1. Introduction
As an essential amino acid and a precursor of histamine, histidine plays a crucial role in human growth, metal ion transport [
1], and neuromodulation [
2]. Abnormal levels of histidine or histidine-rich proteins have been implicated in various diseases. Elevated levels of histidine or histidine-rich proteins have been associated with phenylketonuria [
3], malaria [
4], and thrombotic diseases [
5]. Conversely, low levels of histidine are linked to conditions such as rheumatoid arthritis [
6], epilepsy [
7], chronic kidney disease [
8], and advanced cirrhosis [
9]. For instance, in muscle tissue cells of mice with renal failure, histidine concentration decreased from 1.5 μM to 0.9 μM [
10]. Therefore, it is essential to not only monitor abnormally high histidine levels in living cells but also to have the ability to sensitively detect and signal histidine deficiency or low levels within cells. Such detection can provide valuable insights into the mechanisms of disease development and aid in evaluating the impact of drugs on cellular metabolism. Additionally, it can be used to assess cellular protein supply and nitrogen metabolism.
Various conventional methods, such as chromatography [
11], mass spectrometry [
12], electrochemical analysis [
13], and capillary electrophoresis [
14], have been reported for measuring histidine concentration. However, these destructive techniques are unable to assess histidine levels in living organisms. To address this limitation, several chemical probes have been developed for monitoring histidine in vivo. For example, Un et al. introduced a naphthalene dimaleimide-based fluorescent probe, NPC, capable of selectively detecting histidine in aqueous solutions and enabling intracellular imaging [
15]. Zhu et al. designed a nitrogen-doped carbon nanoparticle (N-CNP) fluorescent probe that offers rapid and selective detection of histidine, suitable for imaging analysis in living cells [
16]. However, their sensitivity is currently restricted to detecting exogenously added histidine. Mao et al. developed a fluorescent probe with chemical selectivity and enantioselectivity, enabling the determination of histidine concentration and enantiomeric composition through fluorescent responses at two different excitation wavelengths [
17]. Despite these advancements, the sensitivity of the probe remains inadequate for detecting endogenous histidine levels.
In addition to the aforementioned chemical sensors, some biosensors have also been developed for fluorescence imaging of histidine in living cells. Hu et al. engineered a genetically encoded fluorescent sensor that utilizes the bacterial periplasmic protein HisJ. Binding of histidine induces conformational changes in HisJ, resulting in alterations in fluorescence intensity when fused with the fluorescent protein cpYFP. This sensor achieves a specific response to histidine with high spatial and temporal resolution [
18]. Corte et al. investigated the structure of the LysG transcription factor from Bacillus subtilis, which naturally binds to three basic amino acids. By mutating LysG at position A219 to L, its binding affinity to L-lysine is reduced while enhancing its specificity to L-histidine. This modification promotes the expression of downstream
EYFP (enhanced yellow fluorescent protein) gene upon binding to L-histidine, thereby creating a L-histidine-sensitive biosensor based on LysG that can screen high-yield strains of L-histidine [
19]. However, these biosensors based on protein-histidine interactions are relatively insensitive to low-level detection of histidine and cannot actively respond to histidine deficiency.
Salmonella is a gram-negative intracellular bacterium known for its ability to invade various host cells, including epithelial cells and macrophages [
20,
21]. Within Salmonella, the biosynthesis of histidine is regulated by the
histidine (
his)operon, which consists of eight genes encoding sequential enzymes responsible for converting phosphoribosyl pyrophosphate (PRPP) into L-histidine [
22]. The transcriptional activity of the
his operon is negatively regulated by the intracellular concentration of histidine in the bacterium. The
his operon consists of an initial promoter known as
hisp1, a leader region, and an attenuator, all of which are located within a genetic locus called
hisO. During transcription, RNA polymerase first transcribes the leader region at
hisp1 and subsequently transcribes through the attenuator-controlled locus before reaching the first gene structure within the operon [
23,
24,
25].
