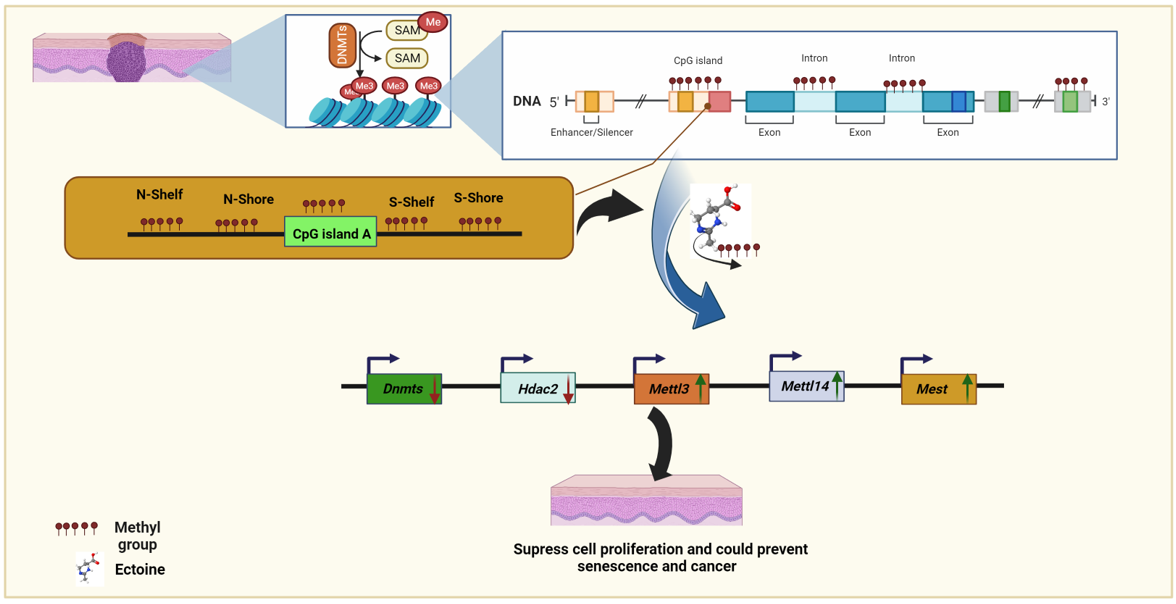
1. Introduction
Epigenetic modifications are mainly driven by DNA hypermethylation and strongly associated with cancer development and aging [
1]. Historically, safe natural compounds have been of interest for the treatment of idiopathic diseases, such as allergies and cancers, because of their safety records. Among these, substantial and naturally produced compatible solutes/osmolytes, including some amino acids and their derivatives, have been proven to be critical in protecting organisms from increased environmental abiotic and biotic stresses [
2]. In 1985, Galinski et al. characterized the well-established flagship and novel amino acid derivative ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidine carboxylic acid) osmolyte as a new component of present in marine microorganisms such as the extremely halophilic bacterial genus
Ectothiorhodospira [
3,
4,
5,
6,
7,
8]. Ectoine binds strongly to water and forms a protective shield against the hydration of DNA, proteins, and other biomolecules [
9,
10]. Owing to the fascinating features of ectoine (low molecular weight, zwitterionic, non-cytotoxic, strong water binding, and cyclic amino acids), it has been used in numerous healthcare applications in the healthcare and medical sectors, such as therapeutic agents for allergies, inflammation, and cancer alleviation [
2,
11,
12,
13,
14,
15,
16].
Epigenetic events such as aberrant DNA methylation have been shown to silence the expression of many genes that suppress malignancy, including skin cancer. DNA cytosine methylation is one of the best-studied epigenetic modifications, and has been proposed to play a key role in multiple biological processes and in the pathogenesis of disease conditions [
17]. The primary mechanism of DNA methylation involves the transfer of methyl groups from SAM metabolites through covalent modification, with one carbon atom attached to the fifth position of cytosine to form 5-methyl cytosine (5mC) [
18]. This covalent modification is mainly driven by the DNA methyltransferase (DNMTs) enzymes DNMT1, DNMT3a, and DNMT3b. DNMT1 maintains the methylation reaction during the replication phase, and the other two DNMTs perform de novo methylation [
19]. Drugs that selectively inhibit DNMT1 using well-known DNA-hypomethylating agents such as 5-aza-deoxycytidine (5-aza-CdR) are used in cancer therapy [
20]. There are several imprinted genes, including methylation markers, such as histone deacetylases (HDACs), and the aberrant expression of these genes is related to various types of cancer development, including melanoma [
21,
22]. Although several epigenetic drugs have been developed to target cancer, several drawbacks prevent their application owing to serious adverse reactions such as HDACi [
1,
23]. Ectoine is considered to be an excellent DNA protectant against environmental biotic and abiotic stresses, UV radiation, and other types of radiation. Another emerging critical methylation mechanism is the methylation of N6-adenosine (m6A) by methyltransferases METTL3 (methyltransferase-like 3) which adds m6A to target RNAs [
24]. Hypermethylation of one of the imprinted genes, mesoderm-specific transcript homologue (MEST) hypermethylation of MEST was observed in males with oligozoospermia and idiopathic male infertility [
25]. Determining the molecular mechanisms underlying the effectiveness of ectoine as a DNA protectant is necessary to highlight the roles of compatible solutes in protecting cells from environmental stress and to better understand senescence and cancer prevention.
In this study, we investigated the roles of ectoine in global DNA methylation in skin cells, 5-mC percentage in skin cells, and regulation of methylation-related genes and markers. We also investigated the effects of ectoine on the viability, proliferation, and functional pathways of skin cancer cells.
