Submitted:
06 August 2023
Posted:
08 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
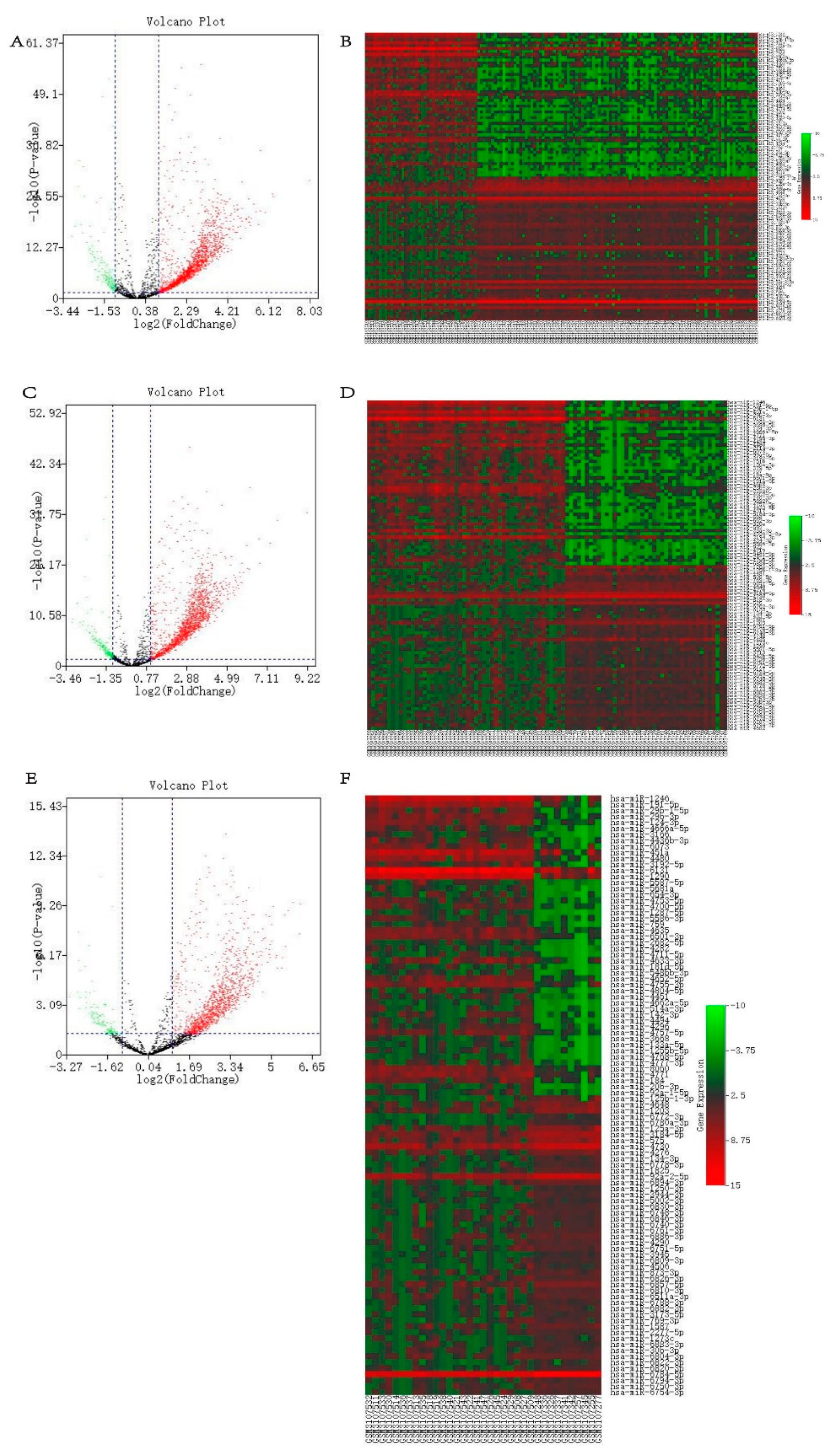
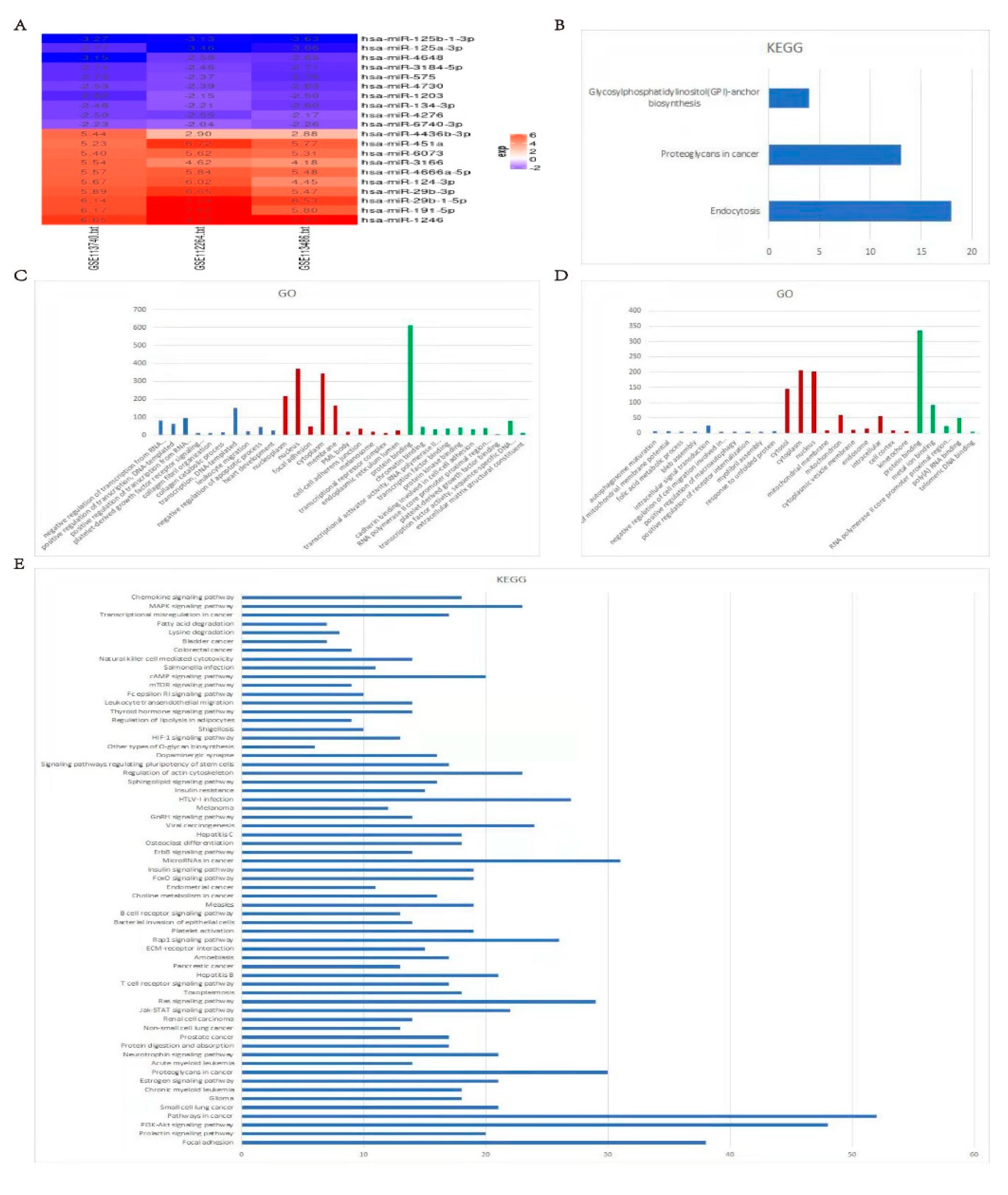
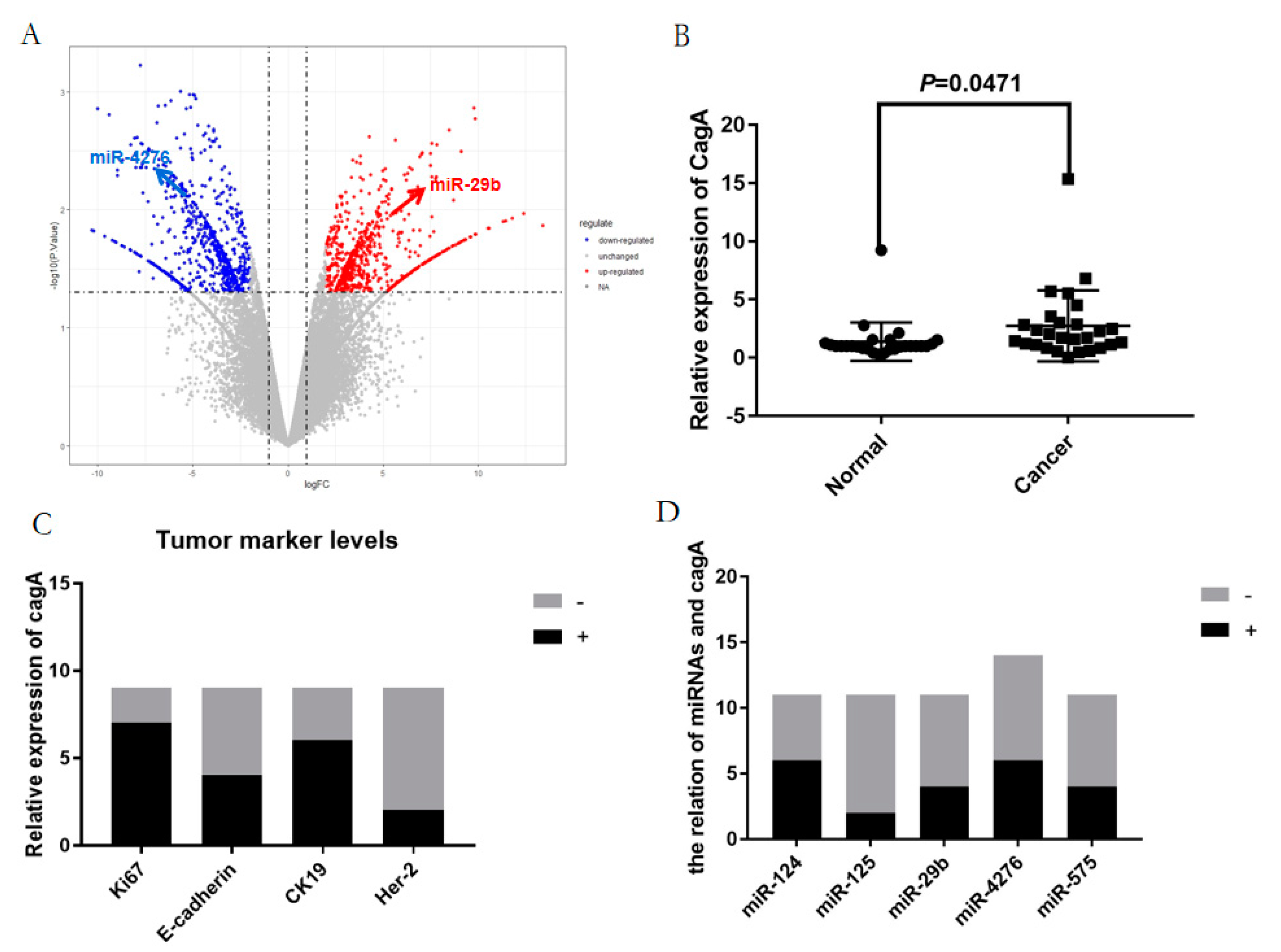