Previous research has demonstrated that negative regulation of histidine levels, along with a loss of histidine biosynthetic function, renders the transcription level of the histidine operon in bacteria highly sensitive to histidine concentration [
26,
27]. Li et al. discovered that the SL1344 strain of Salmonella, which lacks histidine biosynthetic function, showed a significant increase in histidine synthase abundance in macrophages with low histidine concentrations. In contrast, the wild-type 14028s strain with normal histidine biosynthetic function efficiently converted PRPP to histidine, enabling Salmonella to adapt to histidine-deficient environments and maintain stable transcription levels of the histidine operon [
27].
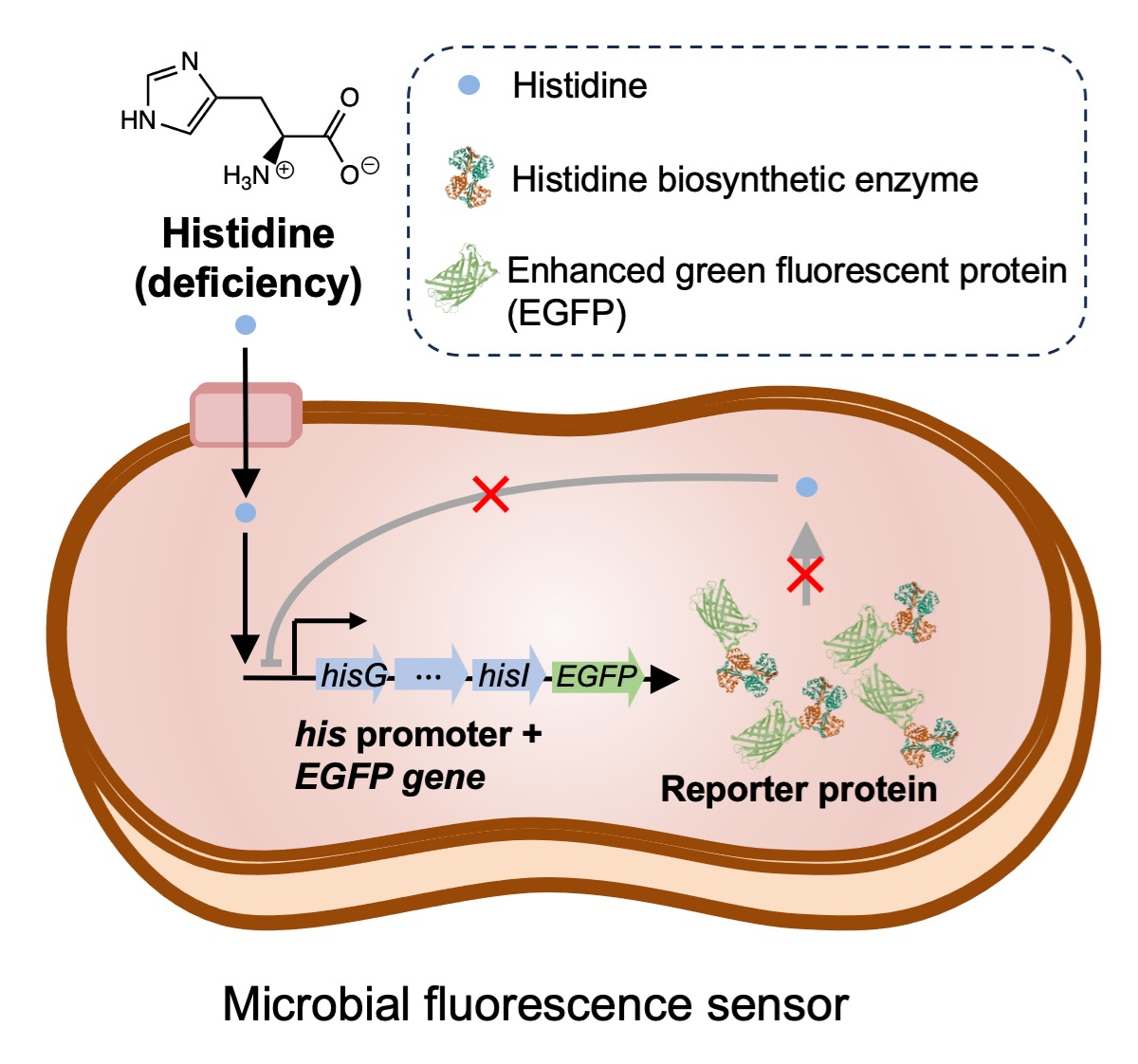
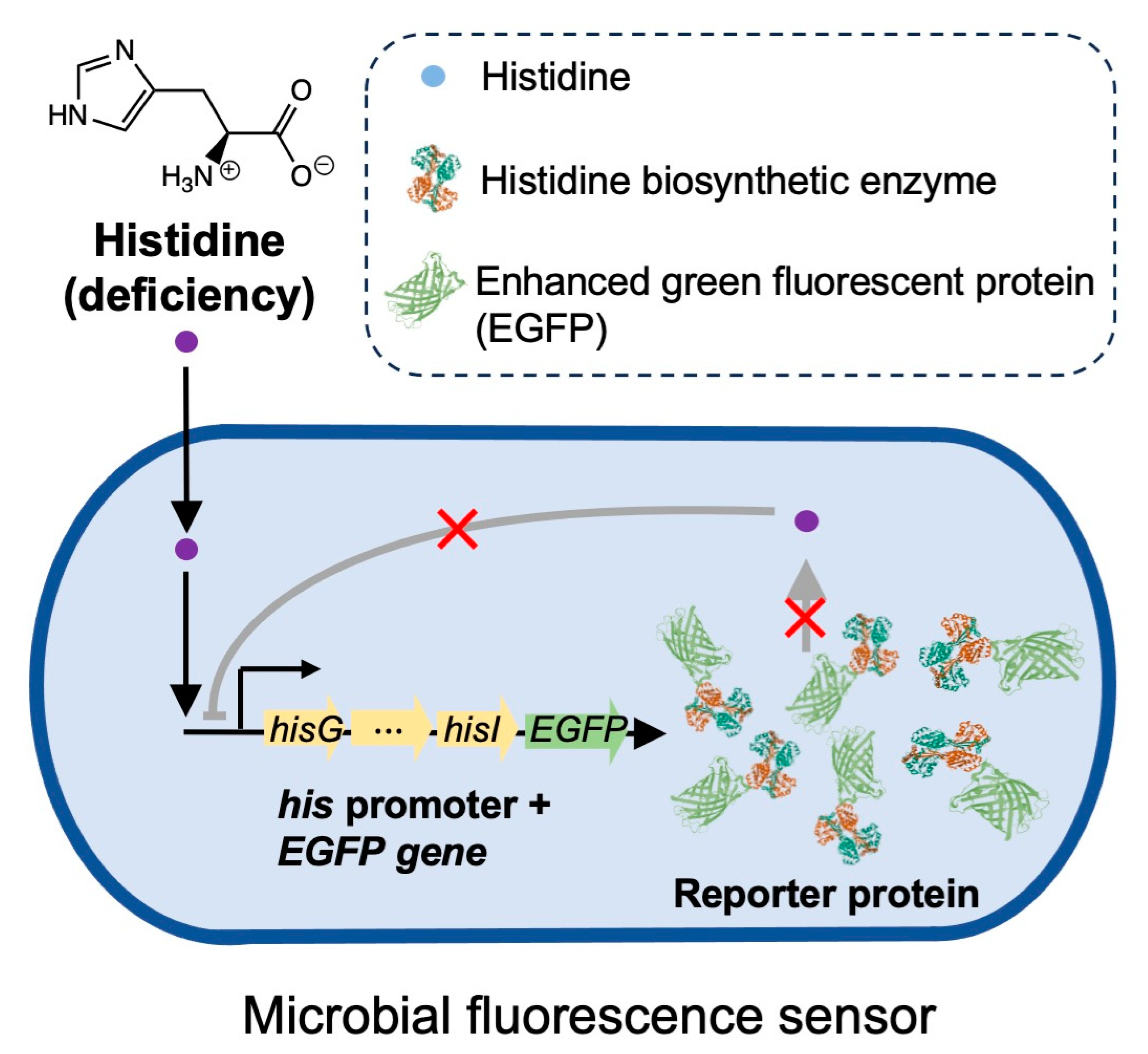
Inspired by these findings, here we introduce a pioneering bacterial fluorescence sensor that exhibits sensitive and specific responses to deficient histidine concentrations. This sensor utilizes the histidine operon of the histidine biosynthetic function-deficient Salmonella SL1344 strain and incorporates the reporter gene enhanced green fluorescent protein (EGFP), resulting in high expression of EGFP in response to histidine deficiency, leading to the emission of strong fluorescent signals. The histidine-specific fluorescence sensor enables sensitive in situ detection of intracellular histidine concentrations in different live cells.
2. Materials and Methods
2.1. Materials
The DNA oligonucleotides were synthesized and purified by HPLC (Sangon Biotech Co., China). DNase/RNase free deionized water purchased from TIANGEN Biotech Co. (Beijing, China) was used with all the experiment. L-histidine and L-arginine were purchased from Energy Chemical Co. (Shanghai, China). D-histidine and L-lysine were purchased from D&B Biotechnology Co. (Shanghai, China). Histamine was purchased from Sigma Aldrich (USA). Taq DNA polymerase and its corresponding reaction buffer were purchased from Yeasen Biotechnology Co. (Shanghai, China). PrimeStar DNA polymerase and its corresponding reaction buffer were purchased from Takara Bio (Beijing, China).
2.2. Bacteria cultivation
The bacterial strains were stored at -80°C in 25% (vol/vol) glycerol. To initiate the cultivation process, frozen bacteria were streaked and cultivated on LB agar plates overnight at 37°C. Subsequently, a single colony was carefully selected and inoculated into 3 mL of LB medium. The culture was allowed to grow overnight. The following morning, the culture was further diluted 1:20 into fresh LB medium and incubated at 37°C until it reached mid-exponential phase, as indicated by an optical density at 600 nm (OD
600) of 0.9. The components of all media used in this study are summarized in
Table 1.
2.3. Acquisition of target DNA fragments by PCR
The primers used in this study are listed in
Table 2. The
EGFP gene was amplified from the pEGFP plasmid using the R1-egfp and egfp-R2 primers. The promoter of the
his operon was amplified from the S. Typhimurium SL1344 strain using the R1-his and his-F primers. Linearized vectors, which contained the origin of replication, antibiotic resistance gene, and its promoter, were amplified from the pGEX-6p-1 plasmid (higher copy number) using the pgex-homo-F/R primers, and from the pSB3313 plasmid using the pSB3313-homo-F/R primers. The PCR reaction was performed using high-fidelity PrimeStar DNA polymerase, with an annealing temperature of 56-58°C, extension temperature of 72°C, and extension time of 1 min per 1000 base pairs. After the PCR reaction, the products were separated and purified using agarose gel electrophoresis, and the target DNA fragments were recovered using a DNA agarose gel recovery kit.
2.4. Homologous recombination
The Hieff Clone Plus One Step Cloning Kit (Yeasen) was utilized for the recombination of sequence H with the linearized vectors. To the reaction system with a total volume of 20 µL, 10 µL of 2x Hieff Clone Enzyme Premix, 1 µL of the amplified insert fragment (188 ng), 1.5 µL of linearized cloning vector (50 ng), and 7.5 µL of water were added. The reaction mixture was incubated at 50°C for 20 min. Subsequently, 10 µL of the cooled reaction mixture was combined with 100 µL of chemically competent DH5α cells. The tube was gently tapped to mix the contents, and then placed on ice for 30 min. Heat shock was performed at 42°C for 90 seconds, followed by a 2-minute incubation on ice. Next, 900 µL of SOC medium was added, and the mixture was incubated at 37°C for 10 min to allow for complete recovery. Finally, 100 µL of the bacterial solution was evenly spread onto LB agar plates containing ampicillin resistance. The plates were inverted and incubated overnight at 37°C. The following day, single colonies were selected for sequencing.