2. Results
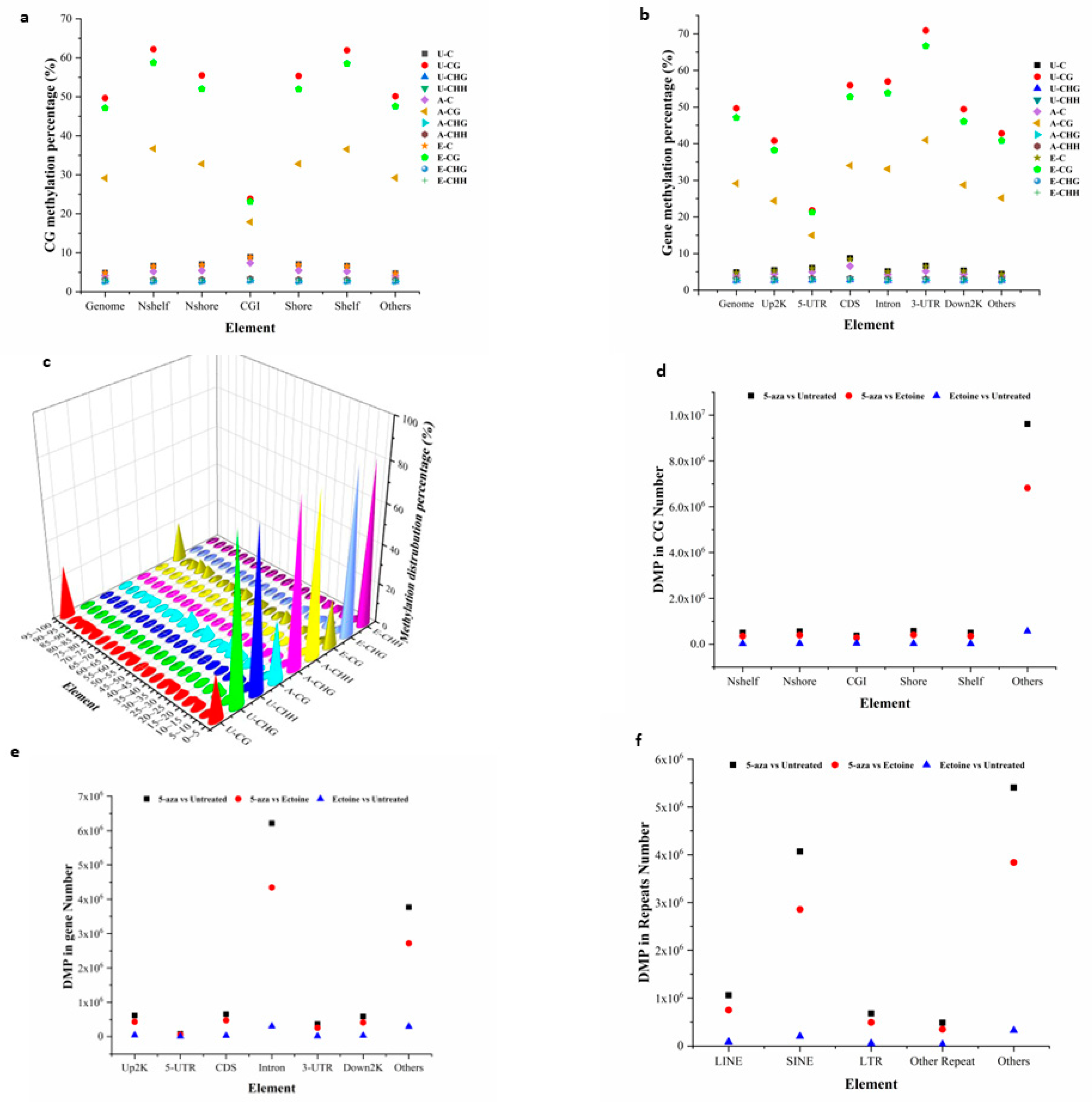
2.1. Global methylation sequencing revealed that ectoine hypomethylates DNA and enriched nucleotide binding ions DEGs.
Ectoine protects DNA from environmental stress and radiation by directly interacting with nearby water molecules [
44]. Methylome analysis of ectoine-treated cells showed that ectoine significantly reduced the methylation levels in almost all regions of the CpG island, except for the CGI region. At the gene methylation level, ectoine-treated cells showed reduced methylation compared with untreated cells. Similarly, our results showed that methylation distribution in the whole genome was reduced compared to that in untreated cells. However, 5-aza-2-deoxycytidine treated cells were remarkably hypomethylated compared to ectoine-and untreated cells. For further analysis, the DMPs between the groups were analyzed, and it was found that the total number of DMPs between ectoine and untreated cells was 689850, while the number of DMPs of 5-aza-2-deoxycytidine was 11687481 compared to untreated cells, and between 5-aza-2-deoxycytidine 8277681 DMPs compared to ectoine-treated cells. Gene DMPs were mostly distributed in introns, while in CG, the CGI region and the other elements were the highest compared to untreated cells (5.9% and 82%, respectively) (
Figure 1A–F and suppl. File 2). Overall, ectoine hypomethylated DNA in skin cells, particularly in CG elements, except for CGI.
Furthermore, GO enrichment functional analysis between functional pathways showed that ectoine-treated cells had a lower expression of BP, MF, and CC pathways related to DEGs than 5-aza vs. untreated cells and 5-aza vs. ectoine DEGs. Interestingly, tree-map GO enrichment analysis showed that ectoine vs. untreated ion binding, small molecule binding, nucleotide binding, and ribonucleotide binding-related DEGs were enriched compared with 5-aza vs. untreated and 5-aza vs. ectoine. Moreover, ectoine improves cellular biological processes such as cell morphogenesis. KEEG functional analysis showed that ectoine reduced the overall expression of genes compared to 5-aza versus untreated and 5-aza versus ectoine. ectoine, and induced morphine addiction (Suppl. Figure 1A-I).
Global DNA methylation analysis in HaCaT cells treated with ectoine compared to that in cells treated with 2-aza-deoxycitidine and untreated cells. Gene methylation analysis was performed using the Bismark software. The data show the methylation level of DNA at the CGI level (a), gene element methylation level (b), and total methylation distribution level (c). The DMPs analysis showed that the number of HaCaT DMPs in the CG elements (d) was similar to the number of DMPs (e) and repeats (f).
