Submitted:
31 July 2023
Posted:
02 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. PCR Amplification Efficiency Using Different Types of DNA Polymerases and dUTPs C5-Modified with Bulky Aromatic Hydrocarbon Substituents
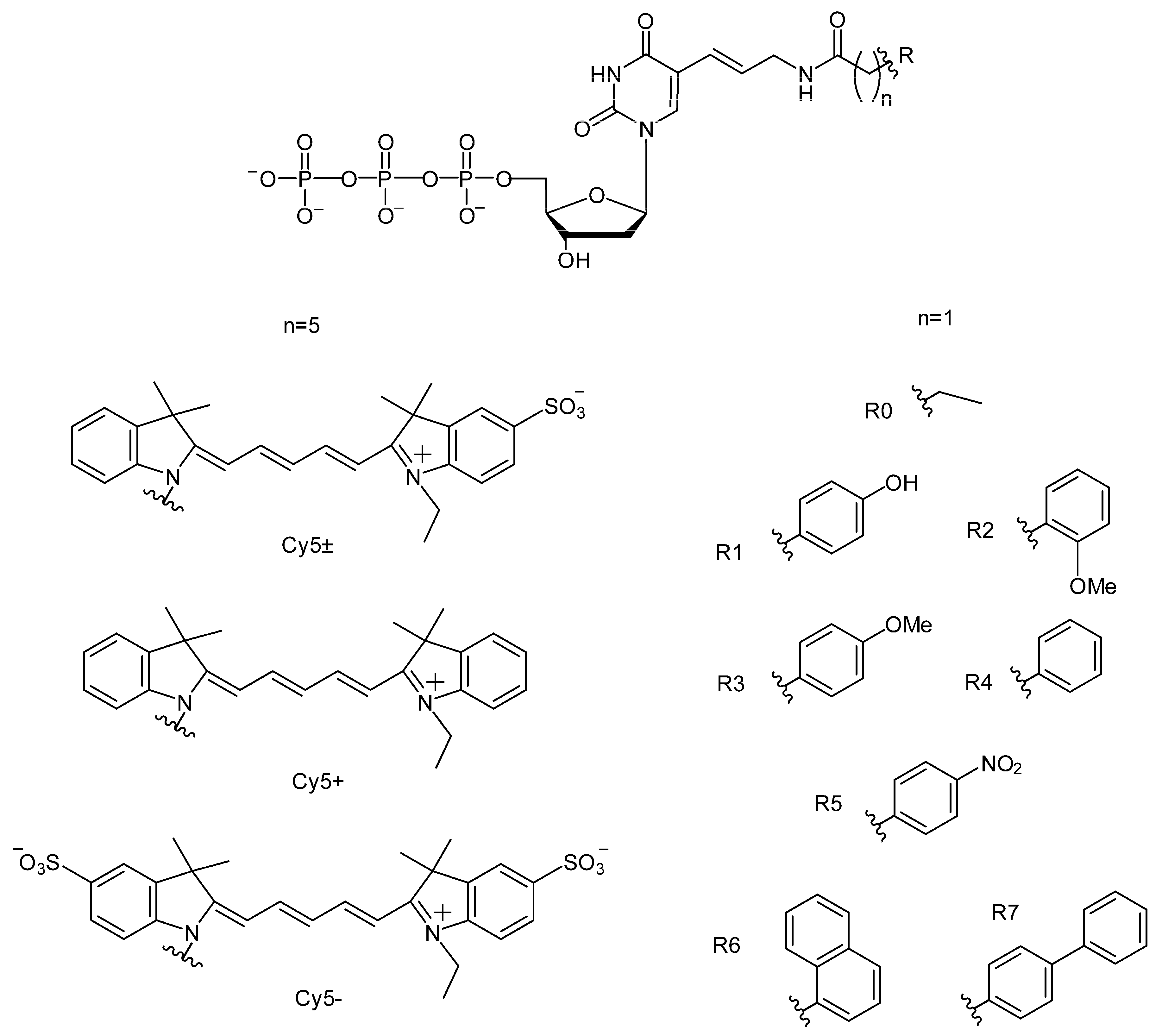
2.1.1. The Structures of C5-Attached dUTP Substituents and Electrophoretic Separation of PCR Products
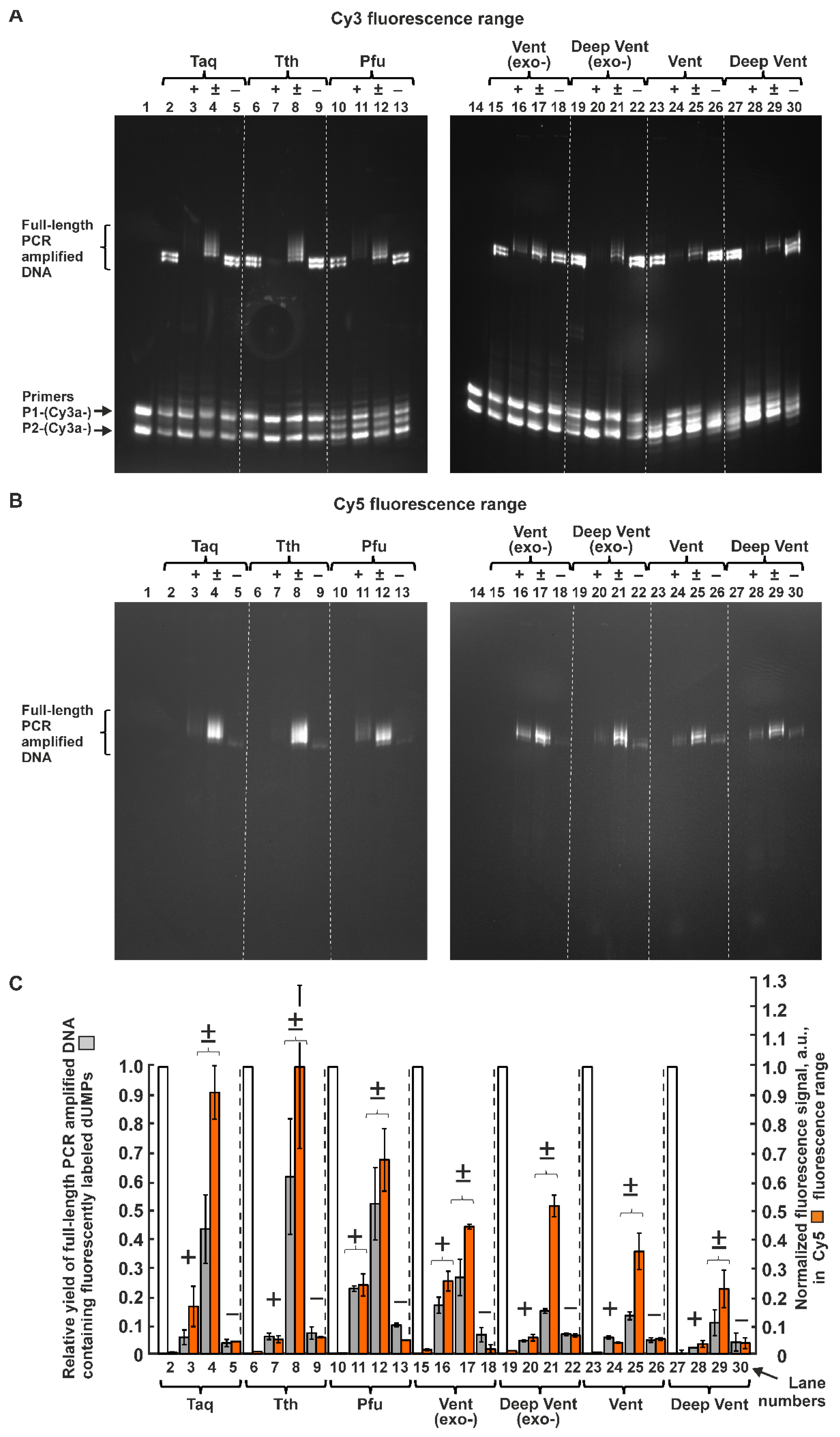
2.1.2. Inhibiting Effect of Modified dUTPs in the Reaction Mixture on the Yield of PCR Product Consisting Only of Natural Nucleotides
2.1.3. PCR Efficiency of Seven A and B Family Polymerases for dUTPs C5-Modified with Bulky Aromatic Hydrocarbon Substituents