2.5. Electrotransformation and biosensor construction
For electrotransformation, 200 ng of plasmid was added to 50 µL of electrocompetent SL1344 strain on ice and pre-mixed for 30 min. The electrotransformation was performed using a MicroPulser Electroporator (Bio-Rad). Immediately after electroporation, the bacterial suspension was added to 1 mL of room temperature SOC medium and incubated at 37°C with shaking for 30 min to facilitate recovery. Subsequently, the bacterial suspension was spread onto LB agar plates containing ampicillin resistance and incubated overnight. The following day, single colonies were selected and cultured in LB medium at 37°C for 8 h. Finally, all bacterial strains were stored at -80°C in 25% (vol/vol) glycerol for long-term preservation.
2.6. Quantification of EGFP in bacterial sensors
The frozen bacterial samples were initially cultured in LB medium overnight at 37°C. The following morning, a specific volume of bacteria was collected through centrifugation and resuspended in an equal volume of M9 medium. The bacteria were then cultured in a shaking incubator at 37°C for 30 min. Subsequently, 1 mL of the bacterial fluorescence sensors was resuspended in 1×PBS and lysed using an ultrasonic cell breaker (Scienze). The fluorescence emission wavelength and peak intensity of the bacterial lysate were measured using an F-7000 fluorescence spectrometer (Hitachi, Japan) with excitation at 488 nm. For SL1344-pSB3313, the spectrometer's excitation slit was set to 7.5 nm, and the emission slit was set to 5.0 nm. For SL1344-pGEX, the excitation slit was set to 5.0 nm, and the emission slit was also set to 5.0 nm. The photomultiplier tube (PMT) voltage of the F-7000 fluorescence spectrometer was set to 950 V.
2.7. Cell culture and bacterial infection
HeLa cells and the murine macrophage cell line RAW 264.7 were cultured in Dulbecco's modified Eagle medium (DMEM; HyClone, USA) supplemented with 10% fetal bovine serum (FBS; PAN-Biotech, Germany) under 5% CO2 conditions at 37°C. Prior to infection, Salmonella bacteria in mid-exponential phase (OD600 = 0.9) were pelleted by centrifugation at 3,000×g and then resuspended in an equal volume of prewarmed (37°C) Hanks' buffered salt solution (HBSS; HyClone, USA) containing 0.3 M NaCl. Infection of the cells was carried out in HBSS with a multiplicity of infection (MOI) of 100 for 30 min. Subsequently, the cell monolayers were washed three times with prewarmed (37°C) HBSS and incubated in prewarmed DMEM containing 100 μg/mL gentamicin for 1 h to eliminate extracellular bacteria. The culture medium was then changed to DMEM supplemented with 10 μg/mL gentamicin.
2.8. Fluorescence Microscopy Imaging
To prepare for confocal laser scanning microscopy, the cell nuclei were stained with a nuclear dye. A 100x concentration of the dye was added to the culture medium, and the cells were incubated for 10 min. Subsequently, the cells were washed with medium to remove any excess dye, and microscopy observations were conducted.
Fluorescence images were acquired using a Laser Scanning Confocal Microscope equipped with a Nikon camera. Raw data were exported as 12-bit TIF files for further analysis using ImageJ software. The color brightness in the images is directly proportional to the intensity of the fluorescent signals across all channels.
3. Results and discussion
3.1. Construction of a bacterial fluorescence sensor responsive to histidine deficiency.
In the wild-type 14028s strain of Salmonella, the histidine biosynthetic function remains normal, and its histidine synthase efficiently converts PRPP to histidine even in histidine-deficient conditions, thereby maintaining stable transcription levels of the histidine operon (Figure S1a). However, in the histidine biosynthetic function-deficient strain SL1344, the expression of histidine synthase, controlled by the histidine operon, undergoes a significant increase under low histidine concentrations (Figure S1b). Leveraging this property, we propose to incorporate the EGFP gene downstream of the his promoter in the S. Typhimurium SL1344 strain, thereby creating a bacterial fluorescence sensor that exhibits high EGFP expression in response to histidine deficiency, leading to the emission of fluorescent signals.
In our preliminary experiments, we attempted to insert the EGFP gene directly after the his promoter in the SL1344 genome using the λ red homologous recombination method. Initially, we transferred the plasmid pKD46, containing the enzymes required for expressing the λ red homologous recombination system, into the SL1344 strain. After inducing enzyme expression, we electroporated the target gene, which contained both upstream and downstream homologous sequences of the insertion site in the bacterial genome, along with the EGFP gene and resistance gene (approximately 2000 bp in length), into the bacteria to insert it at the specific location in the bacterial genome.
However, despite multiple attempts, we observed extremely low transformation efficiency, likely due to the length of the inserted gene, which was around 2000 base pairs, significantly larger than the approximately 800 base pairs target gene fragments that λ red systems usually employ for more efficient recombination. Moreover, the complexity of the bacterial genome itself made it difficult to select homologous sequences with high specificity, thereby hindering the correct recombination of the imported DNA fragments into the bacteria.
As a result, we considered an alternative approach. Instead of modifying the bacterial genome directly, we cloned the
his operon promoter from the S. Typhimurium SL1344 strain and fused it with the reporter gene
EGFP in a plasmid. Subsequently, we introduced this engineered plasmid into the SL1344 strain, resulting in a bacterial strain that exhibits enhanced expression of the reporter gene
EGFP in response to histidine deficiency, as depicted in
Figure 1.
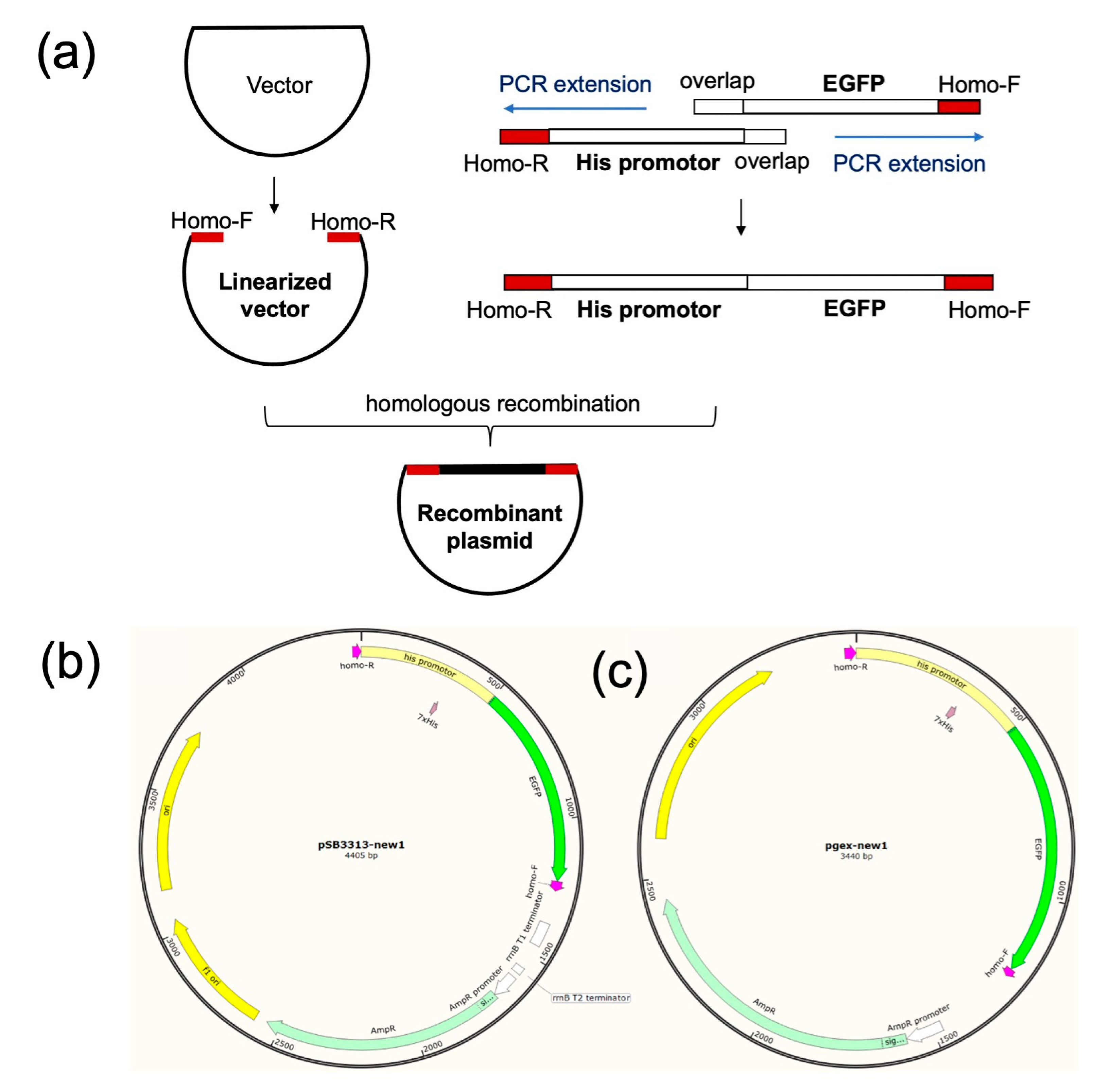
Using pSB3313 and pGEX-6p-1 plasmids as templates, we performed PCR amplification to obtain two different linearized vectors. These vectors contained essential plasmid components, including the origin of replication, resistance gene, and promoter (
Figure 2a).