2.1. The differentially expressed blood miRNAs in gastric carcinoma
2.2. The prediction of target genes directly silenced by the most differentially expressed miRNAs and the enrichment results of GO and KEGG
2.3. The analysis of the proteins interaction network and selection of the key blood miRNAs with the correlant target genes
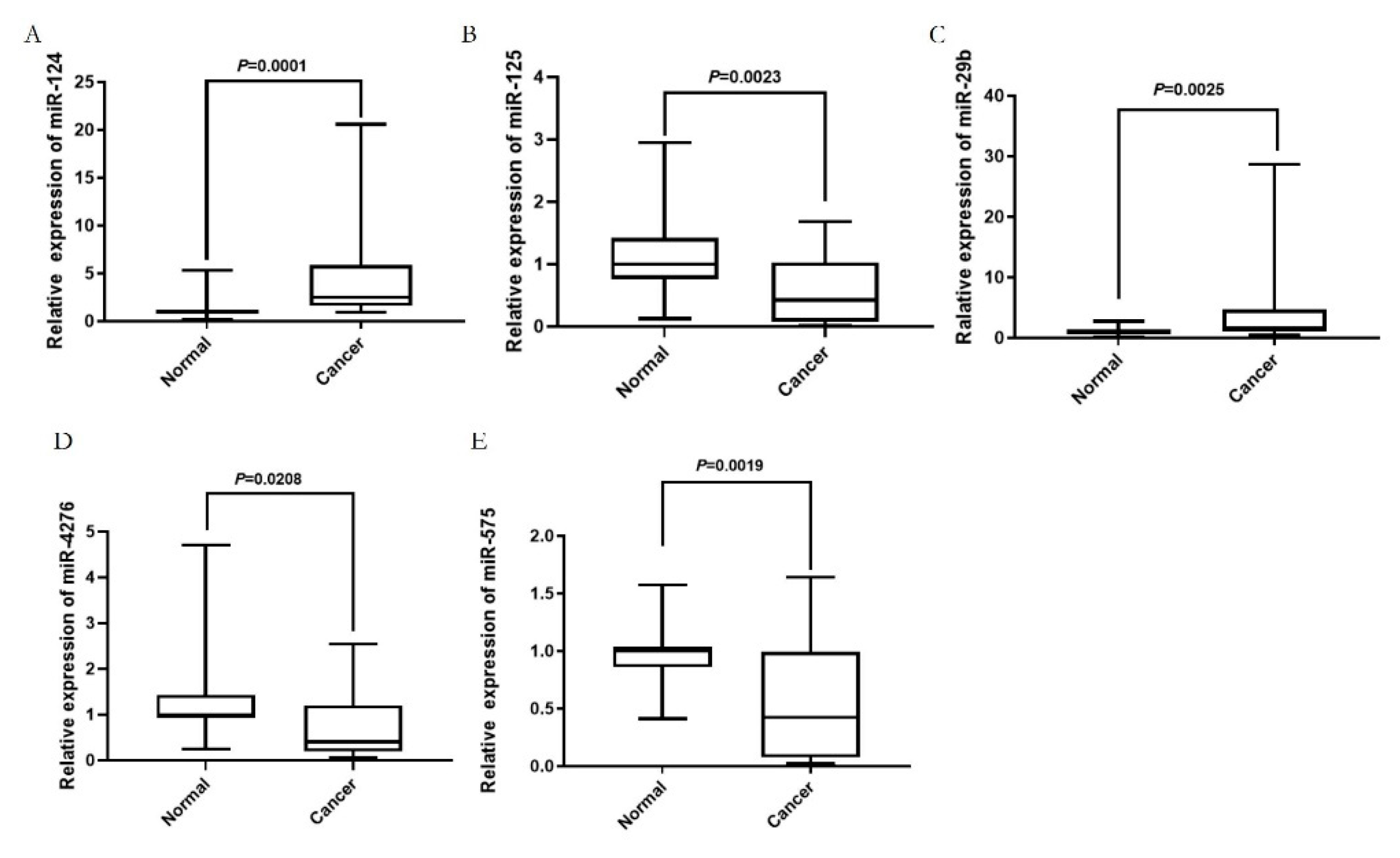