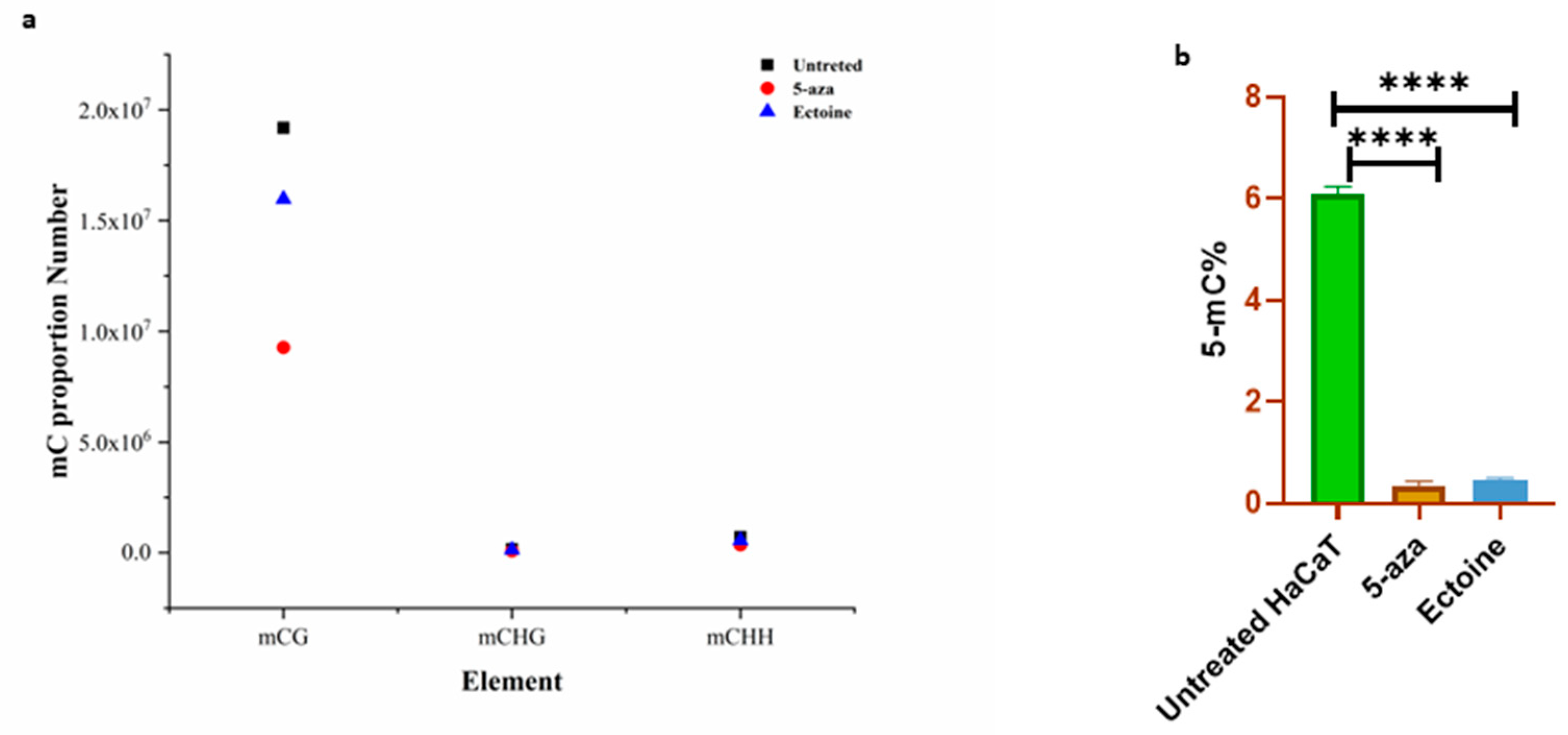
2.2.5. mC Methylation
To validate global DNA methylation, we analyzed the percentage of methylation-targeted 5mC in the DNA. Bioinformatic analysis of the methylome showed that the proportion of mCG, especially DMA, was significantly lower in ectoine-treated cells than in untreated cells (
Figure 2A). Antibodies targeting 5mC showed that ectoine significantly hypomethylated DNA compared to untreated cells (
Figure 2B). However, 5-aza-2-deoxycytidine-treated cells showed marked hypomethylation. Therefore, ectoine is reduced throughout the DNA 5mC level in skin cells.
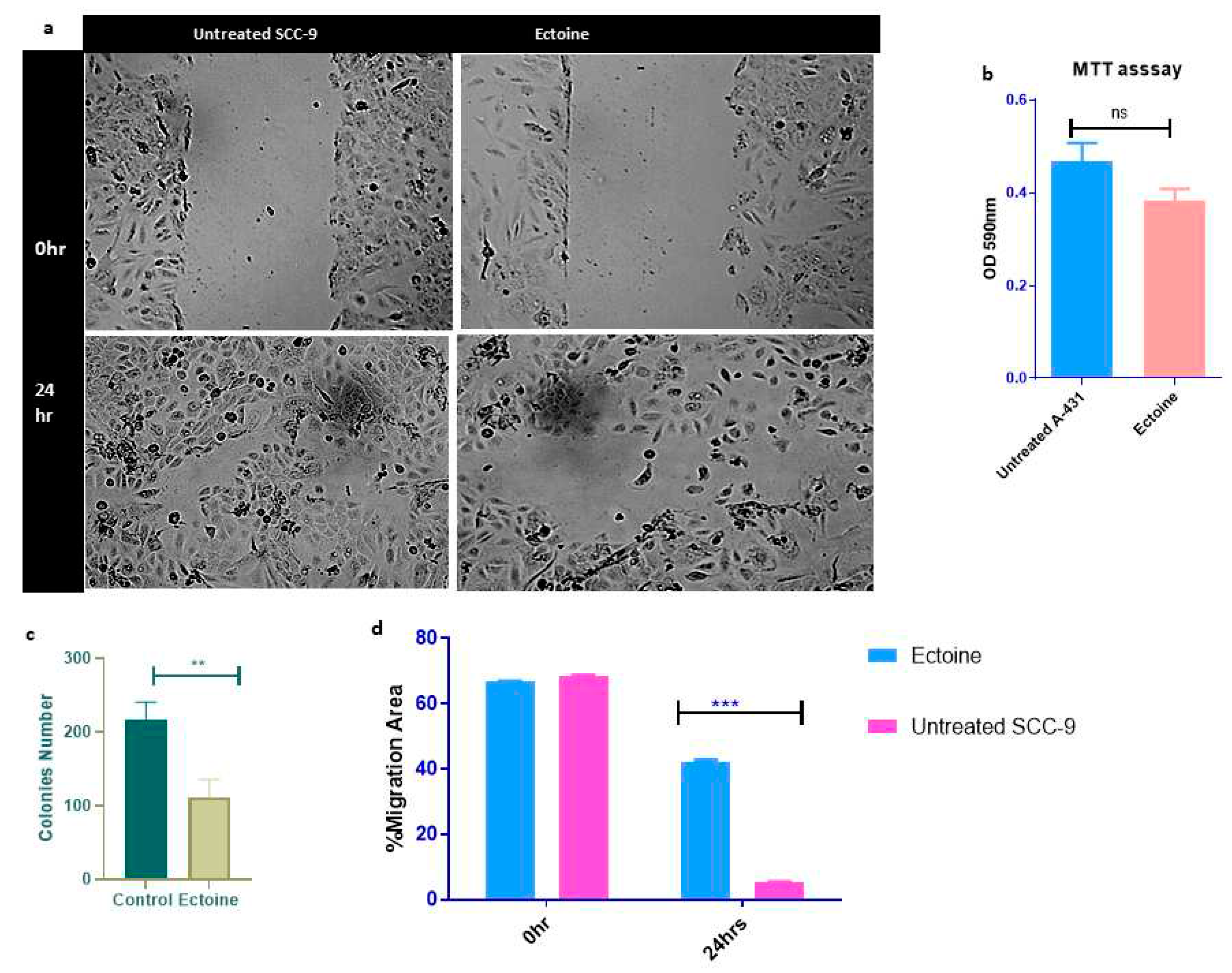
2.3. Ectoine is a non-tumorigenic molecule and reduces cell proliferation.
To study the effects of ectoine on SCC-9 cell migration, we performed a wound-healing test. A wound healing assay showed that ectoine inhibited cell migration compared to untreated cells. The wound healed in untreated cells that were transfected with and the control after 24 h compared to the cells transfected with ectoine, which took longer to heal (data not shown) (
Figure 3A,B). Similarly, A-431 wound healing was slower in ectoine-treated cells than that in untreated cells (Suppl. Figure 2A). To study the effects of ectoine on proliferation and apoptosis of SCC-9 cells, we performed an MTT assay. The results showed that ectoine-treated cell proliferation was lower than that of the untreated control cells, although the difference was not statistically significant. However, ectoine had almost no effect on the cell viability (
Figure 3C). Moreover, the clonogenic assay showed that the number of colonies formed in ectoine-treated cells was reduced compared with that in untreated cells (
Figure 3D and Suppl. Figure 2B). Interestingly, ectoine also suppressed cell proliferation and colony formation. Interestingly, mice injected long-term with HaCaT cells did not develop tumors.