Figure 2b and 2c illustrates the constructed plasmids with different copy numbers. Concurrently, we separately amplified fragments containing the
his promoter and
EGFP gene through PCR. To facilitate the subsequent fusion of these two fragments, we designed a 20 bp sequence at the 5' or 3' end of the corresponding PCR primers, creating a 40 bp overlapping sequence between the fragments. Both fragments were extended by PCR on both ends, generating a larger fragment comprising the
his promoter and
EGFP gene. Finally, we ligated the linearized vector and the inserted fragment through homologous recombination, successfully constructing the plasmid (Figure S2-S4). Specifically, pGEX-6p-1 belongs to the high-copy plasmid category, whereas pSB3313 is not a high-copy plasmid. The Salmonella SL1344 strain carrying the plasmid constructed with pSB3313 as the vector was designated as SL1344-pSB3313, while the strain carrying the plasmid constructed with pGEX-6p-1 as the vector was named SL1344-pGEX.
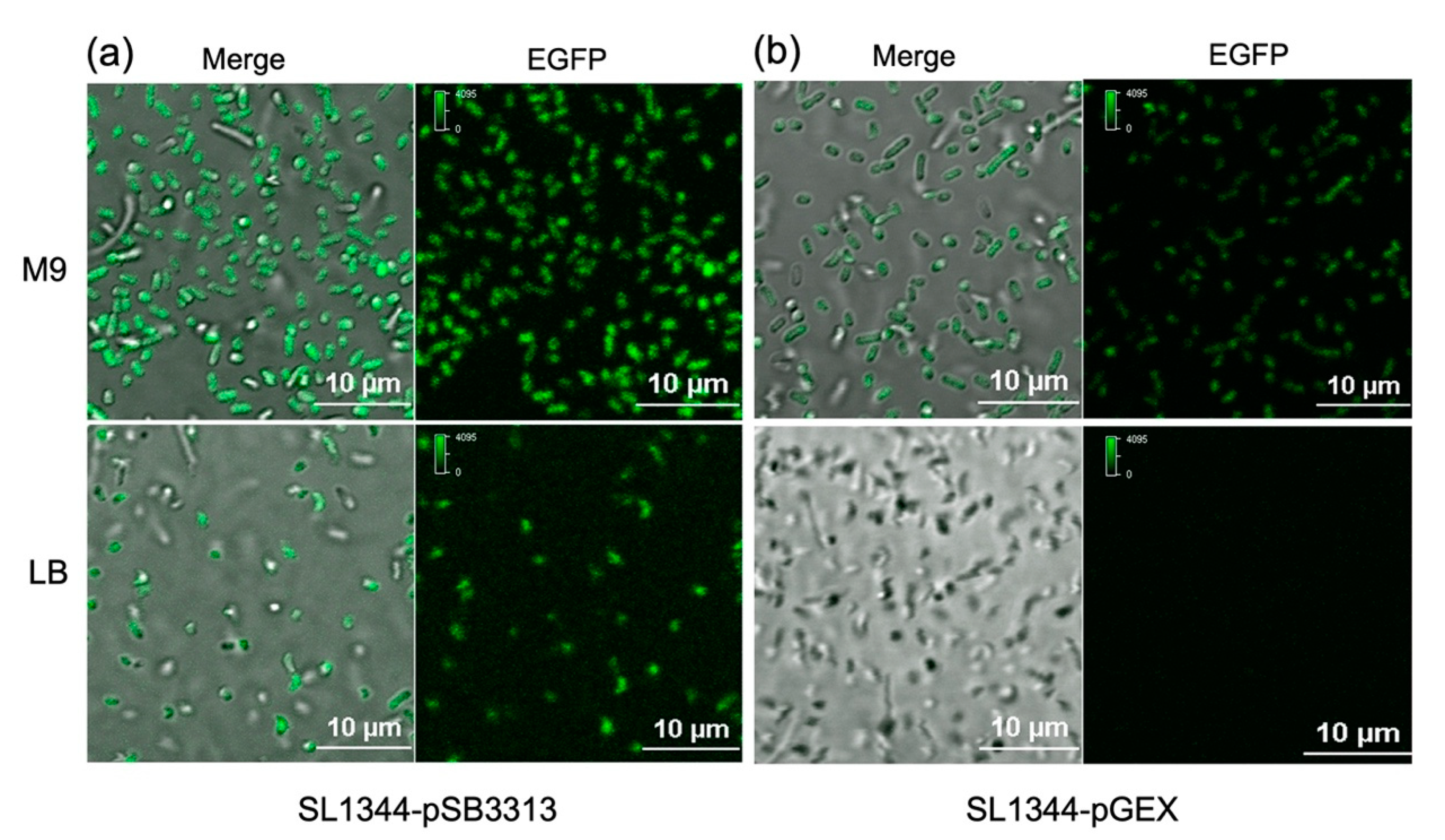
To assess the responsiveness of the developed bacterial fluorescence sensor to varying histidine concentrations, both strains were cultured at 37°C for 30 min in M9 medium (histidine-free) and LB medium (rich in histidine), respectively. The fluorescence intensity of both strains in M9 medium was significantly higher compared to that in LB medium. This observation indicates that in the absence of histidine, the promoter of the
his operon in the plasmid was effectively activated, leading to the expression of the reporter gene
EGFP (
Figure 3). Importantly, it is noteworthy that the two bacterial fluorescence sensors, with different plasmids featuring distinct copy numbers, exhibited diverse responses to the histidine levels in the growth medium. The high-copy-number SL1344-pGEX strain displayed minimal fluorescence in histidine-rich LB medium, whereas it demonstrated strong fluorescence in histidine-free M9 medium, highlighting its heightened sensitivity to histidine deprivation within the cellular growth environment.
3.2. Sensitivity and specificity of the bacterial fluorescence sensors in response to changes in histidine concentration
Histidine exists in two main stereoisomers: L-histidine and D-histidine. L-histidine is the more prevalent form in nature and widely present within organisms [
28]. To assess the response capability of the constructed bacterial fluorescent sensors to histidine concentrations, we cultivated SL1344-pSB3313 in M9 medium with varying L-histidine levels.
