Submitted:
02 August 2023
Posted:
04 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Culture and DNA extraction
Genomic sequencing
Single-nucleotide polymorphism (SNP) and phylogenetic analysis
3. Results
| Count | Percentage (%) | |
|---|---|---|
| Sex | ||
| Male | 39 | 81 |
| Female | 9 | 19 |
| Total | 48 | 100 |
| Age Group | ||
| <21 | 6 | 13 |
| >=21 & <65 | 25 | 52 |
| >=65 | 17 | 35 |
| Total | 48 | 100 |
| Immunosuppressed cases | ||
| Indiana | 1 | 5 |
| Kentucky | 3 | 16 |
| Michigan | 6 | 32 |
| Minnesota | 8 | 42 |
| Wisconsin | 1 | 5 |
| Total | 19 | 100 |
| Specimen Source | ||
| Sputum | 4 | 8 |
| Lymph node | 4 | 8 |
| Bronchial specimen | 17 | 35 |
| Lung tissue | 3 | 6 |
| Blood | 12 | 25 |
| Bone marrow | 3 | 6 |
| Others* | 5 | 10 |
| Total | 48 | 100 |
| States | ||
| Indiana | 2 | 4 |
| Kentucky | 4 | 8 |
| Louisiana | 1 | 2 |
| Michigan | 21 | 44 |
| Minnesota | 13 | 27 |
| Nebraska | 1 | 2 |
| Pennsylvania | 1 | 2 |
| Wisconsin | 5 | 10 |
| Total | 48 | 100 |
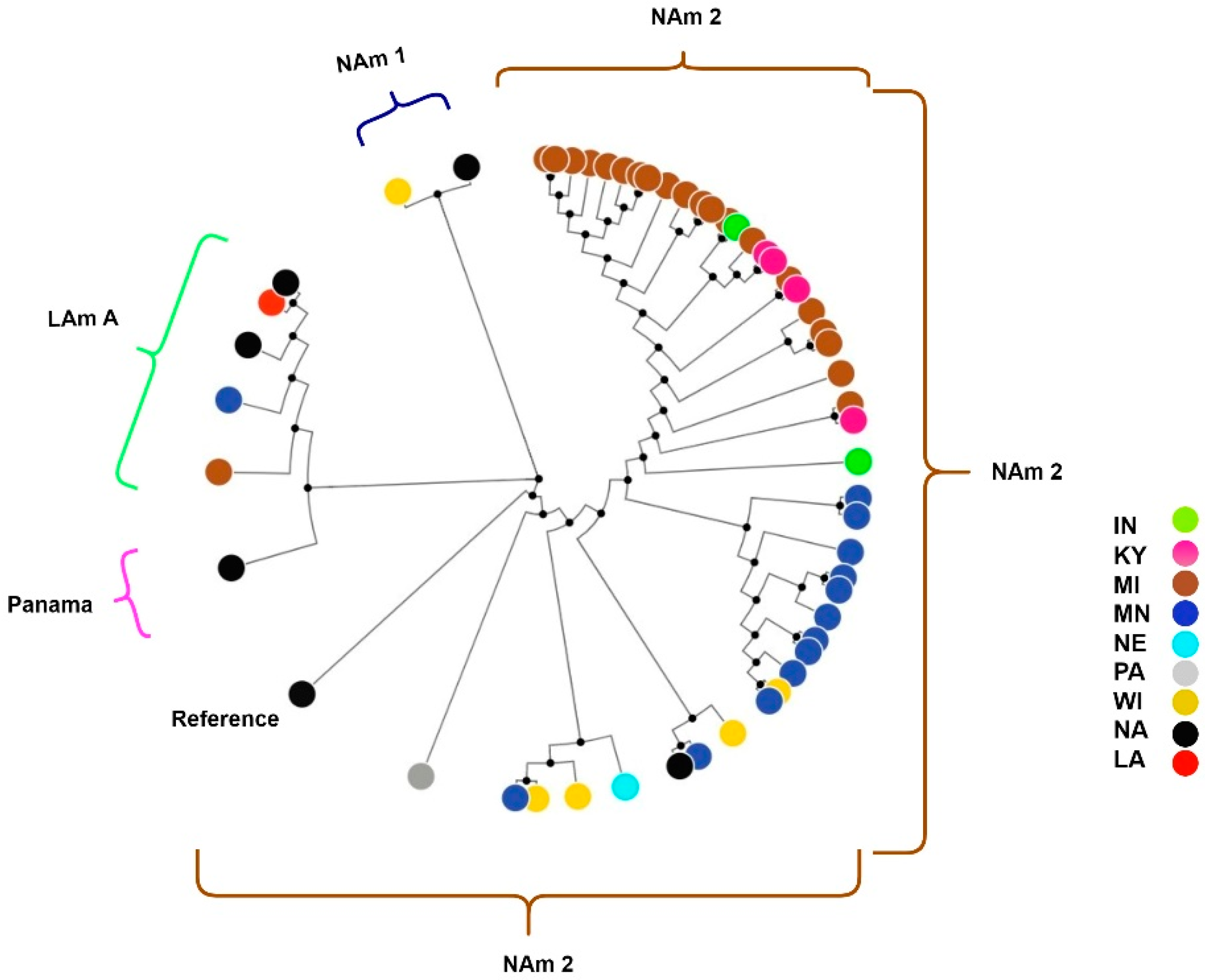
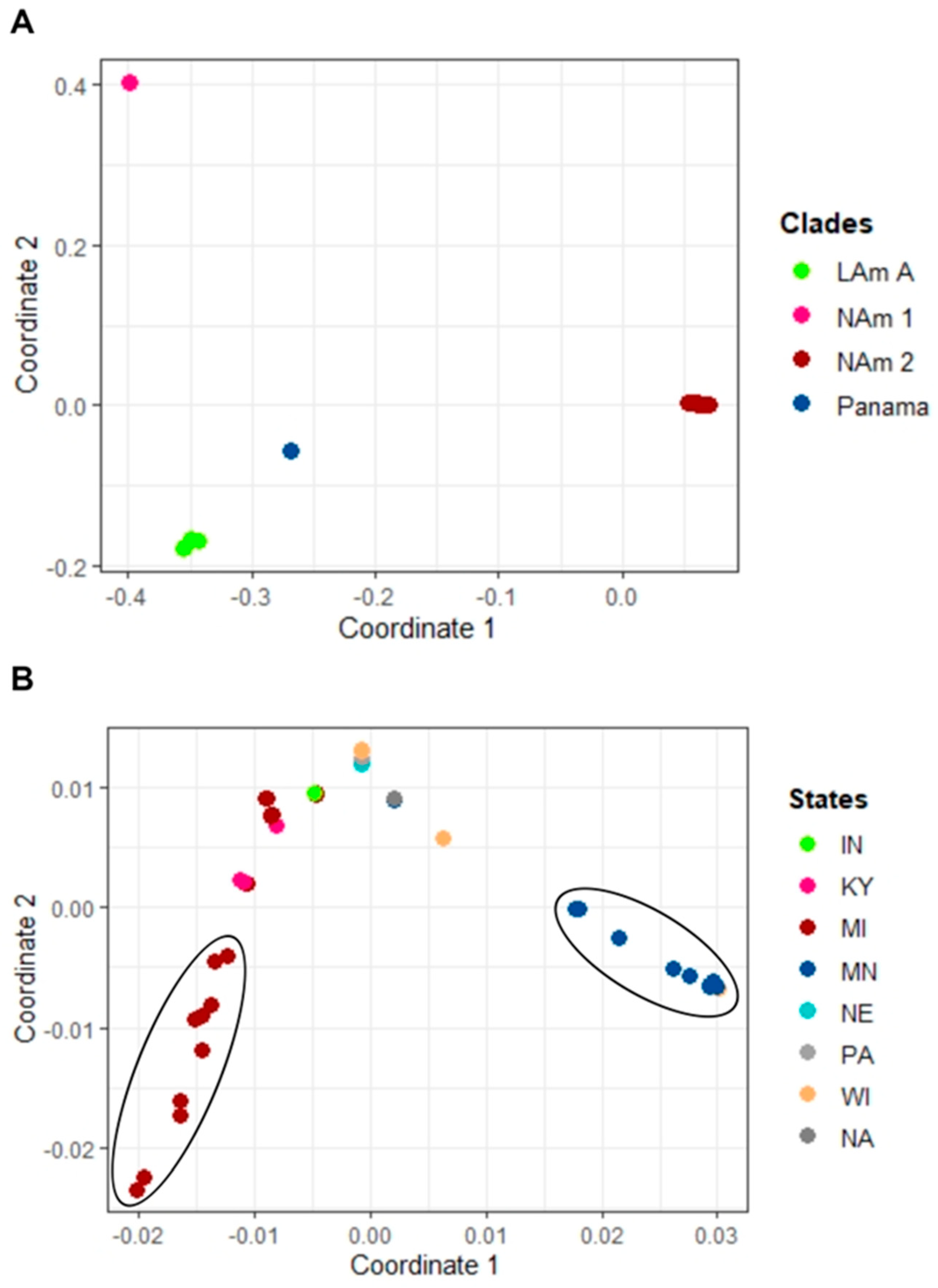
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brömel, Catharina, and Jane E. Sykes. "Histoplasmosis in Dogs and Cats." Clinical Techniques in Small Animal Practice 20, no. 4 (2005): 227-32. [CrossRef]