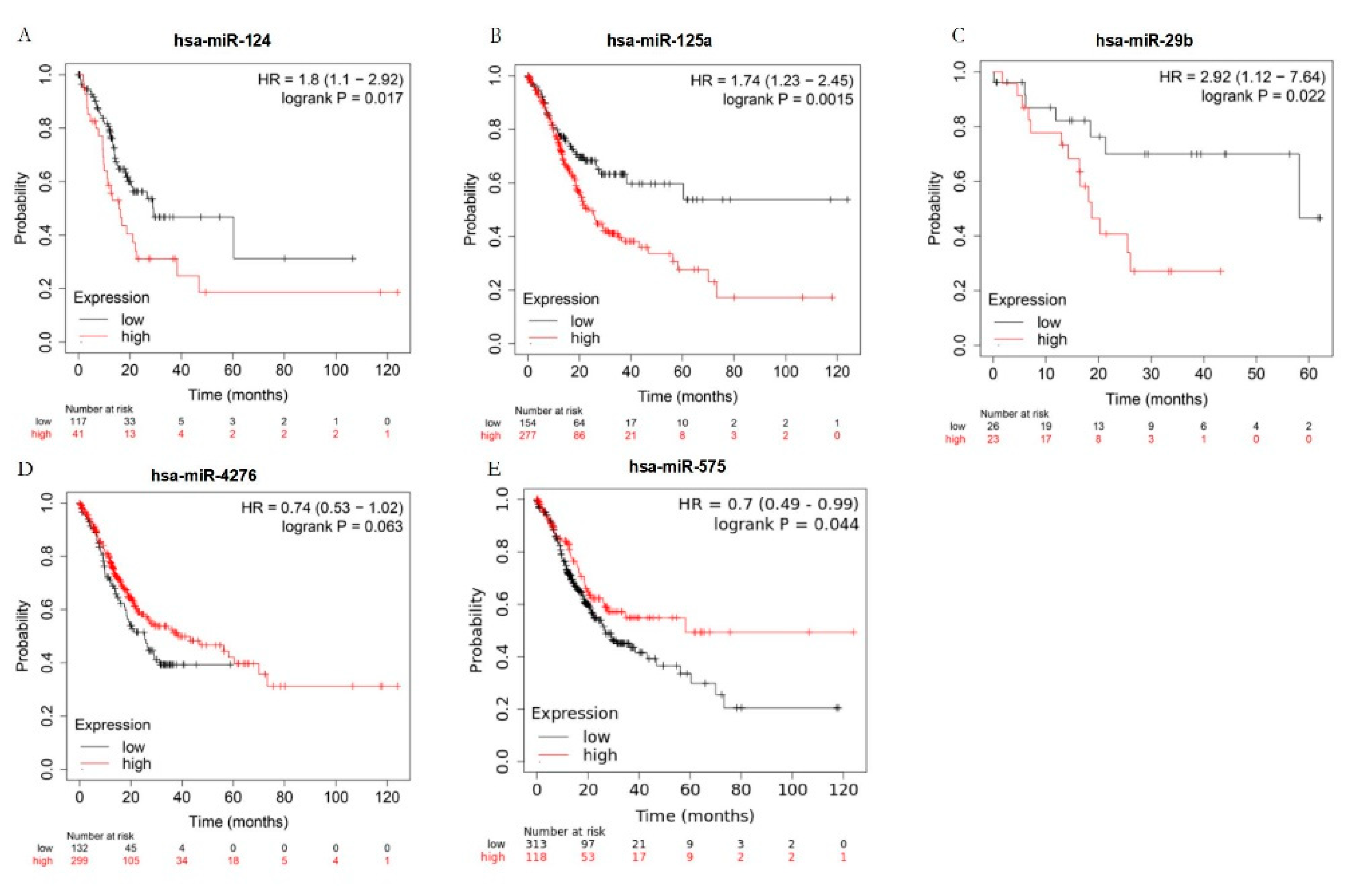
2.4. The key blood miRNAs with the correlant target genes were detected by the examination of the expression levels in human blood species and the survival curve analysis.