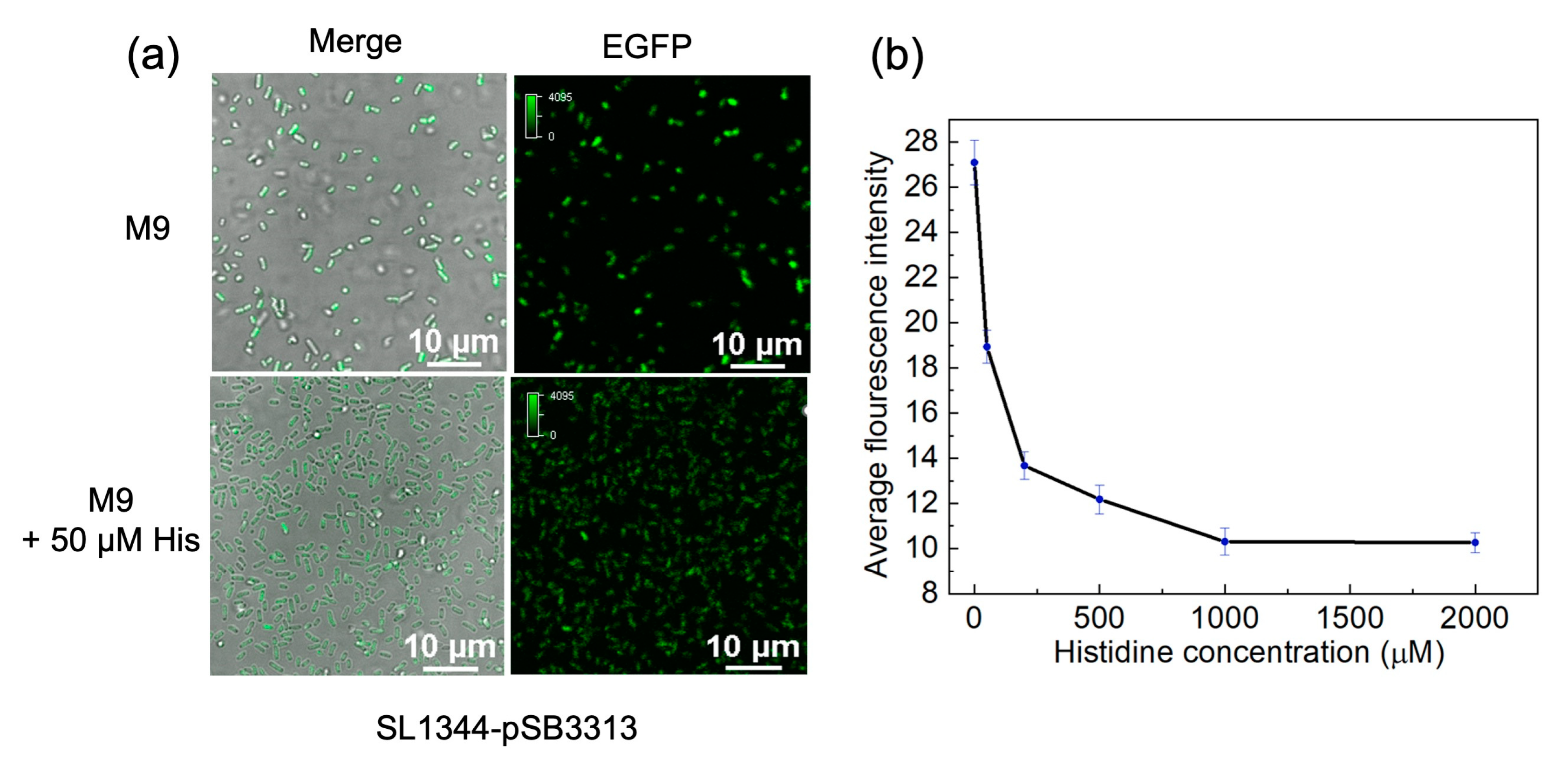
Figure 4a shows a notable decrease in fluorescence intensity with the addition of 50 μM L-histidine. To quantitatively evaluate
EGFP expression under different L-histidine concentration conditions, we determined the bacterial quantity based on OD values and then used ultrasonic lysis to disrupt bacteria, measuring the fluorescence intensity at 488 nm excitation. The average fluorescence intensity displayed an exponential decrease as the L-histidine concentration increased from 0 to 1000 μM (
Figure 4b). Within the range of 0 to 200 μM, the fluorescence decay rate was notably rapid. As L-histidine concentration further increased, the fluorescence gradually declined to background levels.
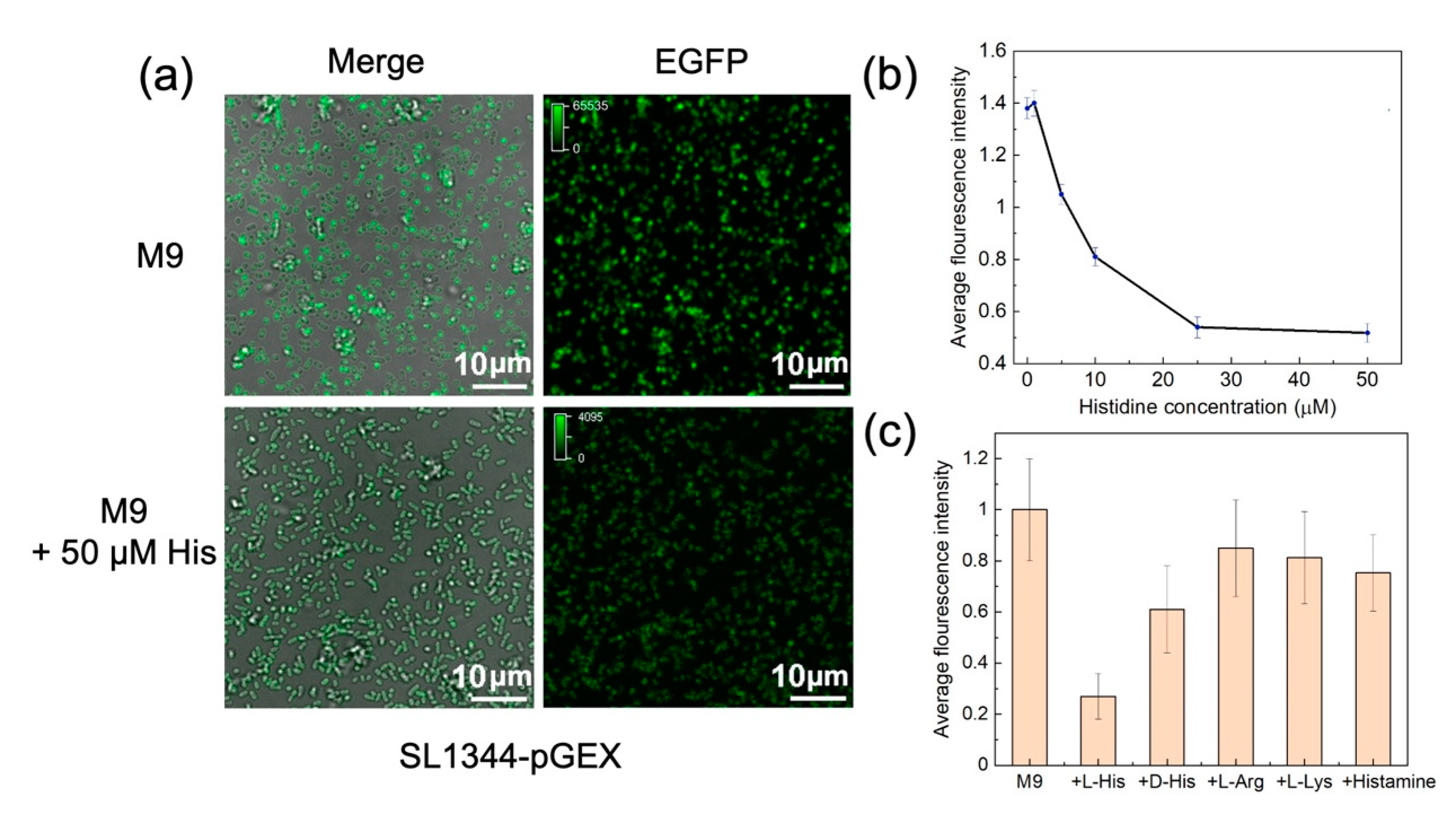
We subsequently evaluated the performance of the SL1344-pGEX strain. A more pronounced reduction in fluorescence intensity was observed upon the addition of 50 μM L-histidine to the M9 medium (
Figure 5a), confirming that the SL1344-pGEX strain is a more sensitive bacterial fluorescence sensor for detecting changes in L-histidine concentration. We also investigated the modulation of EGFP expression in the SL1344-pGEX strain with the addition of varying L-histidine concentrations. As L-histidine concentration increased from 0 to 50 μM, the average fluorescence intensity of the bacterial lysates also exhibited an exponential decrease (
Figure 5b). The most significant variations in fluorescence were observed within the L-histidine concentration range of 1 μM to 10 μM. Notably, even a minimal amount of L-histidine introduced to the M9 medium (approximately 2.5 μM) resulted in discernible reduction in fluorescence.