2.1.4. Efficiency of PCR Amplification by DNA Polymerases of the A and B Families in the Presence of dUTPs C5-Modified with Cy5 Dye Analogs
2.2. Structural Factors Reducing the PCR Efficiency When dUTPs C5-Modified by Various Low-Molecular-Weight Substituents Are Used
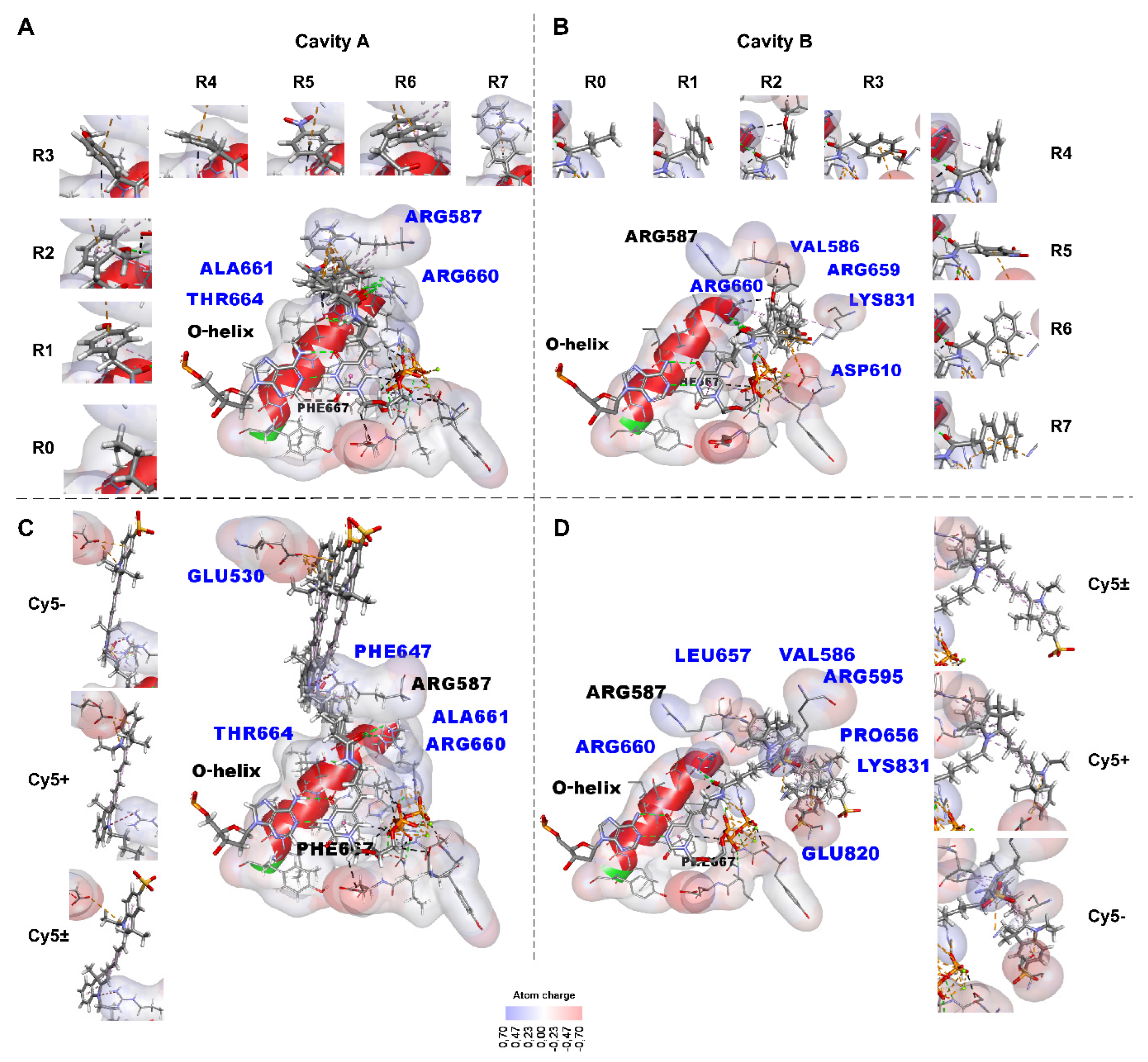
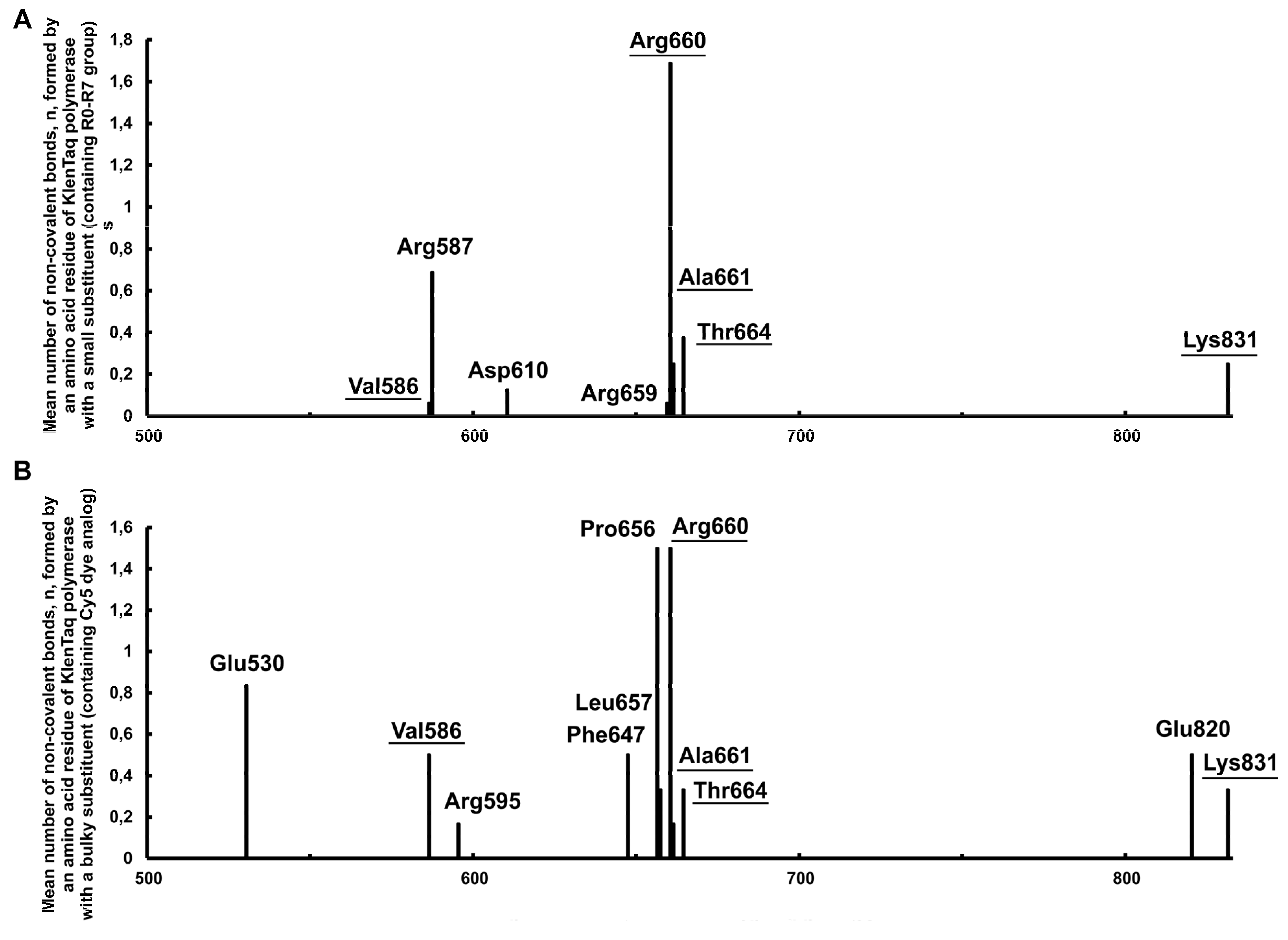
2.2.1. Identifying the Non-Covalent Interactions between the Substituents Containing Small Functional Groups R0-R7 and the KlenTaq Polymerase Amino Acid Residues
- -
- Hydrogen bonds by green dotted lines;
- -
- Carbon and π–donor hydrogen bonds by black dotted lines;
- -
- Electrostatic interactions, including π–cationic and π–anionic, by orange dotted lines;
- -
- Hydrophobic π–alkyl interactions by purple dotted lines;
- -
- Electrostatic attractive charge bonds by orange dotted lines;
- -
- π–lone pair bond by green dotted line;
- -
- π–sulfur bond by yellow dotted line;
- -
- Hydrophobic π–π stacking bond is denoted by a pink dotted line, alkyl bonds by purple dotted lines, and the π–sigma bond by a dark purple dotted line. The bonds can be seen more in the Discovery Studio program [49] using .dsv files with these 3D structures in Supplementary data. An unfavorable positive–positive interaction between the nitrogen atom close to the linker indolyl ring of the substituent and the nitrogen atom of Arg587 is marked by a red dotted line in C.