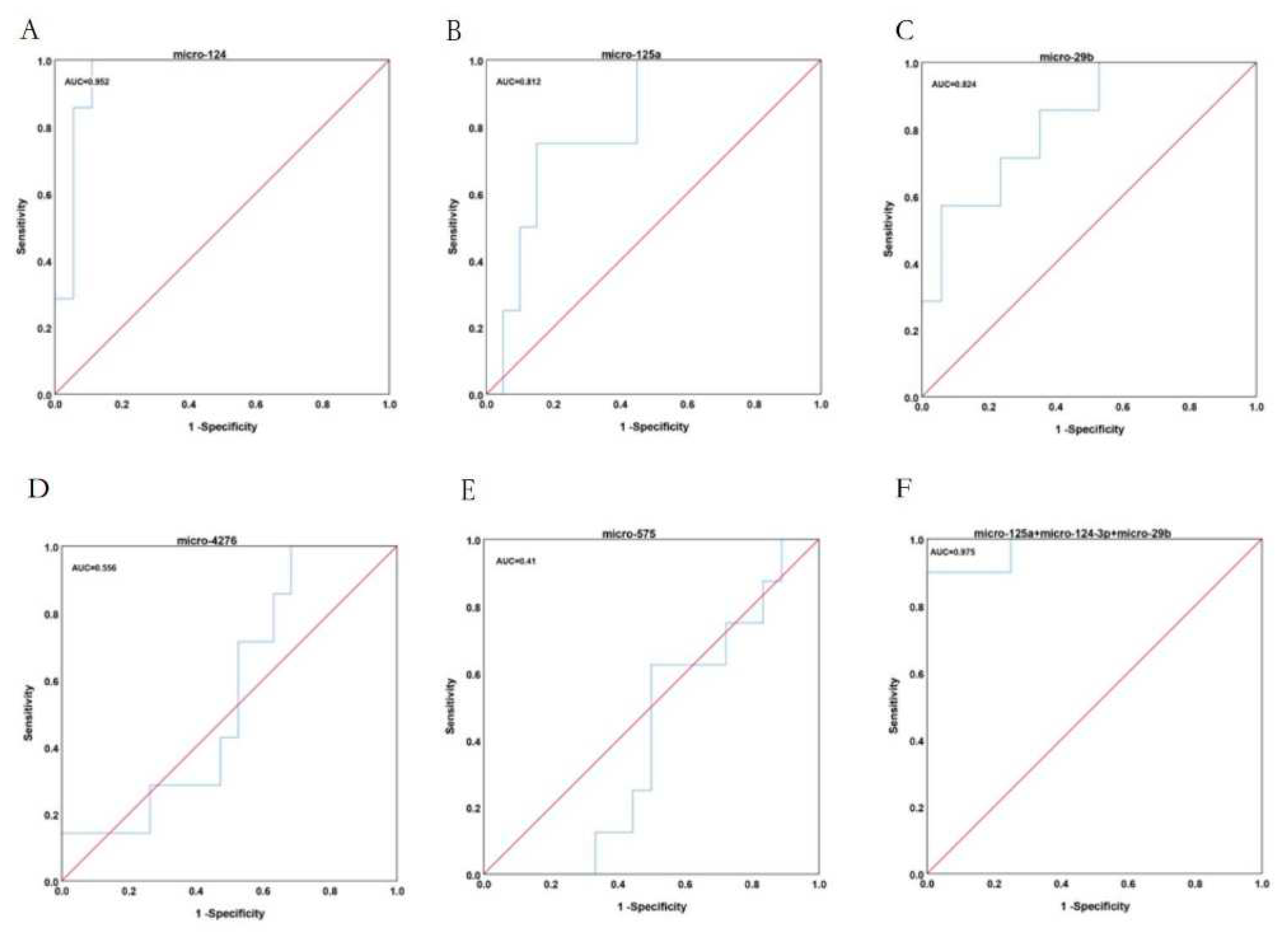
2.5. The diagnostic value of key blood miRNAs in patients with GC.
3. Discussion
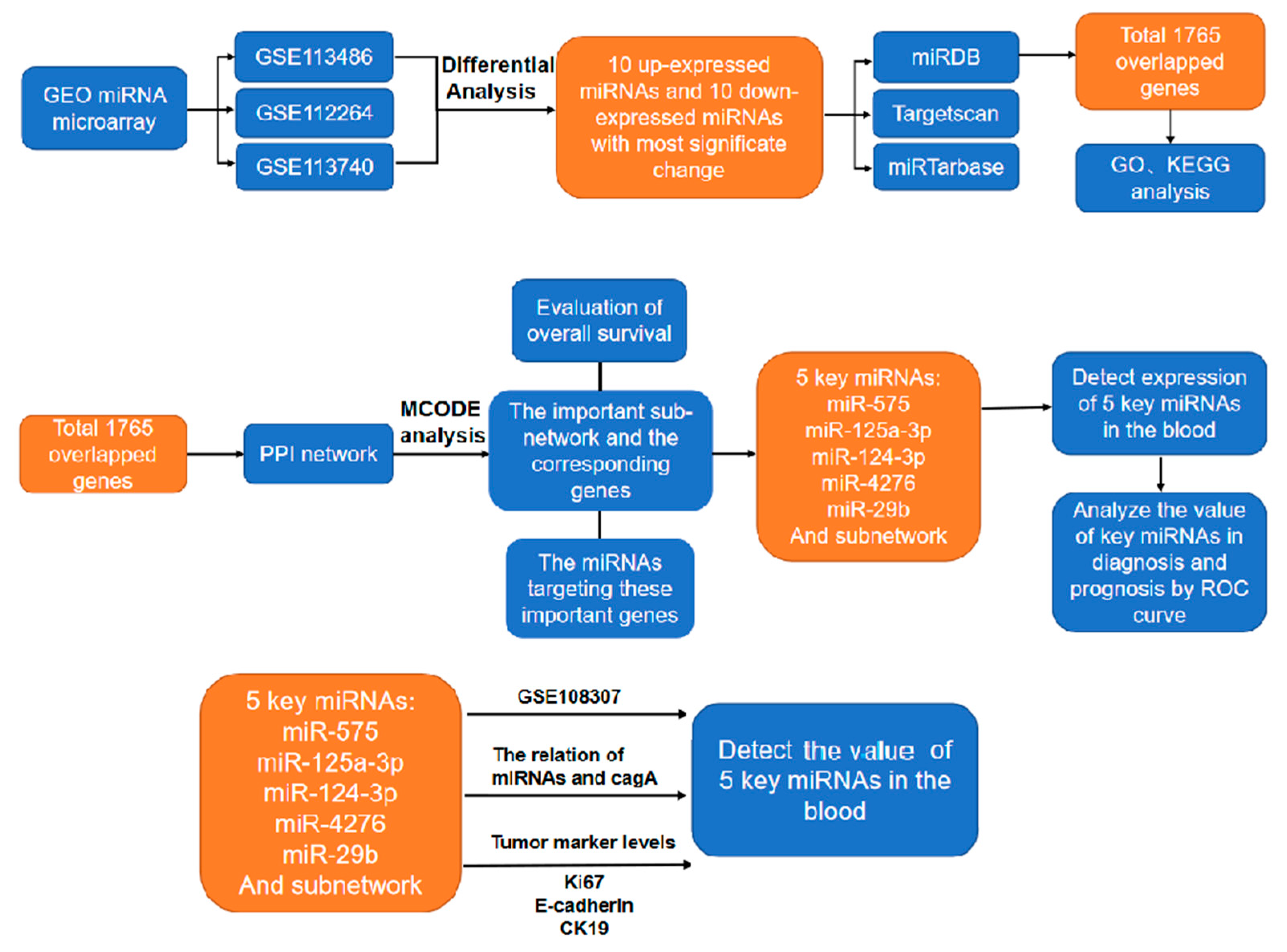
4. Materials and Methods
4.1. Data selection
4.2. Preliminary screening of differentially expressed miRNAs and predicted target genes
4.3. Functional enrichment of differentially expressed genes
4.4. Protein-protein interaction network (PPI network)
4.5. Selection and survival curve analysis of the key miRNAs and relative target genes
4.6. The human blood species and qRT-PCR determination
4.7. ROC analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin. Med J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, G.; Hu, C. Molecular Classification of Gastric Adenocarcinoma. Gastroenterol. Res. 2019, 12, 275–282. [Google Scholar] [CrossRef]
- Hrovatin, K.; Kunej, T. Classification of miRNA-related sequence variations. Epigenomics 2018, 10, 463–481. [PubMed]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Su, F.; Gao, Z.; Liu, Y.; Zhou, G.; Cui, Y.; Deng, C.; Liu, Y.; Zhang, Y.; Ma, X.; Wang, Y.; et al. Integrated Tissue and Blood miRNA Expression Profiles Identify Novel Biomarkers for Accurate Non-Invasive Diagnosis of Breast Cancer: Preliminary Results and Future Clinical Implications. Genes 2022, 13, 1931. [Google Scholar] [CrossRef]
- Scionti F, Arbitrio M, Caracciolo D, Pensabene L, Tassone P, Tagliaferri P, Di Martino MT. Integration of DNA Microarray with Clinical and Genomic Data. Methods Mol Biol. 2022, 2401, 239–248. [Google Scholar]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2014, 13, 8–17. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Van Roosbroeck, K.; Calin, G.A. Cancer Hallmarks and MicroRNAs: The Therapeutic Connection. Adv Cancer Res. 2017, 135, 119–149. [Google Scholar] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, N.; Constantinou, C. Recent Developments in Diagnostic and Prognostic Biomarkers for Colorectal Cancer: A Narrative Review. Oncology 2023, 101, 675–684. [Google Scholar] [CrossRef]