2.4. Ectoine downregulated methylation-related genes.
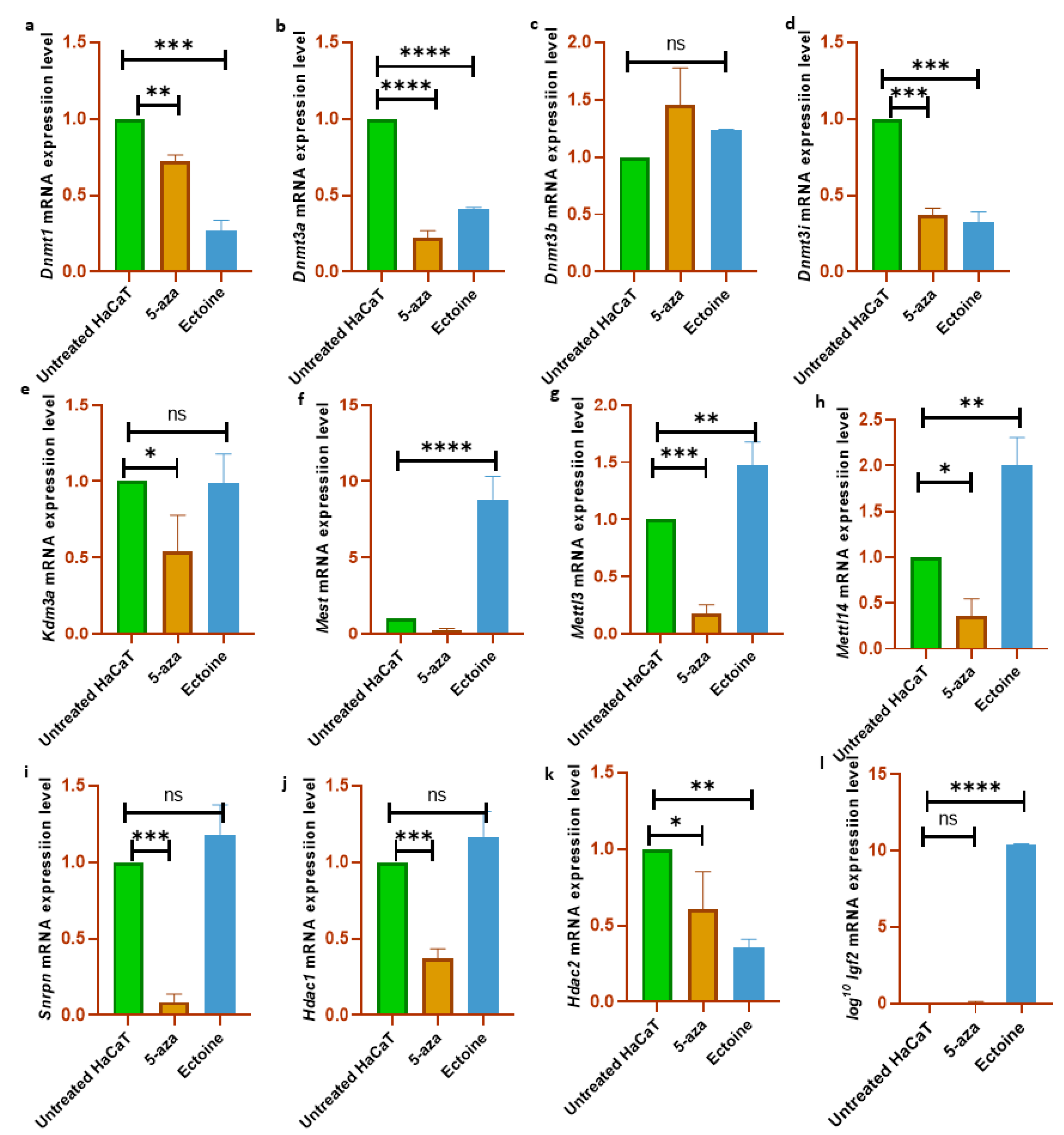
To validate the results of the global methylation analysis, the mRNA expression levels of several methylation-related genes and markers were analyzed (Dnmt1, Dnmt3a, Dnmt3b, Dnmt3i, Hdac1, Hdac2, Kdm3a, Mettl3, Mettl14, Igf2, Snrpn and Mest). We observed that Dnmt1, Dnmt3a, and Dnmt3i were downregulated in cells treated with ectoine for 30 days compared with untreated cells. Similarly, in cells treated with 5-aza-2-deoxycytidine, Dnmt1, Dnmt3a, and Dnmt3i were significantly downregulated compared to those in untreated cells. In contrast, Dnmt3b mRNA expression levels did not change with ectoine or 5-aza-2-deocycitine treatment. Hdac1, Kdm3a, and Snrpn mRNA levels were downregulated in 5-aza-2-deoxycytidine-but not in ectoine-treated cells. Interestingly, Mettl3, Mettl14, Igf2 and Mest mRNA expression levels were upregulated in ectoine-treated cells, whereas they were downregulated in 5-aza-2-deoxycytidine treated cells compared to untreated cells. Hdac2 mRNA levels were downregulated in ectoine-treated cells compared to 5-aza-2-deoxycytidine, and 5-aza-2-deoxycytidine was lower than that in untreated cells (
Figure 4A–K). Thus, ectoine regulates the expression of DNMTs and mRNA expression levels of other DNA methylation markers.
3. Discussion
Epigenetic modifications are primarily driven by DNA hypermethylation, which is strongly associated with cancer development and ageing. Although several epigenetic drugs have been developed to target cancer, several drawbacks prevent their application owing to serious adverse reactions such as HDACi [
1,
23]. Ectoine protects DNA from environmental stress and radiation by directly interacting with nearby water molecules [
44]. In the present study, we analyzed global DNA methylation and 5mC levels in ectoine-treated skin cells, using 5-aza-2-deoxycytidine as a positive control. We also studied the effects of ectoine on skin cell proliferation, tumorigenicity, and the mRNA expression of DNA methylation genes and markers. We observed global ectoine hypomethylation in the skin cells. These results could be explained by another study, which showed that ectoine prevented gene-5-protein (G5P) from bacteriophage Ff from binding to DNA by changing the protein conformation and forming a layer at a distance of 0.5 μm from DNA [
45]. A recent study showed that ectoine binds to DNA-binding G5P without disturbing the protein structure [
46]. This may be because ectoine reduces the binding of DNMTs to DNA strands without disturbing the structure of the protein. This was demonstrated by our GO enrichment analysis, which showed that ectoine-vs. untreated small molecule binding, nucleotide binding, and ribonucleotide binding-related DEGs were enriched. Our methylome analysis of ectoine-treated cells showed that ectoine significantly reduced methylation levels in almost all regions of the CpG island, except for the CGI region, which was consistent with the DMP analysis results. Colorectal cancer cells treated with curcumin showed demethylation at specific CpG loci, but not global hypomethylation, similar to cells treated with 5-aza-2-deoxycytidine [
47]. Similarly, our results showed that methylation distribution in the whole genome was reduced compared to that in untreated cells.