- Cano, MV, and Rana A Hajjeh. "The Epidemiology of Histoplasmosis: A Review." Paper presented at the Seminars in respiratory infections 2001.
- Deepe, G. S., Jr. "Outbreaks of Histoplasmosis: The Spores Set Sail." PLoS Pathog 14, no. 9 (2018): e1007213. [CrossRef]
- Gómez, Luisa F., Myrtha Arango, Juan G. McEwen, Oscar M. Gómez, Alejandra Zuluaga, Carlos A. Peláez, Jose M. Acevedo, María L. Taylor, and María del P. Jiménez. "Molecular Epidemiology of Colombian Histoplasma Capsulatum Isolates Obtained from Human and Chicken Manure Samples." Heliyon 5, no. 7 (2019): e02084. [CrossRef]
- Acir Rachid, Lucila Stange, Rezende, Suzelle Freitas de Moura, Paulo C. Loffy, Francisco Luiz Gomide M. Magalhães. "A Case Study of Disseminated Histoplasmosis Linked to Common Variable Immunodeficiency." Brazilian Journal of Infectious Diseases 7 (2003): 268-72. [CrossRef]
- Assi, Maha A., Mohamad S. Sandid, Larry M. Baddour, Glenn D. Roberts, and Randall C. Walker. "Systemic Histoplasmosis: A 15-Year Retrospective Institutional Review of 111 Patients." Medicine 86, no. 3 (2007): 162-69. [CrossRef]