- Van den Broek, D.; Groen, H.J.M. Screening approaches for lung cancer by blood-based biomarkers: Challenges and opportunities. Tumour Biol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Aalami, A.H.; Aalami, F.; Sahebkar, A.H. Gastric Cancer and Circulating microRNAs: An Updated Systematic Review and Diagnostic Meta-Analysis. Curr. Med. Chem. 2023, 30, 3798–3814. [Google Scholar] [CrossRef]
- Jelski, W.; Mroczko, B. Molecular and Circulating Biomarkers of Gastric Cancer. Int. J. Mol. Sci. 2022, 23, 7588. [Google Scholar] [CrossRef]
- Huang, F.; Wang, M.; Yang, T.; Cai, J.; Zhang, Q.; Sun, Z.; Wu, X.; Zhang, X.; Zhu, W.; Qian, H.; et al. Gastric cancer-derived MSC-secreted PDGF-DD promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2014, 140, 1835–1848. [Google Scholar] [CrossRef]
- Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478. [Google Scholar] [CrossRef]
- Zhang, H.; Schaefer, A.; Wang, Y.; Hodge, R.G.; Blake, D.R.; Diehl, J.N.; Papageorge, A.G.; Stachler, M.D.; Liao, J.; Zhou, J.; Wu, Z.; Akarca, F.G.; de Klerk, L.K.; Derks, S.; Pierobon, M.; Hoadley, K.A.; Wang, T.C.; Church, G.; Wong, K.K.; Petricoin, E.F.; Cox, A.D.; Lowy, D.R.; Der, C.J.; Bass, A.J. Gain-of-Function RHOA Mutations Promote Focal Adhesion Kinase Activation and Dependency in Diffuse Gastric Cancer. Cancer Discov. 2020, 10, 288–30. [Google Scholar] [CrossRef]
- Hu, M.; Zhu, S.; Xiong, S.; Xue, X.; Zhou, X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol. Rep. 2019, 41, 1439–1454. [Google Scholar] [CrossRef]
- Li, Q.; Liu, S.; Yan, J.; Sun, M.-Z.; Greenaway, F.T. The potential role of miR-124-3p in tumorigenesis and other related diseases. Mol. Biol. Rep. 2021, 48, 3579–3591. [Google Scholar] [CrossRef]
- Romero-López, M.J.; Jiménez-Wences, H.; Cruz-De la Rosa, M.I.; Román-Fernández, I.V.; Fernández-Tilapa, G. miR-23b-3p, miR-124-3p and miR-218-5p Synergistic or Additive Effects on Cellular Processes That Modulate Cervical Cancer Progression? A Molecular Balance That Needs Attention. Int J Mol Sci. 2022, 23, 13551. [Google Scholar] [CrossRef]