Our In vitro results demonstrated that ectoine reduced the percentage of global DNA methylation targeting 5-mC, similar to the results of bioinformatics analysis of methylome cells, especially the mCG element in skin cells. A previous study showed that higher 5-mC levels are due to higher DNMTs activity in skin cancer cells than in normal cells, which is involved in the silencing of genes that suppress skin tumor development [
48]. Furthermore, we observed that ectoine improved cell biological processes such as cell morphogenesis. We also observed that ectoine reduced the proliferation of skin cancer cells, but did not induce apoptosis. This is in agreement with a previous study showing that ectoine suppresses the proliferation of head and neck squamous cell carcinoma (HNSCC) [
27]. HaCaT cells are sensitive to conversion to tumor cells using different chemicals and under different conditions, such as temperature treatment [
49,
50,
51]. Interestingly, we observed that mice injected with HaCaT cells treated long-term with 140 mM ectoine for 24 weeks did not develop tumors, which demonstrates its safety.
Moreover, our results demonstrated that ectoine reduced the expression of Dnmt1, Dnmt3a, and Dnmt3i. As discussed above, this may be due to the layer formed by ectoine around the DNA or a change in DNMTs conformation. DNMTs are known to be associated with cancer development, especially hypermethylated tumor suppressor genes and genomic instability [
52]. In contrast, our results demonstrated that Hdac1, Kdm3a, and Snrpn mRNA levels were downregulated in 5-aza-2-deoxycytidine-but not in ectoine-treated cells. Interestingly, our results showed that Mettl3 and Mettl14, mRNA expression levels were upregulated in ectoine-treated cells and downregulated in cells treated with 5-aza-2-deoxycytidine. These two enzyme complexes are important for the repair of UV- and X-ray-exposed DNA-damaged sites and subsequent RNA methyl transfer [
53]. These results indicate that they did not interfere with the cell-critical proteins involved in radiation repair. This agrees with another strategy in which ectoine protects cells against UVA radiation by inhibiting the POMC and tyrosinase pathways in UVA-irradiated HaCaT cells [
28]. Moreover, we observed that the mRNA expression levels of the imprinted genes Igf2 and Mest were upregulated in ectoine-treated cells, whereas they were downregulated in 5-aza-2-deoxycytidine. One study showed that 9.6% of patients with oligospermia had MEST methylation; thus, MEST was significantly hypermethylated compared to the control group (3.5%, p< 0.001) [
54]. HaCaT cells were isolated from human male.
Senescence is a major factor in cancer, nondegenerative diseases, cardiovascular diseases, and several age-related diseases. Epigenetic factors, telomer reduction, DNA damage, and mitochondrial function are the primary drivers of aging [
55,
56]. The ability of ectoine to protect DNA, regulate DNA methylation, and suppress tumor development makes it a potential candidate for anti-aging medicine.
4. Materials and Methods
4.1. Cell cultures and drug treatments
Human HaCaT, SCC-9, and A-431 cells were obtained from the National Center for Cell Sciences (Shanghai, China). These cells were cultivated in Dulbecco’s Modified Eagle’s Medium-F-12 (DMEM-F12) medium enriched with 10% (v/v) decomplemented fetal bovine serum (FBS) (Invitrogen) and 100% 100X anti-mycotic/antibiotic solution containing Amphotericin B, Penicillin, Streptomycin (Gibco, USA). All the samples were incubated under microaerophilic conditions with 100% humidity and 5% CO2 at 37°C. For the HaCaT cell line, the medium was supplemented with 2 mM L-glutamine, 4.5 g/L glucose, whereas the SSC-9 cell culture was supplemented with hydrocortisone (Sigma, USA) at 400ng/ml and 1% sodium bicarbonate [
26]. SCC-9, HaCaT, and A-431 cells were treated with 140 mM (restricted concentration to be used in mouthwash [
27]) ectoine [
28](Sigma, USA) or 5 μM 5-aza-2-deoxycytidine, as modified from a previous study [
29]. Briefly, when the cell lines reached 30-40% confluency, they were treated with 5 μM 5-aza-2-deoxycytidine as a positive control and incubated for 72 h in all experiments. Ectoine dissolved 0.57 g in ectoine/ml of water to reach 4M concertation and 5-aza-deoxycytidine was dissolved 25 mg/ml in DMSO, sterilized it using a 0.22 μm syringe sterile filter, and stored at -20.
4.2. Whole genome bisulfite sequencing and methylation analysis
To investigate the possible effects of ectoine on DNA methylation of the DNA HaCaT cells were treated with ectoine for 24 weeks or with 5-aza-2-deoxycytidine for 72 h. The cells were trypsinized and sent to Wuhan Saiwei Biological Technology Co., Ltd. (Wuhan, China) for whole-genome bisulfite sequencing as previously described [
30]. Briefly, genomic DNA was extracted using a DNeasy kit (QIAGEN, Hilden, Germany) and randomly fragmented by sonication. The ends were repaired by adding methylated adaptors, isolating the correct fragments, and subjecting them to bisulfite PCR amplification. Finally, WGBS-seq DNA libraries were obtained and subjected to HiSeq. Before data analysis, low-quality sequenced data and linker sequences were removed using the Skewer tool [
31] followed by checking the quality control analysis of sequences was processed using FastQC [
32] for the comparison between sequences, the sequenced data were aligned to the human reference genome using the Bismark tool [
33] and sequence comparison was performed using bowtie2 [
34] and only the unique alignments were used for the subsequent analysis. The overall genome methylation and methylation levels of each element of the gene (upstream 2 K, 5-UTR, CDS, intron, 3-UTR, downstream 2 K, and intergenic region) were analyzed to cover the methylated C bases of each gene element. To cover the C bases in the CpG islands, the genome was divided into CpG islands (CGI), Nshelf, Nshore, Sshore, Sshelf, and others, and repeats of CG, CHG, and CHH were calculated. The repeat areas were annotated using ReRepeatMasker, the main categories were SINE, LINE, LTR, and other types of repeats, and the statistics of CG, CHG, and CHH were calculated. The methylated C region was determined from the comparison results of BisMark, using a binomial distribution test. Differentially methylated sites (DMP) and regions (DMR) were analyzed using the bioconductor package DSS [
35]. For gene ontology (GO) and KEGG functional enrichment analyses were performed (Suppl. File 1). The sequences were submitted to the NCBI BioSamples (SAMN34441851, SAMN34441852, and SAMN34441853), BioProject (PRJNA970079), and SRA (SUB13191204).
4.3. Colony formation assay
To study the effects of ectoine on HaCaT colony formation after prolonged treatment, we performed a clonogenic assay as described previously [
36]. Cells were seeded at 100 cells/well in a 6-well plate, treated with ectoine, and treated with culture medium. After 9 days of incubation, the cell colonies were washed with PBS, stained with 0.5% crystal violet in water (Sigma, USA), and washed with ddH2O.