We further evaluated the specificity of the SL1344-pGEX bacterial fluorescence sensor.D-histidine is the enantiomer of L-histidine. Histamine emerges as a product resulting from the enzymatic conversion of histidine. L-arginine and L-lysine share structural and property similarities with L-histidine, particularly as alkaline cationic amino acids. Our investigation concentrated on appraising the SL1344-pGEX bacterial fluorescence sensor's response to these four compounds using consistent conditions. As depicted in
Figure 5c, the introduction of 50 μM L-histidine into the M9 culture medium led to a substantially more pronounced reduction in fluorescence than the other four compounds tested. The addition of 50 μM L-arginine or L-lysine to the M9 culture medium scarcely influenced the fluorescence signal of the SL1344-pGEX strain. On the other hand, the inclusion of D-histidine resulted in a minor fluorescence response, while introducing histamine at the same concentration yielded a conspicuously smaller fluorescence signal compared to L-histidine.
Previous reports indicate that certain extracellular macromolecules, such as peptidoglycan and teichoic acids, function as principal sources of D-amino acids in bacteria [
29]. Furthermore, even bacteria that naturally do not synthesize D-amino acids can utilize externally supplied D-amino acids for peptidoglycan synthesis [
30]. Drawing from this, we postulate that bacteria may employ D-histidine and histamine as mechanisms to alleviate some of the survival pressures resulting from the absence of L-histidine in their environment [
31].
3.3. Application of the bacterial fluorescence sensor for real-time monitoring of histidine levels within live cells.
Salmonella is a common foodborne pathogenic bacterium that primarily infects epithelial cells and macrophages in animal hosts [
32,
33]. Using the isolation and extraction method coupled with LC/MS/MS quantification, Li et al. found that the histidine concentration in RAW264.7 macrophage cell extracts (approximately 60 μM) was much lower than that in HeLa epithelial cell extracts (around 200 μM) [
27].
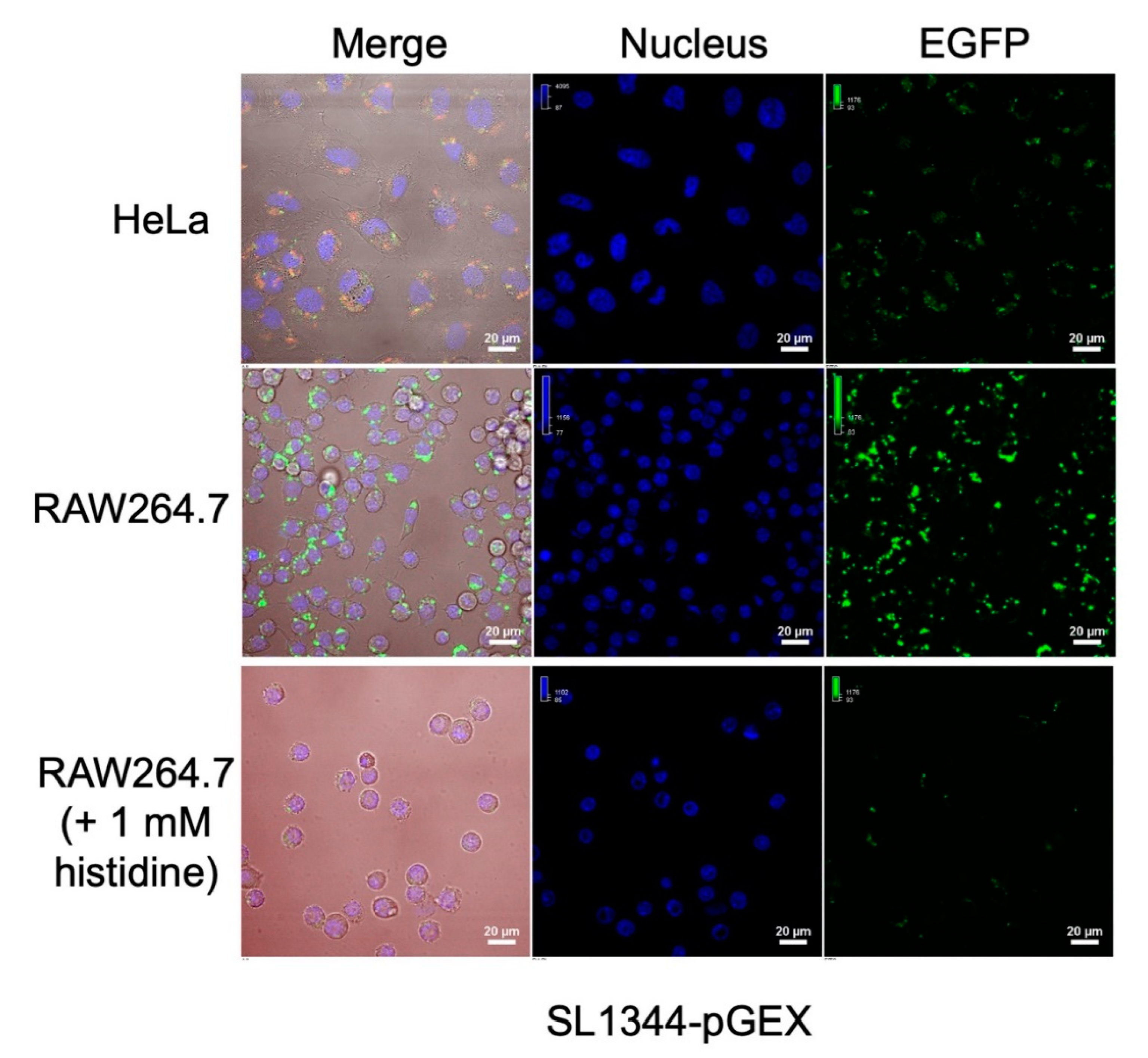
To evaluate the real-time response of the constructed bacterial fluorescence sensors to intracellular histidine concentration, we infected RAW264.7 macrophage cells and HeLa epithelial cells with the SL1344-pGEX sensor. After approximately 1 h of infection, we observed the fluorescence signals as shown in
Figure 6.
The SL1344-pGEX sensor exhibited significantly higher EGFP expression in macrophages with lower histidine concentration compared to HeLa cells with higher histidine concentration. This indicates that the lower histidine concentration in macrophages triggered an up-regulation of the histidine-responsive promoter in the bacterial sensor, leading to increased EGFP expression. When supplemented with 1 mM L-histidine in the macrophage culture, a noticeable decrease in EGFP expression by the bacterial sensor was observed. These findings demonstrate the bacterial fluorescent sensors' capability for in-situ response to the variation of intracellular histidine concentration. The SL1344-pGEX sensor exhibited significantly higher EGFP expression in macrophages with lower histidine concentration compared to HeLa cells with higher histidine concentration. This indicates that the lower histidine concentration in macrophages triggered an up-regulation of the histidine-responsive promoter in the bacterial sensor, leading to increased EGFP expression. When supplemented with 1 mM L-histidine in the macrophage culture, a noticeable decrease in EGFP expression by the bacterial sensor was observed. These findings demonstrate the bacterial fluorescent sensors' capability for in-situ response to the variation of intracellular histidine concentration.