- Peng, B.; Guo, C.; Guan, H.; Liu, S.; Sun, M.-Z. Annexin A5 as a potential marker in tumors. Clin. Chim. Acta 2014, 427, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dai, Y.; Zhao, Y.; Li, M.; Zhang, J.; Ci, Y.; Wang, H.; Li, X. AnnexinA5 Might Suppress the Phenotype of Human Gastric Cancer Cells via ERK Pathway. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, G.; Wu, W.; Zhang, J.; Xia, X.; Jiang, T.; Cao, J.; Huang, K.; Qiu, Z.; Huang, C. Decreased Expression of Caveolin-1 and E-Cadherin Correlates with the Clinicopathologic Features of Gastric Cancer and the EMT Process. Recent Patents Anti-Cancer Drug Discov. 2016, 11, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Rong, Z.; Zhang, J.; Zhu, Z.; Yu, Z.; Li, T.; Fu, Z.; Qiu, Z.; Huang, C. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol. Cancer 2020, 19, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.K.; Wang, Z.; Li, G. MicroRNA-125 in immunity and cancer. Cancer Lett. 2019, 454, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Potenza, N. Antiproliferative Activity of microRNA-125a and its Molecular Targets. MicroRNA 2019, 8, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Amiri-Dashatan, N.; Koushki, M.; Naghi-Zadeh, M.; Razzaghi, M.R.; Shalmani, H.M. Prognostic value of microRNA-125a/b family in patients with gastric cancer: a meta-analysis. . 2021, 14, S1–S9. [Google Scholar]
- Boi, M.; Zucca, E.; Inghirami, G.; Bertoni, F. PRDM1/BLIMP1: a tumor suppressor gene in B and T cell lymphomas. Leuk Lymphoma. 2015, 56, 1223–8. [Google Scholar]
- Shen, L.; Chen, Q.; Yang, C.; Wu, Y.; Yuan, H.; Chen, S.; Ou, S.; Jiang, Y.; Huang, T.; Ke, L.; et al. Role of PRDM1 in Tumor Immunity and Drug Response: A Pan-Cancer Analysis. Front. Pharmacol. 2020, 11, 593195. [Google Scholar] [CrossRef] [PubMed]
- Martin-Arruti, M.; Vaquero, M.; de Otazu, R.D.; Zabalza, I.; Ballesteros, J.; Roncador, G.; García-Orad, A. Bcl-2 and BLIMP-1 expression predict worse prognosis in gastric diffuse large B cell lymphoma (DLCBL) while other markers for nodal DLBCL are not useful. Histopathology 2012, 60, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Shim, H.G.; Hwang, T.; Kim, H.; Kang, S.H.; Dho, Y.S.; Park, S.H.; Kim, S.J.; Park, C.K. Restoration of miR-29b-3p exerts anti-cancer effects on glioblastoma. Cancer Cell Int. 2017, 17, 104. [Google Scholar] [CrossRef]