4.4. Wound Healing Assay
The treated cell lines (1.6 x106 cells per well) were seeded in 6-well plates until the cells reached a confluent state, and the cell layer was scratched with a sterile 200-μl pipette tip. The medium and cell debris were aspirated and replaced with 2 ml of fresh DMEM-F12 without antibiotics or FBS. Images of the wounded areas were captured at 0 h and 24 h using a DM13000B light microscope (Leica Microsystems GmbH). The wound healing area was calculated as the difference in the area between 0 and 24 h divided by the height of the wound using the ImageJ software [
37].
4.5. MTT Assay
A modified version of a previous study was used to study the effects of ectoine on skin cell viability, a modified version of a previous study [
28]. SCC-9 and A-431 cells were seeded in 96 well plates at 7500 cells/well, treated with 140 mM ectoine and untreated cells in complete medium, and incubated for 72 h. The medium was removed carefully, and 20 µL of 5 mg/ml MTT was added to each well. Includes one set of wells with MTT, but no cells (control). The plate was incubated for 3.5 hours at 37 °C in a culture hood, the media was removed carefully, and 100 μl DMSO was added. The plate was covered with tinfoil, agitated on an orbital shaker for 30 min, and the absorbance was read at 590 nm with a reference filter of 620 nm using a Tecan microplate reader Infinite® F50 (Tecan, Switzerland).
4.6. Tumorigenicity in vivo experiments
Ectoine has been used in various skincare products. To study the possibility of ectoine-induced tumors in HaCaT keratinocytes, we treated cells with ectoine for 30 weeks and xenografted them into athymic nude mice. This experiment was modified from the previous studies [
38,
39]. Approximately 5 × 106 HaCaT cells treated with ectoine for 24 weeks were subcutaneously injected into the dorsal flank of athymic nude mice. Mice were divided (ectoine-treated group, 5-aza-2-deoxycytidine treated group and control groups). Ectoine was prepared according to the weight of the mice at 200 mg/kg in ultrapure water and administered every three days. Similar to deoxycytidine dissolved in DMSO 2.5ng/kg was injected intraperitoneally every three days and continued for 40 days. Mice were euthanized by CO2 asphyxiation, as described previously [
40,
41]. Briefly, the mice were kept in a CO2 chamber in small groups and CO2 gas cylinder medical grade A (3.5 L/min CO2 chamber) for 2-3 minutes until a lack of respiration and faded eyes were observed, followed by 1 min after respiration ceased. The experiments were performed at the Jiangsu University Animal Center, and the experimental procedures were performed in accordance with the principles of the Declaration of Helsinki regulations and guidelines, and approved by the Animal Ethics Committee of Jiangsu University.
4.7. DNA Methylation Analysis
To validate DNA methylation, HaCaT cells were treated with either ectoine or 5-aza-2-deoxycytidine and subjected to DNA methylation analysis as described previously [
42]. Briefly, genomic DNA was extracted from treated HaCaT cells and controls using Axygen® AxyPrep (Axygen, USA), according to the manufacturer’s instructions. DNA quality was checked using agarose gel, and the concentration was measured using a Nanodrop spectrophotometer (Thermo Scientific, USA). An amount of 100ng from each sample used genomic DNA per reaction to quantify the methylated DNA was quantified using a Methylated DNA Quantification Kit (Colorimetric) (Abcam, ab117128) according to the manufacturer’s instructions. The absorbance was measured using a Tecan microplate reader (Infinite® F50, Tecan, Switzerland) at 450 nm after 10 min.
4.8. Quantitative PCR and gene expression analysis.
qRT-PCR analysis was performed as previously described [
43]. Briefly, RNA was isolated from HaCaT cells treated with ectoine or 5-aza-2-deoxycytidine using the TRIzol reagent (Invitrogen, USA). Afterward, 3μg of RNA was converted to cDNA using SuperScript III (Invitrogen, USA) and TA primers, according to the manufacturer’s instructions. For qRT-PCR, 40 ng of the first transcribed DNA strand was amplified using SYBR Fast qPCR Mix (TaKaRa, Japan) with primers targeting
Dnmt1, Dnmt3a, Dnmt3b, Dnmt3i, Hdac1, Hdac2, Kdm3a, Mettl3, Mettl14, Igf2, Snrpn and
Mest genes, and
GAPDH as an internal control. Primer sequences used in this study are listed (Suppl. Table 1).
4.9. Statistical analysis
Statistical analyses were performed using one-way analysis of variance (ANOVA), followed by Tukey’s multiple comparison test. The data are presented as the mean ± standard error (of the mean) from three independent experiments.
5. Conclusions
Ectoine globally demethylates DNA, CpG islands, and genes, which is another advantage for ectoine therapeutic applications. DNA methylation was mild compared with that in skin cells treated with 5-aza-deoxycytidine. Furthermore, in vitro studies of skin cells treated with ectoine have shown mild suppression of skin cells and cancer cell proliferation. Interestingly, long-term treatment of skin cells with ectoine and its use in vivo did not induce tumorigenicity. Moreover, ectoine significantly regulated the expression of DNA methylation genes and markers. Future studies should focus on molecular mechanisms and biophysics to elucidate the manner in which ectoine binds to DNA methylation regulators and ultimately prevents DNA hypermethylation.
6. Patents
The patent related to this study has been filed waiting for the final results.
Author Contributions
Conceptualization, Majjid Qaria and Daochen Zhu; Data curation, Majjid Qaria, Chunyan Xu, Hu Ran and Roua A. Alsubki; Formal analysis, Majjid Qaria, Hu Ran and Sethupathy Sivasamy; Funding acquisition, Kotb A. Attia and Daochen Zhu; Investigation, Majjid Qaria, Chunyan Xu and Hu Ran; Methodology, Majjid Qaria and Chunyan Xu; Resources, Mohamed Ali; Software, Majjid Qaria and Chunyan Xu; Supervision, Daochen Zhu; Validation, Mohamed Ali and Sethupathy Sivasamy; Visualization, Majjid Qaria, Chunyan Xu and Roua A. Alsubki; Writing – original draft, Majjid Qaria; Writing – review & editing, Majjid Qaria, Chunyan Xu, Hu Ran, Roua A. Alsubki, Mohamed Ali, Sethupathy Sivasamy, Kotb A. Attia and Daochen Zhu.
Funding
This study was supported by the Key Research and Development Program of Jiangsu Province (Grant No. BE2021691), Natural Science Foundation of Jiangsu Province (Carbon Neutralization, BK20220003), and Jiangsu Collaborative Innovation Center of Technology and Material of Water Treatment, Suzhou University of Science and Technology, Suzhou 215009, China. We also extend our acknowledgment to the Researchers Supporting Project number (RSP-2023 R369), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
The experiments were performed at the Jiangsu University animal center and the experimental procedures were performed in accordance with the principles of the Declaration of Helsinki regulations and guidelines and approved by the Animal Ethics Committee of Jiangsu University.