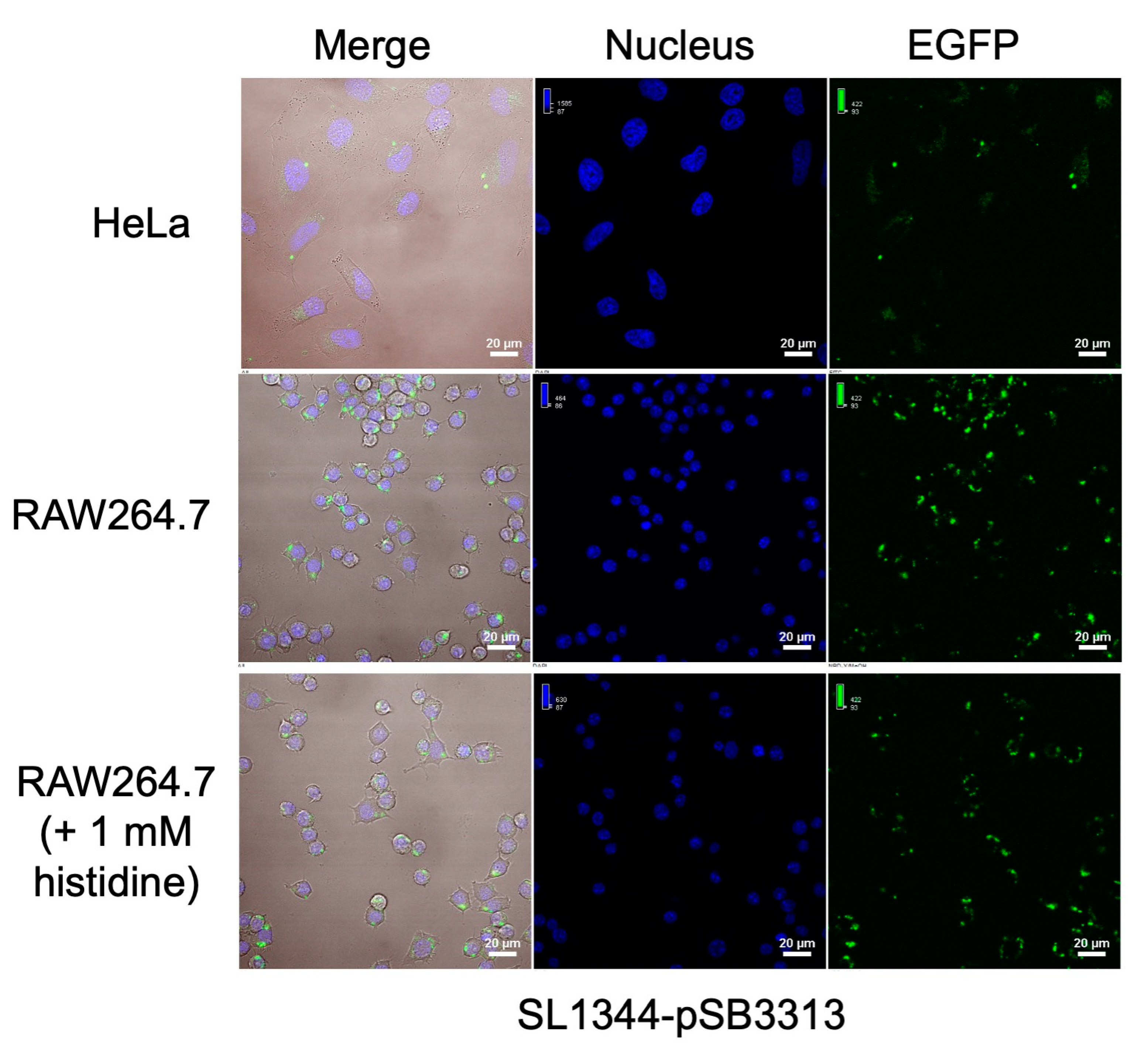
We also conducted tests involving the infection of RAW264.7 macrophage cells and HeLa epithelial cells using the SL1344-pSB3313 sensor. As illustrated in
Figure 7, the SL1344-pSB3313 sensor also demonstrated notably elevated EGFP expression in macrophages with lower histidine concentrations, while displaying subdued fluorescence signals in HeLa cells. Upon introducing 1 mM L-histidine into the macrophage culture, a noticeable reduction in EGFP expression by the bacterial sensor was also observed. In contrast to SL1344-pGEX, the SL1344-pSB3313 sensor features a broader histidine response range, thereby facilitating enhanced differentiation among cellular environments with varying histidine levels, including both higher and lower concentrations.
Our bacterial fluorescence sensor, designed to detect intracellular histidine deficiency, is crucial for preserving vital physiological functions. Detecting such deficiency is crucial, as it could negatively impact vital physiological functions in organisms. Histidine is essential for fundamental growth during infancy and cognitive progress. Insufficient histidine can disrupt neurotransmitter synthesis and transmission, potentially leading to neurological issues. Detecting and addressing intracellular histidine deficiency is crucial to maintain these functions and mitigate health risks.
Histidine deficiency also affects amino acid oxidation and protein turnover, highlighting its importance for healthy adults. Moreover, histidine status impacts skin health, potentially influencing skin dysfunction and diseases due to decreased hyaluronan levels [
34,
35]. Our bacterial fluorescence sensor offers a valuable tool to monitor and address histidine deficiency, providing insights into its multifaceted implications and potential therapeutic applications across physiological contexts.
4. Conclusion
In summary, our study has innovatively engineered a bacterial fluorescence sensor that actively responds to histidine deficiency, emitting detectable fluorescence signals. This sensor is created by constructing plasmids containing the his operon promoter and the reporter gene EGFP. The design of this microbial fluorescence sensor facilitates straightforward assembly, and circumvents the complexities linked to genetic modifications in bacteria. Moreover, it offers adaptability in fine-tuning the specific response range and sensitivity of the reporter gene through manipulation of the plasmid's copy number.
The high-copy-number SL1344-pGEX sensor, characterized by its heightened sensitivity, triggers signal changes even in response to minimal histidine concentration fluctuations. Conversely, the low-copy-number SL1344-pSB3313 sensor exhibits a broader response range, capable of tracking more extensive shifts in histidine concentrations. Both bacterial fluorescence sensors enable efficient in-situ monitoring of histidine variations within both culture mediums and live cells. These sensors not only facilitate investigations into histidine metabolism and its implications across diverse biological processes but also hold potential for broader applications. For instance, they can be employed to monitor intracellular histidine levels during pathogenic bacterial infections and evaluate the impact of drugs on cellular metabolism, offering substantial convenience for related research.
Furthermore, this approach to constructing bacterial fluorescence sensors can be expanded to encompass other amino acids regulated by synthetic attenuators, such as tryptophan, leucine, and others, opening avenues for broader applications beyond histidine.
Author Contributions
Conceptualization, Z.L. and M.Z.; methodology, Z.L. and M.Z.; formal analysis, X.L. and Z.L.; investigation, X.L.; writing—original draft preparation, X.L.; writing—review and editing, M.Z.; supervision, M.Z.; project administration, M.Z.; funding acquisition, M.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the National Natural Science Foundation of China (No. 21974005 and No. 22174005).
Data Availability Statement
Data will be available on request.