- Ulivi, P.; Canale, M.; Passardi, A.; Marisi, G.; Valgiusti, M.; Frassineti, G.L.; Calistri, D.; Amadori, D.; Scarpi, E. Circulating Plasma Levels of miR-20b, miR-29b-3p and miR-155 as Predictors of Bevacizumab Efficacy in Patients with Metastatic Colorectal Cancer. Int J Mol Sci. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Amirian, M.; Jafari-Nozad, A.M.; Darroudi, M.; Farkhondeh, T.; Samarghandian, S. Overview of the miR-29 family members' function in breast cancer. Int. J. Biol. Macromol. 2023, 230, 123280. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Li, Z.; Zheng, S.; Wang, Z.; Li, W.; Bi, Z.; Li, L.; Jiang, Y.; Luo, Y.; Lin, Q.; Fu, Z.; Rufu, C. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med. 2018, 22, 655–667. [Google Scholar] [CrossRef]
- Bloomfield, J.; Sabbah, M.; Castela, M.; Mehats, C.; Uzan, C.; Canlorbe, G. Clinical Value and Molecular Function of Circulating MicroRNAs in Endometrial Cancer Regulation: A Systematic Review. Cells 2022, 11, 1836. [Google Scholar] [CrossRef]
- Guo, X.; Li, K.; Jiang, W.; Hu, Y.; Xiao, W.; Huang, Y.; Feng, Y.; Pan, Q.; Wan, R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol. Cancer 2020, 19, 1–19. [Google Scholar] [CrossRef]
- Othumpangat, S.; Noti, J.D.; Beezhold, D.H. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 2014, 468-470, 256–264. [Google Scholar] [CrossRef]
- Zhang, S.; Zang, D.; Cheng, Y.; Li, Z.; Yang, B.; Guo, T.; Liu, Y.; Qu, X.; Che, X. Identification of Key Gene and Pathways for the Prediction of Peritoneal Metastasis of Gastric Cancer by Co-expression Analysis. J. Cancer 2020, 11, 3041–3051. [Google Scholar] [CrossRef]
- Ma, X.; Wang, J.; Zhao, M.; Huang, H.; Wu, J. Increased expression of methionine sulfoxide reductases B3 is associated with poor prognosis in gastric cancer. Oncol. Lett. 2019, 18, 465–471. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