- Manos, Nicholas E., Shirley H. Ferebee, and Winifred F. Kerschbaum. "Geographic Variation in the Prevalence of Histoplasmin Sensitivity." Diseases of the chest 29, no. 6 (1956): 649-68. [CrossRef]
- Benedict, Kaitlin, Stephanie McCracken, Kimberly Signs, Malia Ireland, Victoria Amburgey, Jose Antonio Serrano, Natalie Christophe, Suzanne Gibbons-Burgener, Sara Hallyburton, Kimberly A Warren, Alison Keyser Metobo, Racheal Odom, Matthew R Groenewold, and Brendan R Jackson. "Enhanced Surveillance for Histoplasmosis—9 States, 2018–2019." Open Forum Infectious Diseases 7, no. 9 (2020). [CrossRef]
- Dallas J. Smith; Samantha L. Williams; Endemic Mycoses State Partners Group; Kaitlin M. Benedict; Brendan R. Jackson; Mitsuru Toda. "Surveillance for Coccidioidomycosis, Histoplasmosis, and Blastomycosis — United States, 2019." MMWR Morb Mortal Wkly Rep 71, no. 7 (2022): 1-14. [CrossRef]
- Mazi, P. B., J. M. Sahrmann, M. A. Olsen, A. Coler-Reilly, A. M. Rauseo, M. Pullen, J. C. Zuniga-Moya, W. G. Powderly, and A. Spec. "The Geographic Distribution of Dimorphic Mycoses in the United States for the Modern Era." Clin Infect Dis 76, no. 7 (2023): 1295-301. [CrossRef]
- Kwon-Chung, K. J., and J. E. Bennett. "Medical Mycology." Revista do Instituto de Medicina Tropical de São Paulo 34 (1992).