2.2.2. Localization of Bulky Aromatic Hydrocarbon Substituents (Containing Cy5 Dye Analogs) in KlenTaq Polymerase–DNA–(Modified dUTP) 3D Complexes
2.2.3. Non-Covalent Interactions between the Bulky Aromatic Substituents and the KlenTaq Polymerase Amino Acid Residues
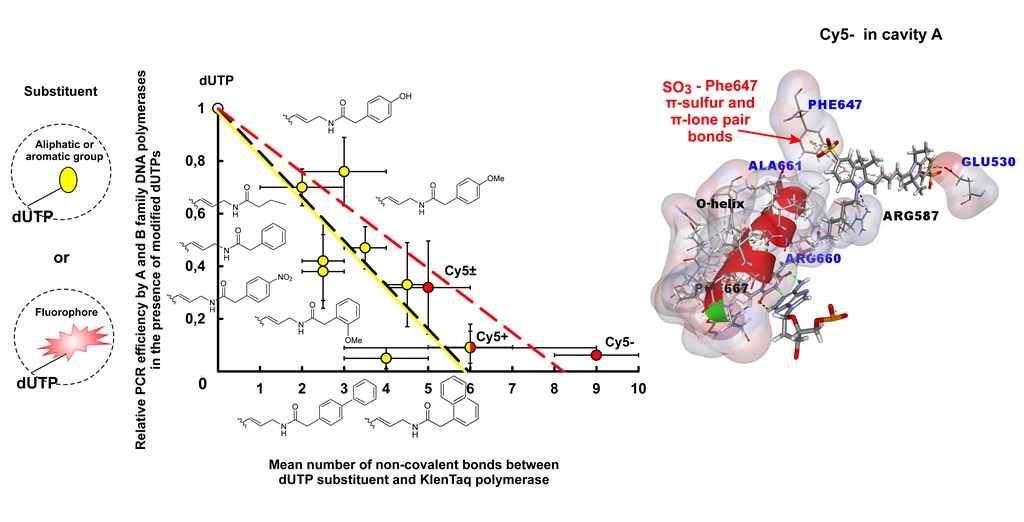
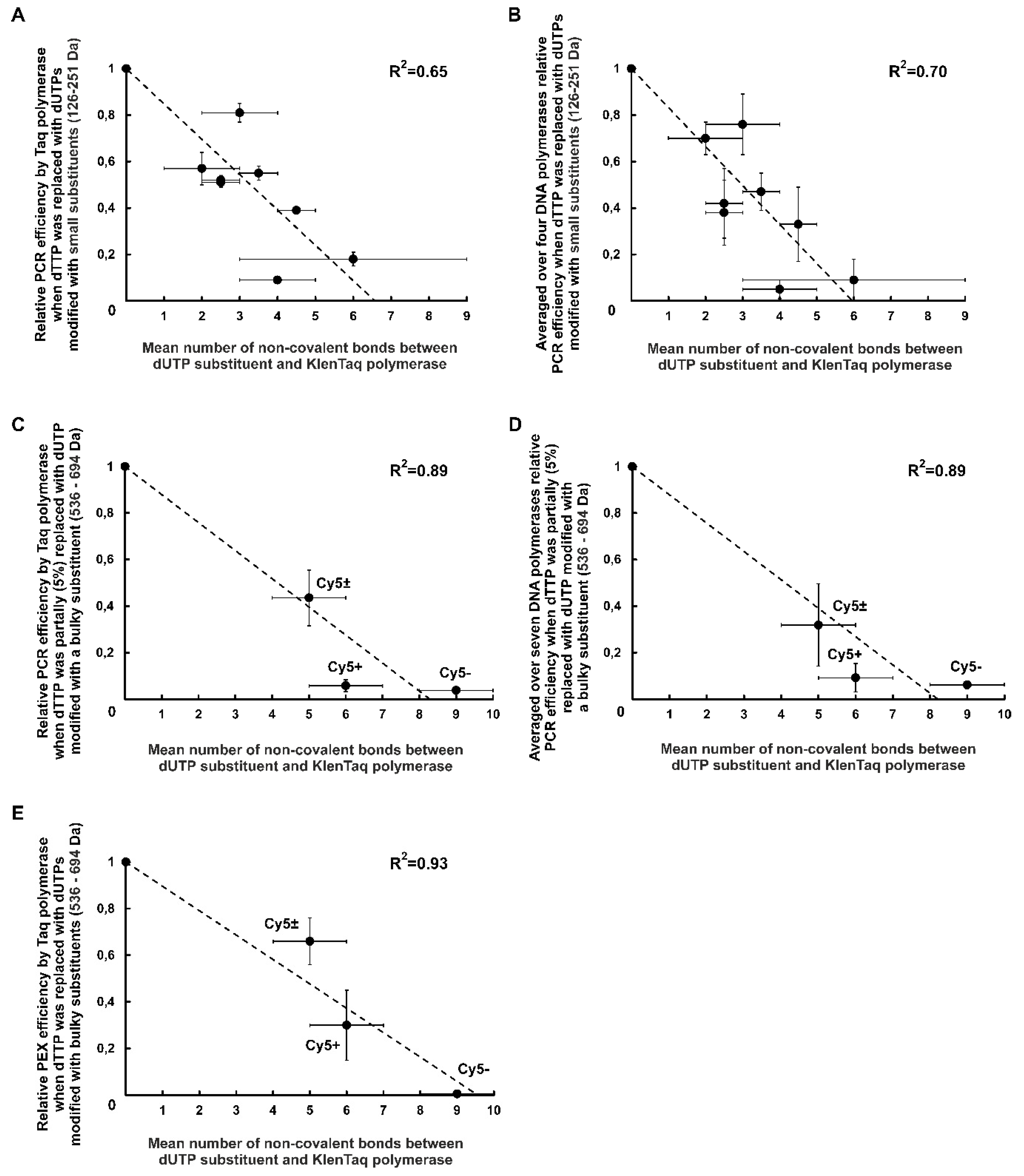
2.2.4. Negative Correlation between PCR Efficiency in the Presence of Modified dUTPs and the Number of Non-Covalent Bonds between the dUTP C5-Substituents and DNA Polymerase
2.2.4.1. PCR Compatibility of dUTPs C5-Modified with R0-R7 Functional Groups
2.2.4.2. PCR Compatibility of dUTPs C5-Modified with the Bulky Substituents
2.2.5. Negative Correlation between the Number of Non-Covalent Bonds Formed by dUTP C5-Substituents with the Taq Polymerase and PEX Efficiency in the Presence of dUTPs Modified with Bulky Chromophore-Containing Substituents
2.2.6. The Similarity of the Local Environments of the dUTP C5-Substituents Localized in the Active Center of Different Polymerases
2.2.7. Analysis of the Linker Parts of the dUTP-Attached Substituents in Non-Covalent Interactions with Taq Polymerase Amino Acid Residues
2.2.8. Analysis of the Functional Groups R0-R7 of the dUTP-Attached Substituents in Non-Covalent Interactions with Taq Polymerase Amino Acid Residues