Data Availability Statement
All data included in this study are available for public use.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Skvortsova, K.; Stirzaker, C.; Taberlay, P. , The DNA methylation landscape in cancer. Essays in biochemistry 2019, 63, 797–811. [Google Scholar] [PubMed]
- Becker, J.; Wittmann, C. , Microbial production of extremolytes—high-value active ingredients for nutrition, health care, and well-being. Curr. Opin. Biotechnol. 2020, 65, 118–128. [Google Scholar] [CrossRef]
- Galinski, E.A.; PFEIFFER, H.P.; Trüper, H.G. , 1, 4, 5, 6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid: A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. European Journal of Biochemistry 1985, 149, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Boas Lichty, K.E.; Gregory, G.J.; Boyd, E.F. , NhaR, LeuO, and H-NS Are Part of an Expanded Regulatory Network for Ectoine Biosynthesis Expression. Applied and Environmental Microbiology 2023, e00479–23. [Google Scholar] [CrossRef] [PubMed]
- Kadam, P.; Kajale, S.; Sharma, A.; Dhotre, D.; Barvkar, V.; Shouche, Y.; Zinjarde, S. , Whole-Genome Sequencing of the Tropical Marine Bacterium Nocardiopsis dassonvillei NCIM 5124, Containing the Ectoine Biosynthesis Gene Cluster ectABC. Microbiology Resource Announcements 2022, 11, e00435–22. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Lee, B.; Kim, J.A.; Kim, M.-S.; Kim, C.H. , Identification and characterization of an ectoine biosynthesis gene cluster from Aestuariispira ectoiniformans sp. nov., isolated from seawater. Microbiological Research 2022, 254, 126898. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, S.; Thume, K.; Wirgenings, M.; Pohnert, G. , Ectoine from bacterial and algal origin is a compatible solute in microalgae. Marine drugs 2020, 18, 42. [Google Scholar] [CrossRef]
- Azizah, M.; Pohnert, G. , Orchestrated Response of Intracellular Zwitterionic Metabolites in Stress Adaptation of the Halophilic Heterotrophic Bacterium Pelagibaca bermudensis. Marine Drugs 2022, 20, 727. [Google Scholar] [CrossRef]
- Zaccai, G.; Bagyan, I.; Combet, J.; Cuello, G.J.; Demé, B.; Fichou, Y.; Gallat, F.-X.; Josa, V.M.G.; von Gronau, S.; Haertlein, M. , Neutrons describe ectoine effects on water H-bonding and hydration around a soluble protein and a cell membrane. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Kolp, S.; Pietsch, M.; Galinski, E.A.; Gütschow, M. , Compatible solutes as protectants for zymogens against proteolysis. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 2006, 1764, 1234–1242. [Google Scholar] [CrossRef]
- Kanapathipillai, M.; Lentzen, G.; Sierks, M.; Park, C.B. , Ectoine and hydroxyectoine inhibit aggregation and neurotoxicity of Alzheimer’s β-amyloid. FEBS Lett. 2005, 579, 4775–4780. [Google Scholar] [CrossRef] [PubMed]
- Unfried, K.; Sydlik, U.; Bierhals, K.; Weissenberg, A.; Abel, J. , Carbon nanoparticle-induced lung epithelial cell proliferation is mediated by receptor-dependent Akt activation. American Journal of Physiology-Lung Cellular and Molecular Physiology 2008, 294, L358–L367. [Google Scholar]
- Eichel, A.; Wittig, J.; Shah-Hosseini, K.; Mösges, R. , A prospective, controlled study of SNS01 (ectoine nasal spray) compared to BNO-101 (phytotherapeutic dragées) in patients with acute rhinosinusitis. Curr. Med. Res. Opin. 2013, 29, 739–746. [Google Scholar] [CrossRef]
- Bilstein, A.; Heinrich, A.; Rybachuk, A.; Mösges, R. , Ectoine in the Treatment of Irritations and Inflammations of the Eye Surface. BioMed Research International 2021, 2021. [Google Scholar] [CrossRef]
- Kauth, M.; Trusova, O.V. , Topical Ectoine Application in Children and Adults to Treat Inflammatory Diseases Associated with an Impaired Skin Barrier: A Systematic Review. Dermatology and Therapy 2022, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wellmerling, J.; Rayner, R.E.; Chang, S.W.; Kairis, E.L.; Kim, S.H.; Sharma, A.; Boyaka, P.N.; Cormet-Boyaka, E. , Targeting the EGFR-ERK axis using the compatible solute ectoine to stabilize CFTR mutant F508del. FASEB J. 2022, 36, e22270. [Google Scholar] [CrossRef]
- Sun, W.; Zang, L.; Shu, Q.; Li, X. , From development to diseases: the role of 5hmC in brain. Genomics 2014, 104, 347–351. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. , Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef]
- Lyko, F. , The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nature Reviews Genetics 2018, 19, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, K.; Datta, J.; Majumder, S.; Bai, S.; Kutay, H.; Motiwala, T.; Jacob, S.T. , 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Molecular and cellular biology 2005, 25, 4727–4741. [Google Scholar] [CrossRef]
- Shetty, M.G.; Pai, P.; Deaver, R.E.; Satyamoorthy, K.; Babitha, K.S. , Histone deacetylase 2 selective inhibitors: A versatile therapeutic strategy as next generation drug target in cancer therapy. Pharmacological Research 2021, 170, 105695. [Google Scholar] [CrossRef]
- Yeon, M.; Kim, Y.; Jung, H.S.; Jeoung, D. , Histone deacetylase inhibitors to overcome resistance to targeted and immuno therapy in metastatic melanoma. Frontiers in cell and developmental biology 2020, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Yang, Y.; Li, W.; Yang, H.; Zhang, Y.; Ge, B.; Zhang, S.; Du, G.; Wang, J. , Recent advances in epigenetic anticancer therapeutics and future perspectives. Frontiers in Genetics 2022, 13, 3658. [Google Scholar] [CrossRef]
- Xu, Z.; Ma, A.; Ma, Y.C. , RNA methylation preserves ES cell identity by chromatin silencing of retrotransposons. Signal Transduction and Targeted Therapy 2021, 6, 258. [Google Scholar] [CrossRef]
- Huntriss, J.D.; Hemmings, K.E.; Hinkins, M.; Rutherford, A.J.; Sturmey, R.G.; Elder, K.; Picton, H.M. , Variable imprinting of the MEST gene in human preimplantation embryos. European Journal of Human Genetics 2013, 21, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Rieckmann, T.; Gatzemeier, F.; Christiansen, S.; Rothkamm, K.; Münscher, A. , The inflammation-reducing compatible solute ectoine does not impair the cytotoxic effect of ionizing radiation on head and neck cancer cells. Scientific reports 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rieckmann, T.; Gatzemeier, F.; Christiansen, S.; Rothkamm, K.; Münscher, A. , The inflammation-reducing compatible solute ectoine does not impair the cytotoxic effect of ionizing radiation on head and neck cancer cells. Scientific Reports 2019, 9, 6594. [Google Scholar] [CrossRef]
- Hseu, Y.-C.; Chen, X.-Z.; Vudhya Gowrisankar, Y.; Yen, H.-R.; Chuang, J.-Y.; Yang, H.-L. , The skin-whitening effects of ectoine via the suppression of α-MSH-stimulated melanogenesis and the activation of antioxidant Nrf2 pathways in UVA-irradiated keratinocytes. Antioxidants 2020, 9, 63. [Google Scholar] [CrossRef]
- Yoshinaga-Sakurai, K.; Rossman, T.G.; Rosen, B.P. , Regulation of arsenic methylation: identification of the transcriptional region of the human AS3MT gene. Cell Biology and Toxicology 2022, 38, 765–780. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M. , Human DNA methylomes at base resolution show widespread epigenomic differences. nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. , Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC bioinformatics 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Andrews, S. , FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom: 2010.