- Kasuga, T., T. J. White, G. Koenig, J. McEwen, A. Restrepo, E. Castañeda, C. Da Silva Lacaz, E. M. Heins-Vaccari, R. S. De Freitas, R. M. Zancopé-Oliveira, Z. Qin, R. Negroni, D. A. Carter, Y. Mikami, M. Tamura, M. L. Taylor, G. F. Miller, N. Poonwan, and J. W. Taylor. "Phylogeography of the Fungal Pathogen Histoplasma Capsulatum." Mol Ecol 12, no. 12 (2003): 3383-401. [CrossRef]
- Teixeira, Marcus de M., José S. L. Patané, Maria L. Taylor, Beatriz L. Gómez, Raquel C. Theodoro, Sybren de Hoog, David M. Engelthaler, Rosely M. Zancopé-Oliveira, Maria S. S. Felipe, and Bridget M. Barker. "Worldwide Phylogenetic Distributions and Population Dynamics of the Genus Histoplasma." PLOS Neglected Tropical Diseases 10, no. 6 (2016): e0004732. [CrossRef]
- Sepúlveda, Victoria E., Roberto Márquez, David A. Turissini, William E. Goldman, and Daniel R. Matute. "Genome Sequences Reveal Cryptic Speciation in the Human Pathogen <I>Histoplasma Capsulatum</I>." mBio 8, no. 6 (2017): e01339-17. [CrossRef]
- Jofre, Gaston I., Ashutosh Singh, Heidi Mavengere, Gandhi Sundar, Emmanuel D’Agostino, Anuradha Chowdhary, and Daniel R. Matute. "An Indian Lineage of Histoplasma with Strong Signatures of Differentiation and Selection." Fungal Genetics and Biology 158 (2022): 103654. [CrossRef]
- Robert, V., D. Vu, A. B. Amor, N. van de Wiele, C. Brouwer, B. Jabas, S. Szoke, A. Dridi, M. Triki, S. Ben Daoud, O. Chouchen, L. Vaas, A. de Cock, J. A. Stalpers, D. Stalpers, G. J. Verkley, M. Groenewald, F. B. Dos Santos, G. Stegehuis, W. Li, L. Wu, R. Zhang, J. Ma, M. Zhou, S. P. Gorjón, L. Eurwilaichitr, S. Ingsriswang, K. Hansen, C. Schoch, B. Robbertse, L. Irinyi, W. Meyer, G. Cardinali, D. L. Hawksworth, J. W. Taylor, and P. W. Crous. "Mycobank Gearing up for New Horizons." IMA Fungus 4, no. 2 (2013): 371-9. [CrossRef]
- Köser, Claudio U., Matthew J. Ellington, Edward J. P. Cartwright, Stephen H. Gillespie, Nicholas M. Brown, Mark Farrington, Matthew T. G. Holden, Gordon Dougan, Stephen D. Bentley, Julian Parkhill, and Sharon J. Peacock. "Routine Use of Microbial Whole Genome Sequencing in Diagnostic and Public Health Microbiology." PLOS Pathogens 8, no. 8 (2012): e1002824. [CrossRef]
- Roetzer, Andreas, Roland Diel, Thomas A. Kohl, Christian Rückert, Ulrich Nübel, Jochen Blom, Thierry Wirth, Sebastian Jaenicke, Sieglinde Schuback, Sabine Rüsch-Gerdes, Philip Supply, Jörn Kalinowski, and Stefan Niemann. "Whole Genome Sequencing Versus Traditional Genotyping for Investigation of a Mycobacterium Tuberculosis Outbreak: A Longitudinal Molecular Epidemiological Study." PLOS Medicine 10, no. 2 (2013): e1001387. [CrossRef]
- Ujwal R. Bagal, John Phan, Rory M. Welsh, Elizabeth Misas, Darlene Wagner, Lalitha Gade, Anastasia P. Litvintseva, Christina A. Cuomo, Nancy A. Chow. "Mycosnp: A Portable Workflow for Performing Whole-Genome Sequencing Analysis of Candida Auris." In Candida Auris: Methods and Protocols, edited by Alexander Lorenz: Humana New York, NY, 2022. [CrossRef]
- Delcher, A. L., S. L. Salzberg, and A. M. Phillippy. "Using Mummer to Identify Similar Regions in Large Sequence Sets." Curr Protoc Bioinformatics Chapter 10 (2003): Unit 10.3. [CrossRef]
- Li, H., B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, and R. Durbin. "The Sequence Alignment/Map Format and Samtools." Bioinformatics 25, no. 16 (2009): 2078-9. [CrossRef]
- Picard Toolkit: Repository. Http://Broadinstitute.Github.Io/Picard/.
