. [Google Scholar] [CrossRef]
- Avitabile, C.; Netti, F.; Orefice, G.; Palmieri, M.; Nocerino, N.; Malgieri, G.; D'Andrea, L.D.; Capparelli, R.; Fattorusso, R.; Romanelli, A. Design, structural and functional characterization of a Temporin-1b analog active against Gram-negative bacteria. Biochimica et Biophysica Acta (BBA)-General Subjects 2013, 1830, 3767–3775. [Google Scholar] [CrossRef]
- Kumar, S.D.; Shin, S.Y. Antimicrobial and anti-inflammatory activities of short dodecapeptides derived from duck cathelicidin: Plausible mechanism of bactericidal action and endotoxin neutralization. European journal of medicinal chemistry 2020, 204, 112580. [Google Scholar] [CrossRef]
- Jantaruk, P.; Roytrakul, S.; Sitthisak, S.; Kunthalert, D. Potential role of an antimicrobial peptide, KLK in inhibiting lipopolysaccharide-induced macrophage inflammation. Plos One 2017, 12, e0183852. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, I.; Hirota, S.; Niyonsaba, F.; Hirata, M.; Adachi, Y.; Tamura, H.; Tanaka, S.; Heumann, D. Augmentation of the lipopolysaccharide-neutralizing activities of human cathelicidin CAP18/LL-37-derived antimicrobial peptides by replacement with hydrophobic and cationic amino acid residues. Clinical and Vaccine Immunology 2002, 9, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.M.; Inácio, R.G.; Raimundo, J.M.; Martins, M.; Castanho, M.A.; Santos, N.C. Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Peptide Science 2012, 98, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Edition, A.S.-T. CLSI document M07-A10 [2015]. Clinical and Laboratory Standards Institute 950.
- Basak, A.; Abouelhassan, Y.; Zuo, R.; Yousaf, H.; Ding, Y.; Huigens, R.W. Antimicrobial peptide-inspired NH125 analogues: bacterial and fungal biofilm-eradicating agents and rapid killers of MRSA persisters. Organic & biomolecular chemistry 2017, 15, 5503–5512. [Google Scholar]
- Harrison, J.J.; Stremick, C.A.; Turner, R.J.; Allan, N.D.; Olson, M.E.; Ceri, H. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nature protocols 2010, 5, 1236–1254. [Google Scholar] [CrossRef]
- Wood, S.J.; Miller, K.A.; David, S.A. Anti-endotoxin agents. 1. Development of a fluorescent probe displacement method optimized for the rapid identification of lipopolysaccharide-binding agents. Combinatorial chemistry & high throughput screening 2004, 7, 239–249. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).