2.2.9. Analysis of Bulky dUTP-Attached Substituents in Non-Covalent Interactions with Taq Polymerase
2.2.10. Analysis of Amino Acid Residues Capable of Forming Non-Covalent Bonds with the dUTP-Attached Substituents
3. Discussion
3.1. Negative Correlation between the PCR Amplification Efficiency in the Presence of the Modified Dutps and the Number of Non-Covalent Bonds between the dUTP-Attached Substituents and DNA Polymerase Amino Acid Residues
3.2. Commonality of Seven Polymerases from the Families A and B to Use the dUTPs Modified with Small or Bulky Aromatic Substituents in PCR
3.3. The Role of Non-Covalent Interactions between Low-Molecular-Weight Substituents Attached to the C5 Position of the Pyrimidine ring of dUTPs and the Amino Acid Residues of Taq DNA Polymerase in PCR Efficiency
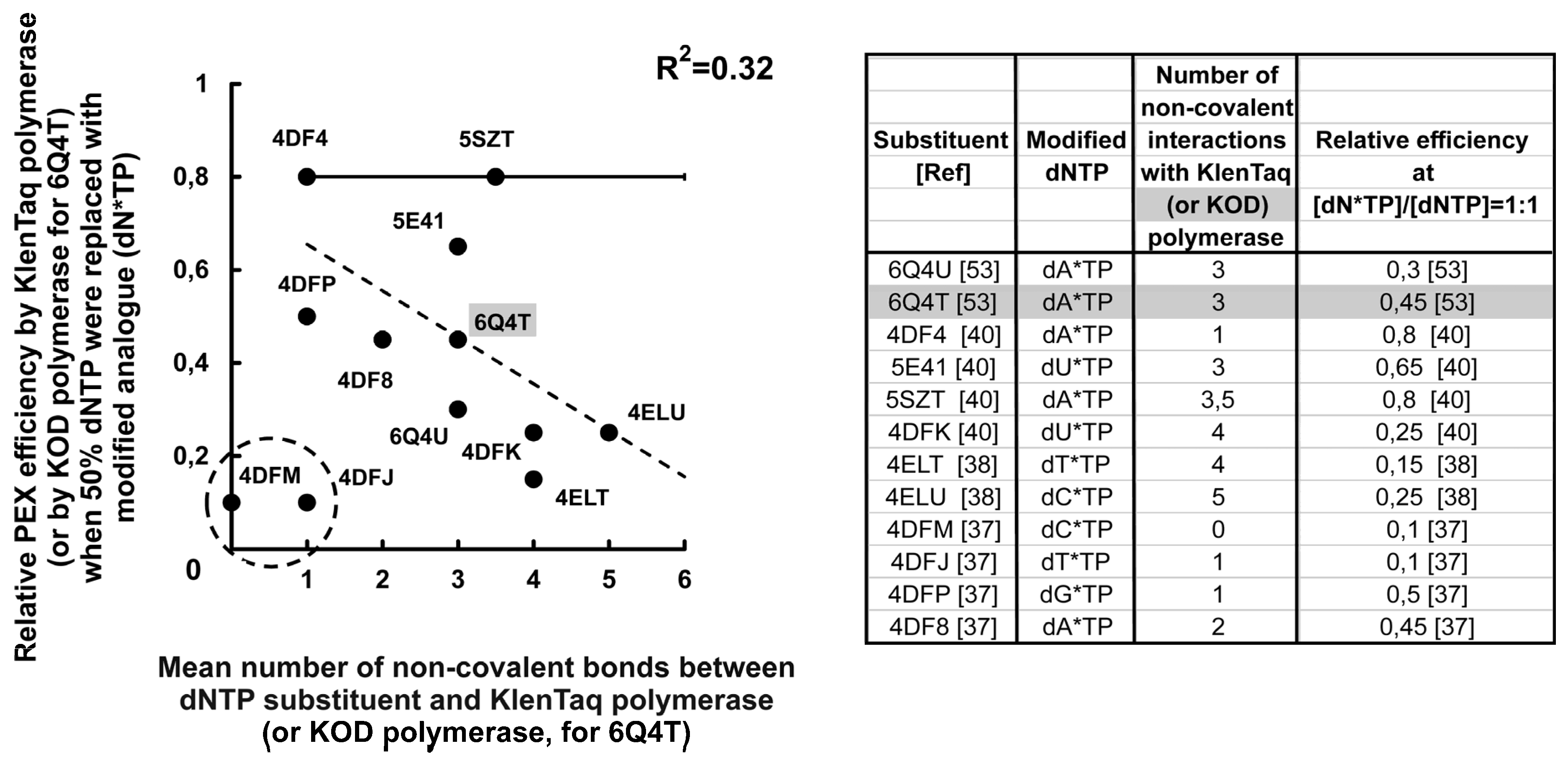
3.3.1. Incorporation Efficiency of dTspinMP and dTdendMP and X ray Structural Data Obtained by Marx A. et al. Analyzed in the Discovery Studio Program
3.3.2. Competitive Incorporation of the Modified dNTPs and the X-ray Structural Data for KlenTaq Polymerase–DNA–(Modified dNTP) Complexes
3.4. Analysis of Amino Acid Residues of the KlenTaq Polymerase Capable of Forming Non-Covalent Bonds with the dUTP C5-Attached Substituents