- Krueger, F.; Andrews, S.R. , Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. , Fast gapped-read alignment with Bowtie 2. Nat Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Park, Y.; Wu, H. , Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 2016, 32, 1446–1453. [Google Scholar] [CrossRef]
- Strus, P.; Borensztejn, K.; Szczepankiewicz, A.A.; Lisiecki, K.; Czarnocki, Z.; Nieznanska, H.; Wojcik, C.; Bialy, L.P.; Mlynarczuk-Bialy, I. , Novel podophyllotoxin and benzothiazole derivative induces transitional morphological and functional changes in HaCaT cells. Toxicology in Vitro 2021, 73, 105144. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. , ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Alexander, E.T.; Mariner, K.; Borodyanskaya, Y.; Minton, A.; Gilmour, S.K. , Polyamine-stimulation of arsenic-transformed keratinocytes. Carcinogenesis 2019, 40, 1042–1051. [Google Scholar] [CrossRef]
- Merckx, C.; Zschüntzsch, J.; Meyer, S.; Raedt, R.; Verschuere, H.; Schmidt, J.; De Paepe, B.; De Bleecker, J.L. , Exploring the Therapeutic Potential of Ectoine in Duchenne Muscular Dystrophy: Comparison with Taurine, a Supplement with Known Beneficial Effects in the mdx Mouse. International Journal of Molecular Sciences 2022, 23, 9567. [Google Scholar] [CrossRef]
- Keeter, W.C.; Moriarty, A.K.; Akers, R.; Ma, S.; Mussbacher, M.; Nadler, J.L.; Galkina, E.V. , Neutrophil-specific STAT4 deficiency attenuates atherosclerotic burden and improves plaque stability via reduction in neutrophil activation and recruitment into aortas of Ldlr (-/-) mice. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yamaga, S.; Murao, A.; Ma, G.; Brenner, M.; Aziz, M.; Wang, P. , Radiation upregulates macrophage TREM-1 expression to exacerbate injury in mice. Front Immunol 2023, 14, 1151250. [Google Scholar] [CrossRef]
- Schmidt, S.; Luecken, M.D.; Trümbach, D.; Hembach, S.; Niedermeier, K.M.; Wenck, N.; Pflügler, K.; Stautner, C.; Böttcher, A.; Lickert, H.; et al. Primary cilia and SHH signaling impairments in human and mouse models of Parkinson’s disease. Nat Commun 2022, 13, 4819. [Google Scholar] [CrossRef]
- Qaria, M.A.; Kumar, N.; Hussain, A.; Qumar, S.; Doddam, S.N.; Sepe, L.P.; Ahmed, N., Roles of Cholesteryl-α-Glucoside Transferase and Cholesteryl Glucosides in Maintenance of Helicobacter pylori Morphology, Cell Wall Integrity, and Resistance to Antibiotics. mBio 2018, 9, (6).
- Hahn, M.B.; Smales, G.J.; Seitz, H.; Solomun, T.; Sturm, H. , Ectoine interaction with DNA: influence on ultraviolet radiation damage. Physical Chemistry Chemical Physics 2020, 22, 6984–6992. [Google Scholar] [CrossRef]
- Hahn, M.B.; Solomun, T.; Wellhausen, R.; Hermann, S.; Seitz, H.; Meyer, S.; Kunte, H.-J. r.; Zeman, J.; Uhlig, F.; Smiatek, J. , Influence of the compatible solute ectoine on the local water structure: implications for the binding of the protein G5P to DNA. The Journal of Physical Chemistry B 2015, 119, 15212–15220. [Google Scholar] [CrossRef]
- Hallier, D.C.; Smales, G.J.; Seitz, H.; Hahn, M.B. , Bio-SAXS of single-stranded DNA-binding proteins: radiation protection by the compatible solute ectoine. Physical Chemistry Chemical Physics 2023. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Balaguer, F.; Shen, Y.; Lozano, J.J.; Leung, H.-C. E.; Boland, C.R.; Goel, A. , Curcumin modulates DNA methylation in colorectal cancer cells. PloS one 2013, 8, e57709. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, V.; Vaid, M.; Tollefsbol, T.O.; Katiyar, S.K. , RETRACTED: Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis 2011, 32, 597–604. [Google Scholar] [CrossRef]
- Boukamp, P.; Popp, S.; Bleuel, K.; Tomakidi, E.; Bürkle, A.; Fusenig, N.E. , Tumorigenic conversion of immortal human skin keratinocytes (HaCaT) by elevated temperature. Oncogene 1999, 18, 5638–5645. [Google Scholar] [CrossRef] [PubMed]
- Valerio, H.P.; Ravagnani, F.G.; Ronsein, G.E.; Di Mascio, P. , A single dose of Ultraviolet-A induces proteome remodeling and senescence in primary human keratinocytes. Scientific Reports 2021, 11, 23355. [Google Scholar] [CrossRef]
- Yuan, P.; Dou, G.; Liu, T.; Guo, X.; Bai, Y.; Chu, D.; Liu, S.; Chen, X.; Jin, Y. , On-demand manipulation of tumorigenic microenvironments by nano-modulator for synergistic tumor therapy. Biomaterials 2021, 275, 120956. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. , DNA methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef]
- Yu, D.; Horton, J.R.; Yang, J.; Hajian, T.; Vedadi, M.; Sagum, C.A.; Bedford, M.T.; Blumenthal, R.M.; Zhang, X.; Cheng, X. , Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic acids research 2021, 49, 11629–11642. [Google Scholar] [CrossRef]
- Poplinski, A.; Tüttelmann, F.; Kanber, D.; Horsthemke, B.; Gromoll, J. , Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. International journal of andrology 2010, 33, 642–649. [Google Scholar] [PubMed]
- Field, A.E.; Robertson, N.A.; Wang, T.; Havas, A.; Ideker, T.; Adams, P.D. , DNA methylation clocks in aging: categories, causes, and consequences. Mol. Cell 2018, 71, 882–895. [Google Scholar] [CrossRef] [PubMed]
- McHugh, D.; Gil, J. , Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 2018, 217, 65–77. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).