- Lo, Chien-Chi, and Patrick S. G. Chain. "Rapid Evaluation and Quality Control of Next Generation Sequencing Data with Faqcs." BMC Bioinformatics 15, no. 1 (2014): 366. [CrossRef]
- Li, Heng. "Aligning Sequence Reads, Clone Sequences and Assembly Contigs with Bwa-Mem." arXiv (2013): 1303.3997.
- Cheng, A. Y., Y. Y. Teo, and R. T. Ong. "Assessing Single Nucleotide Variant Detection and Genotype Calling on Whole-Genome Sequenced Individuals." Bioinformatics 30, no. 12 (2014): 1707-13. [CrossRef]
- Kumar, S., G. Stecher, and K. Tamura. "Mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets." Mol Biol Evol 33, no. 7 (2016): 1870-4. [CrossRef]
- Price, Morgan N., Paramvir S. Dehal, and Adam P. Arkin. "Fasttree 2 – Approximately Maximum-Likelihood Trees for Large Alignments." PLOS ONE 5, no. 3 (2010): e9490. [CrossRef]
- R: A Language and Environment for Statistical Computing. Vienna, Austria.
- Hepler, S. A., K. A. Kaufeld, K. Benedict, M. Toda, B. R. Jackson, X. Liu, and D. Kline. "Integrating Public Health Surveillance and Environmental Data to Model Presence of Histoplasma in the United States." Epidemiology 33, no. 5 (2022): 654-9. [CrossRef]
- Taylor, Maria Lucia, María del Rocío Reyes-Montes, Daniel A. Estrada-Bárcenas, Rosely M. Zancopé-Oliveira, Gabriela Rodríguez-Arellanes, and José Antonio Ramírez. "Considerations About the Geographic Distribution of <I>Histoplasma</I> Species." Applied and Environmental Microbiology 88, no. 7 (2022): e02010-21. [CrossRef]
- Dingle, Tanis C., Matthew A. Croxen, Sumana Fathima, Sandy Shokoples, Ashlesha Sonpar, Lynora Saxinger, and Ilan S. Schwartz. "Histoplasmosis Acquired in Alberta, Canada: An Epidemiological and Genomic Study." The Lancet Microbe 2, no. 5 (2021): e191-e97. [CrossRef]
- Almeida-Silva, Fernando, Marcus de Melo Teixeira, Daniel R. Matute, Marcela de Faria Ferreira, Bridget M. Barker, Rodrigo Almeida-Paes, Allan J. Guimarães, and Rosely M. Zancopé-Oliveira. "Genomic Diversity Analysis Reveals a Strong Population Structure in Histoplasma Capsulatum Lama (Histoplasma Suramericanum)." Journal of Fungi 7, no. 10 (2021): 865. [CrossRef]
- Gusa, Asiya, and Sue Jinks-Robertson. "Mitotic Recombination and Adaptive Genomic Changes in Human Pathogenic Fungi." Genes 10, no. 11 (2019): 901. [CrossRef]
- Oltean, H. N., K. A. Etienne, C. C. Roe, L. Gade, O. Z. McCotter, D. M. Engelthaler, and A. P. Litvintseva. "Utility of Whole-Genome Sequencing to Ascertain Locally Acquired Cases of Coccidioidomycosis, Washington, USA." Emerg Infect Dis 25, no. 3 (2019): 501-06. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