4. Materials and Methods
4.2. The DNA Template and Primers.
4.3. PCR and Electrophoresis
4.4. Quantitative Analysis of Electrophoretic Bands Containing PCR-Amplified Full-Length DNA Fragments
4.5. Computer Modeling of 3D Structures of Triple Complexes Consisting of KlenTaq Polymerase–DNA–(Modified dUTP)
4.6. KlenTaq Polymerase Amino Acid Residues' Ability to Form Non-Covalent Interactions with the dUTP Substituents Attached at the C5 Position of the Pyrimidine Ring Localized in the Active Center of the Enzyme
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walsh, J.M.; Beuning, P.J. Synthetic Nucleotides as Probes of DNA Polymerase Specificity. J. Nucleic Acids. 2012, 2012, 530963. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. Nucleoside triphosphates—building blocks for the modification of nucleic acids. Molecules. 2012, 17, 13569–13591. [Google Scholar] [CrossRef]
- Wu, W.-J.; Yang, W.; Tsai, M.-D. How DNA polymerases catalyse replication and repair with contrasting fidelity. Nat. Rev. Chem. 2017, 1, 0068. [Google Scholar] [CrossRef]
- Perlíková, P.; Hocek, M. Pyrrolo[2,3-d]pyrimidine (7-deazapurine) as a privileged scaffold in design of antitumor and antiviral nucleosides. Med Res Rev. 2017, 37, 1429–1460. [Google Scholar] [CrossRef] [PubMed]
- Tokarenko, A.; Lišková, B.; Smoleń, S.; Táborská, N.; Tichý, M.; Gurská, S.; Perlíková, P.; Frydrych, I.; Tloušt'ová, E.; Znojek, P.; Mertlíková-Kaiserová, H.; Poštová Slavětínská, L.; Pohl, R.; Klepetářová, B.; Khalid, N.U.; Wenren, Y.; Laposa, R.R.; Džubák, P.; Hajdúch, M.; Hocek, M. Synthesis and Cytotoxic and Antiviral Profiling of Pyrrolo- and Furo-Fused 7-Deazapurine Ribonucleosides. J Med Chem. 2018, 61, 9347–9359. [Google Scholar] [CrossRef]
- Tichý, M.; Pohl, R.; Xu, H.Y.; Chen, Y.L.; Yokokawa, F.; Shi, P.Y.; Hocek, M. Synthesis and antiviral activity of 4,6-disubstituted pyrimido[4,5-b]indole ribonucleosides. Bioorg Med Chem. 2012, 20, 6123–6133. [Google Scholar] [CrossRef]
- Fleuti, M.; Bártová, K.; Slavětínská, L.P.; Tloušt'ová, E.; Tichý, M.; Gurská, S.; Pavliš, P.; Džubák, P.; Hajdúch, M.; Hocek, M. Synthesis and Biological Profiling of Pyrazolo-Fused 7-Deazapurine Nucleosides. J Org Chem. 2020, 85, 10539–10551. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; Flather, D.; Forbes, A.; Foreman, T.; Fowler, C.; Gawande, B.; Goss, M.; Gunn, M.; Gupta, S.; Halladay, D.; Heil, J.; Heilig, J.; Hicke, B.; Husar, G.; Janjic, N.; Jarvis, T.; Jennings, S.; Katilius, E.; Keeney, T.R.; Kim, N.; Koch, T.H.; Kraemer, S.; Kroiss, L.; Le, N.; Levine, D.; Lindsey, W.; Lollo, B.; Mayfield, W.; Mehan, M.; Mehler, R.; Nelson, S.K.; Nelson, M.; Nieuwlandt, D.; Nikrad, M.; Ochsner, U.; Ostroff, R.M.; Otis, M.; Parker, T.; Pietrasiewicz, S.; Resnicow, D.I.; Rohloff, J.; Sanders, G.; Sattin, S.; Schneide, D.; Singer, B.; Stanton, M.; Sterkel, A.; Stewart, A.; Stratford, S.; Vaught, J.D.; Vrkljan, M.; Walker, J.J.; Watrobka, M.; Waugh, S.; Weiss, A.; Wilcox, S.K.; Wolfson, A.; Wolk, S.K.; Zhang, C.; Zichi, D. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010, 5, e15004. [Google Scholar] [CrossRef]
- Kuwahara, M; Sugimoto, N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules. 2010, 15, 5423–5444. [CrossRef]
- Gold, L.; Walker, J.J.; Wilcox, S.K.; Williams, S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol. 2012, 29, 543–549. [Google Scholar] [CrossRef]
- Famulok, M.; Mayer, G. Aptamers and SELEX in Chemistry & Biology. Chem Biol. 2014, 21, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Drolet, D.W.; Green, L.S.; Gold, L.; Janjic, N. Fit for the Eye: Aptamers in Ocular Disorders. Nucleic Acid Ther. 2016, .26, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Gawande, B. N.; Rohloff, J. C.; Carter, J. D.; von Carlowitz, I.; Zhang, C.; Schneider, D. J.; Janjic, N. Selection of DNA aptamers with two modified bases. Proc. Natl. Acad. Sci. 2017, 114, 2898–2903. [Google Scholar] [CrossRef]
- Ladju, R.B.; Pascut, D.; Massi, M.N.; Tiribelli, C.; Sukowati, C.H. Aptamer: A potential oligonucleotide nanomedicine in the diagnosis and treatment of hepatocellular carcinoma. Oncotarget. 2017, 9, 2951–2961. [Google Scholar] [CrossRef]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug. Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Rose, K.M.; Alves Ferreira-Bravo, I.; Li, M.; Craigie, R.; Ditzler, M.A.; Holliger, P; DeStefano, J.J. Selection of 2'-Deoxy-2'-Fluoroarabino Nucleic Acid (FANA) Aptamers That Bind HIV-1 Integrase with Picomolar Affinity. ACS Chem Biol. 2019, 14, 2166–2175. [CrossRef]