Acknowledgments
The measurements of Optical Spectroscopy were performed at the Analytical Instrumentation Center of Peking University (PKUAIC). We thank Yan Guan at PKUAIC for her excellent technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sundberg, R.J.; Martin, R.B. Interactions of histidine and other imidazole derivatives with transition-metal ions in chemical and biological-systems. Chem. Rev. 1974, 74, 471–517. [Google Scholar] [CrossRef]
- Haas, H.; Panula, P. The role of histamine and the tuberomamillary nucleus in the nervous system. Nat. Rev. Neurosci. 2003, 4, 121–130. [Google Scholar] [CrossRef]
- Ghadimi, H.; Partington, M.W.; Hunter, A. A familial disturbance of histidine metabolism. N. Engl. J. Med. 1961, 265, 221–224. [Google Scholar] [CrossRef]
- Sullivan, D.J.; Gluzman, I.Y.; Goldberg, D.E. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science. 1996, 271, 219–222. [Google Scholar] [CrossRef]
- Engesser, L.; Kluft, C.; Briet, E.; Brommer, E.J.P. Familial elevation of plasma histidine-rich glycoprotein in a family with thrombophilia. Brit. J. Haematol. 1987, 67, 355–358. [Google Scholar] [CrossRef]
- Gerber, D.A. Low free serum histidine concentration in rheumatoid-arthritis - measure of disease activity. J. Clin. Invest. 1975, 55, 1164–1173. [Google Scholar] [CrossRef]
- Rao, M.L.; Stefan, H.; Scheid, C.; Kuttler, A.D.S.; Froscher, W. Serum amino-acids, liver status, and antiepileptic drug-therapy in epilepsy. Epilepsia. 1993, 34, 347–354. [Google Scholar] [CrossRef]
- Watanabe, M.; Suliman, M.E.; Qureshi, A.R.; Garcia-Lopez, E.; Barany, P.; Heimburger, O.; Stenvinkel, P.; Lindholm, B. Consequences of low plasma histidine in chronic kidney disease patients: associations with inflammation, oxidative stress, and mortality. Am. J. Clin. Nutr. 2008, 87, 1860–1866. [Google Scholar] [CrossRef]
- Saito, H.; Goodnough, L.T.; Boyle, J.M.; Heimburger, N. Reduced histidine-rich glycoprotein levels in plasma of patients with advanced liver-cirrhosis - possible implications for enhanced fibrinolysis. Am. J. Med. 1982, 73, 179–182. [Google Scholar] [CrossRef]
- Schmid, G.; Fricke, L.; Lange, H.W.; Heidland, A. Intracellular histidine content of various tissues (brain, striated-muscle and liver) in experimental chronic renal-failure. Klin. Wschr. 1977, 55, 583–585. [Google Scholar] [CrossRef]
- Tateda, N.; Matsuhisa, K.; Hasebe, K.; Kitajima, N.; Miura, T. High-performance liquid chromatographic method for rapid and highly sensitive determination of histidine using postcolumn fluorescence detection with o-phthaldialdehyde. J. Chromatogr. B. 1998, 718, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, M.; Nakazawa, T. Determination of pK(a) values of individual histidine residues in proteins using mass spectrometry. Anal. Chem. 2008, 80, 6481–6487. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.B.; Kumar, D.; Madhuri, R.; Tiwari, M.P. Metal ion mediated imprinting for electrochemical enantioselective sensing of L-histidine at trace level. Biosens. Bioelectron. 2011, 28, 117–126. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Sun, M.X. Determination of histamine and histidine by capillary zone electrophoresis with pre-column naphthalene-2,3-dicarboxaldehyde derivatization and fluorescence detection. J. Chromatogr. A. 2004, 1040, 133–140. [Google Scholar] [CrossRef]
- Un, H.I.; Wu, S.; Huang, C.B.; Xu, Z.; Xu, L. A naphthalimide-based fluorescent probe for highly selective detection of histidine in aqueous solution and its application in in vivo imaging. Chem. Commun. 2015, 51, 3143–3146. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.H.; Zhao, T.B.; Nie, Z.; Miao, Z.; Liu, Y.; Yao, S.Z. Nitrogen-doped carbon nanoparticle modulated turn-on fluorescent probes for histidine detection and its imaging in living cells. Nanoscale. 2016, 8, 2205–2211. [Google Scholar] [CrossRef]
- Mao, Y.; Abed, M.A.; Lee, N.B.; Wu, X.; Du, G.; Pu, L. Determining the concentration and enantiomeric composition of histidine using one fluorescent probe. Chem. Commun. 2021, 57, 587–590. [Google Scholar] [CrossRef]
- Hu, H.; Gu, Y.; Xu, L.; Zou, Y.; Wang, A.; Tao, R.; Chen, X.; Zhao, Y.; Yang, Y. A genetically encoded toolkit for tracking live-cell histidine dynamics in space and time. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Della Corte, D.; van Beek, H.L.; Syberg, F.; Schallmey, M.; Tobola, F.; Cormann, K.U.; Schlicker, C.; Baumann, P.T.; Krumbach, K.; Sokolowsky, S.; et al. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Gonzalez-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef]
- Mastroeni, P.; Grant, A.; Restif, O.; Maskell, D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 2009, 7, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.E.; Ramos-Montanez, S. Biosynthesis of Histidine. EcoSal. Plus. 2009, 3, 1–49. [Google Scholar] [CrossRef]
- Johnston, H.M.; Barnes, W.M.; Chumley, F.G.; Bossi, L.; Roth, J.R. Model for regulation of the histidine operon of salmonella. Proc. Nat. Acad. Sci. India Sect. B-Biol. Sci. 1980, 77, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Kasai, T. Regulation of expression of histidine operon in salmonella-typhimurium. Nature. 1974, 249, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.A.; Ames, B.N. Histidine regulation in salmonella-typhimurium.11. percentage of transfer rnahis charges in-vivo and its relation to repression of histidine operon. J. Mol. Biol. 1972, 66, 31–42. [Google Scholar] [CrossRef]
- Henry, T.; Garcia-del Portillo, F.; Gorvel, J.P. Identification of Salmonella functions critical for bacterial cell division within eukaryotic cells. Mol. Microbiol. 2005, 56, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Y.; Fu, J.; Zhang, B.; Cheng, S.; Wu, M.; Wang, Z.; Jiang, J.; Chang, C.; Liu, X. Salmonella Proteomic Profiling during Infection Distinguishes the Intracellular Environment of Host Cells. Msystems. 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Nagao, T.; Nakayama-Imaohji, H.; Elahi, M.; Tada, A.; Toyonaga, E.; Yamasaki, H.; Okazaki, K.; Miyoshi, H.; Tsuchiya, K.; Kuwahara, T. L histidine augments the oxidative damage against Gram negative bacteria by hydrogen peroxide, Int. J. Mol. Med. 2018, 41, 2847–2854. [Google Scholar] [CrossRef]
- Radkov, A.D.; Moe, L.A. Bacterial synthesis of D-amino acids. Appl. Microbiol. Biotechnol. 2014, 98, 5363–5374. [Google Scholar] [CrossRef]
- Cava, F.; de Pedro, M.A.; Lam, H.; Davis, B.M.; Waldor, M.K. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011, 30, 3442–3453. [Google Scholar] [CrossRef]
- LaRue, T.A.; Spencer, J.F.T. The utilization of imidazoles by yeasts. Can. J. Microbiol. 2011, 13, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Ohl, M.E.; Miller, S.I. Salmonella: A model for bacterial pathogenesis. Annu. Rev. Med. 2001, 52, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Monack, D.M.; Mueller, A.; Falkow, S. Persistent bacterial infections: The interface of the pathogen and the host immune system. Nat. Rev. Microbiol. 2004, 2, 747–765. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Amino acids: metabolism, functions, and nutrition. Amino Acids, 2009, 37, 1–17. [Google Scholar] [CrossRef]
- Moro, J.; Tomé, D.; Schmidely, P.; Demersay, T.-C.; Azzout-Marniche, D. Histidine: A Systematic Review on Metabolism and Physiological Effects in Human and Different Animal Species. Nutrients, 2020, 12, 1414. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).