- Minagawa, H.; Kataoka, Y.; Kuwahara, M.; Horii, K.; Shiratori, I.; Waga, I. A high affinity modified DNA aptamer containing base-appended bases for human β-defensin. Anal Biochem. 2020, 594, 113627. [Google Scholar] [CrossRef]
- Minagawa, H.; Kataoka, Y.; Fujita, H.; Kuwahara, M.; Horii, K.; Shiratori, I.; Waga, I. Modified DNA Aptamers for C-Reactive Protein and Lactate Dehydrogenase-5 with Sub-Nanomolar Affinities. Int J Mol Sci. 2020, 21, 2683. [Google Scholar] [CrossRef]
- Schmitz, A.; Weber, A.; Bayin, M.; Breuers, S.; Fieberg, V.; Famulok, M.; Mayer, G. A SARS-CoV-2 Spike Binding DNA Aptamer that Inhibits Pseudovirus Infection by an RBD-Independent Mechanism. Angew Chem Weinheim Bergstr Ger. 2021, 133, 10367–10373. [Google Scholar] [CrossRef]
- McKenzie, L.K.; El-Khoury, R.; Thorpe, J.D.; Damha, M.J.; Hollenstein, M. Recent progress in non-native nucleic acid modifications. Chem Soc Rev. 2021, 50, 5126–5164. [Google Scholar] [CrossRef]
- Jager, S.; Rasched, G.; Kornreich-Leshem, H.; Engeser, M.; Thum, O.; Famulok, M. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 2005, 127, 15071–15082. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Nagashima, J.; Hasegawa, M.; Tamura, T.; Kitagata, R.; Hanawa, K.; Hososhima, S.; Kasamatsu, T.; Ozaki, H.; Sawai, H. Systematic characterization of 2’-deoxynucleoside-5’-triphosphate analogs as substrates for DNA polymerases by polymerase chain reaction and kinetic studies on enzymatic production of modified DNA. Nucl. Acids Res. 2006, 34, 5383–5394. [Google Scholar] [CrossRef] [PubMed]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef]
- Hocek, M. Synthesis of base-modified 2′-deoxyribonucleoside triphosphates and their use in enzymatic synthesis of modified DNA for applications in bioanalysis and chemical biology. J. Org. Chem. 2014, 79, 9914–9921. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. Generation of long, fully modified, and serum-resistant oligonucleotides by rolling circle amplification. Org Biomol Chem. 2015, 13, 9820–9824. [Google Scholar] [CrossRef] [PubMed]
- Ménová, P.; Cahová, H.; Vrábel, M.; Hocek, M. Synthesis of Base-Modified dNTPs Through Cross-Coupling Reactions and Their Polymerase Incorporation to DNA. Methods Mol Biol. 2019, 1973, 39–57. [Google Scholar] [CrossRef]
- Röthlisberger, P.; Levi-Acobas, F.; Leumann, C.J.; Hollenstein, M. Enzymatic synthesis of biphenyl-DNA oligonucleotides. Bioorg Med Chem. 2020, 28, 115487. [Google Scholar] [CrossRef]
- Zasedateleva, O.A.; Surzhikov, S.A.; Shershov, V.E.; Miftakhov, R.A.; Yurasov, D.A.; Kuznetsova, V.E.; Chudinov, A.V. PCR incorporation of dUMPs modified with aromatic hydrocarbon substituents of different hydrophilicities: Synthesis of C5-modified dUTPs and PCR studies using Taq, Tth, Vent (exo-) and Deep Vent (exo-) polymerases. Bioorg Chem. 2020, 99, 103829. [Google Scholar] [CrossRef]
- Ondruš, M.; Sýkorová, V.; Bednárová, L.; Pohl, R.; Hocek, M. Enzymatic synthesis of hypermodified DNA polymers for sequence-specific display of four different hydrophobic groups. Nucleic Acids Res. 2020, 48, 11982–11993. [Google Scholar] [CrossRef]
- Ollis, D.L.; Brick, P.; Hamlin, R.; Xuong, N.G.; Steitz, T.A. Structure of the large fragment of Escherichia coli DNA polymerase I complexed with TMP. Nature. 1985, 313, 762–766. [Google Scholar] [CrossRef]
- Kim, Y.; Eom, S.H.; Wang, J.; Lee, D.S.; Suh, S.W.; Steitz, T.A. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995, 376, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Korolev, S.; Waksman, G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998, 17, 7514–7525. [Google Scholar] [CrossRef]
- Li, Y.; Mitaxov, V.; Waksman, G. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl. Acad. Sci. USA. 1999, 96, 9491–9496. [Google Scholar] [CrossRef]
- Steitz, T.A. DNA Polymerases: Structural Diversity and Common Mechanisms. J Biol Chem. 1999, 274, 17395–17398. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, P.J.; Waksman, G. Structure and mechanism of DNA polymerases, in Advances in protein chemistry; Squire, J.M.; Parry, D.A.D. (Eds.); Elsevier, Amsterdam, Netherlands, 2005, Volume 71, pp. 401-440. [CrossRef]
- Obeid, S.; Baccaroa, A.; Welteb, W.; Diederichsb, K.; Marx, A. Structural basis for the synthesis of nucleobase modified DNA by Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA. 2010, 107, 21327–21331. [Google Scholar] [CrossRef] [PubMed]
- Bergen, K.; Steck, A.-L.; Strütt, S.; Baccaro, A.; Welte, W.; Diederichs, K.; Marx, A. Structures of KlenTaq DNA polymerase caught while incorporating C5-modified pyrimidine and C7-modified 7-deazapurine nucleoside triphosphates. J Am Chem Soc. 2012, 134, 11840–11843. [Google Scholar] [CrossRef]
- Obeid, S.; Busskamp, H.; Welte, W.; Diederichs, K.; Marx, A. Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site. Chem Commun (Camb). 2012, 48, 8320–8322. [Google Scholar] [CrossRef]
- Hottin, A.; Marx, A. Structural Insights into the processing of nucleobase-modified nucleotides by DNA polymerases. Acc Chem Res. 2016, 49, 418–427. [Google Scholar] [CrossRef]
- Hottin, A.; Betz, K.; Diederichs, K.; Marx, A. Structural Basis for the KlenTaq DNA Polymerase Catalysed Incorporation of Alkene- versus Alkyne-Modified Nucleotides. Chemistry. 2017, 23, 2109–2118. [Google Scholar] [CrossRef]
- Kropp, H.M.; Durr, S.L.; Peter, C.; Diederichs, K.; Marx, A. Snapshots of a modified nucleotide moving through the confines of a DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 9992–9997. [Google Scholar] [CrossRef]
- Cahová, H.; Panattoni, A.; Kielkowski, P.; Jindřich Fanfrlík, J.; Hocek, M. 5-Substituted Pyrimidine and 7-Substituted 7-Deazapurine dNTPs as Substrates for DNA Polymerases in Competitive Primer Extension in the Presence of Natural dNTPs. ACS Chem. Biol. 2016, 11, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.; Jemth, A.S.; Brown, A.; Crampton, N.; Dear, P.; Holliger, P. CyDNA: synthesis and replication of highly Cy-dye substituted DNA by an evolved polymerase. J. Am. Chem. Soc. 2010, 132, 5096–5104. [Google Scholar] [CrossRef]
- Wynne, S.A.; Pinheiro, V.B.; Holliger, P.; Leslie, A.G. Structures of an apo and a binary complex of an evolved archeal B family DNA polymerase capable of synthesizing highly Cy-dye labelled DNA. PLoS One. 2013, 8, e70892. [Google Scholar] [CrossRef]
- Ivancová, I.; Leone, D.L.; Hocek, M. Reactive modifications of DNA nucleobases for labelling, bioconjugations, and cross-linking. Curr Opin Chem Biol. 2019, 52, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Kuba, M.; Kraus, T.; Pohl, R.; Hocek, M. Nucleotide-Bearing Benzylidene-Tetrahydroxanthylium Near-IR Fluorophore for Sensing DNA Replication, Secondary Structures and Interactions. Chemistry. 2020, 26, 11950–11954. [Google Scholar] [CrossRef] [PubMed]
- Zasedateleva, O.A.; Vasiliskov, V.A.; Surzhikov, S.A.; Kuznetsova, V.E.; Shershov, V.E.; Guseinov, T.O.; Smirnov, I.P.; Yurasov, R.A.; Spitsyn, M.A.; Chudinov, A.V. dUTPs conjugated with zwitterionic Cy3 or Cy5 fluorophore analogues are effective substrates for DNA amplification and labelling by Taq polymerase. Nucleic Acids Res. 2018, 46, e73. [Google Scholar] [CrossRef]
- Mader, O.; Reiner, K.; Egelhaaf, H.J.; Fischer, R; Brock R. Structure property analysis of pentamethine indocyanine dyes: identification of a new dye for life science applications. Bioconjug. Chem. 2004, 15, 70–78. [CrossRef]
- BIOVIA, Dassault Systèmes, Discovery Studio, 2020, San Diego: Dassault Systèmes, 2020. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 31 July 2023).
- https://www.rcsb.org Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N. and Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [CrossRef]
- Shershov, V.E.; Kuznetsova, V.E.; Lysov, Y.P.; Guseinov, T.O.; Barsky, V.E.; Spitsyn, M.A.; Zasedateleva, O.A.; Vasiliskov, V.A.; Surzhikov, S.A.; Zasedatelev, A.S.; Chudinov, A.V. The effect of chromophore charge on the incorporation efficiency of fluorescence-labeled nucleotides catalyzed by Taq DNA polymerase in template synthesis. Biophysiks, 2015, 60, 1013–1015. [Google Scholar] [CrossRef]
- Aschenbrenner, J.; Marx, A. DNA polymerases and biotechnological applications. Curr Opin Biotechnol. 2017, 48, 187–195. [Google Scholar] [CrossRef]
- Kropp, H.M.; Diederichs, K.; Marx, A. The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide. Angew Chem Int Ed Engl. 2019, 58, 5457–5461. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




