Submitted:
08 August 2023
Posted:
09 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Granger causality basic theory
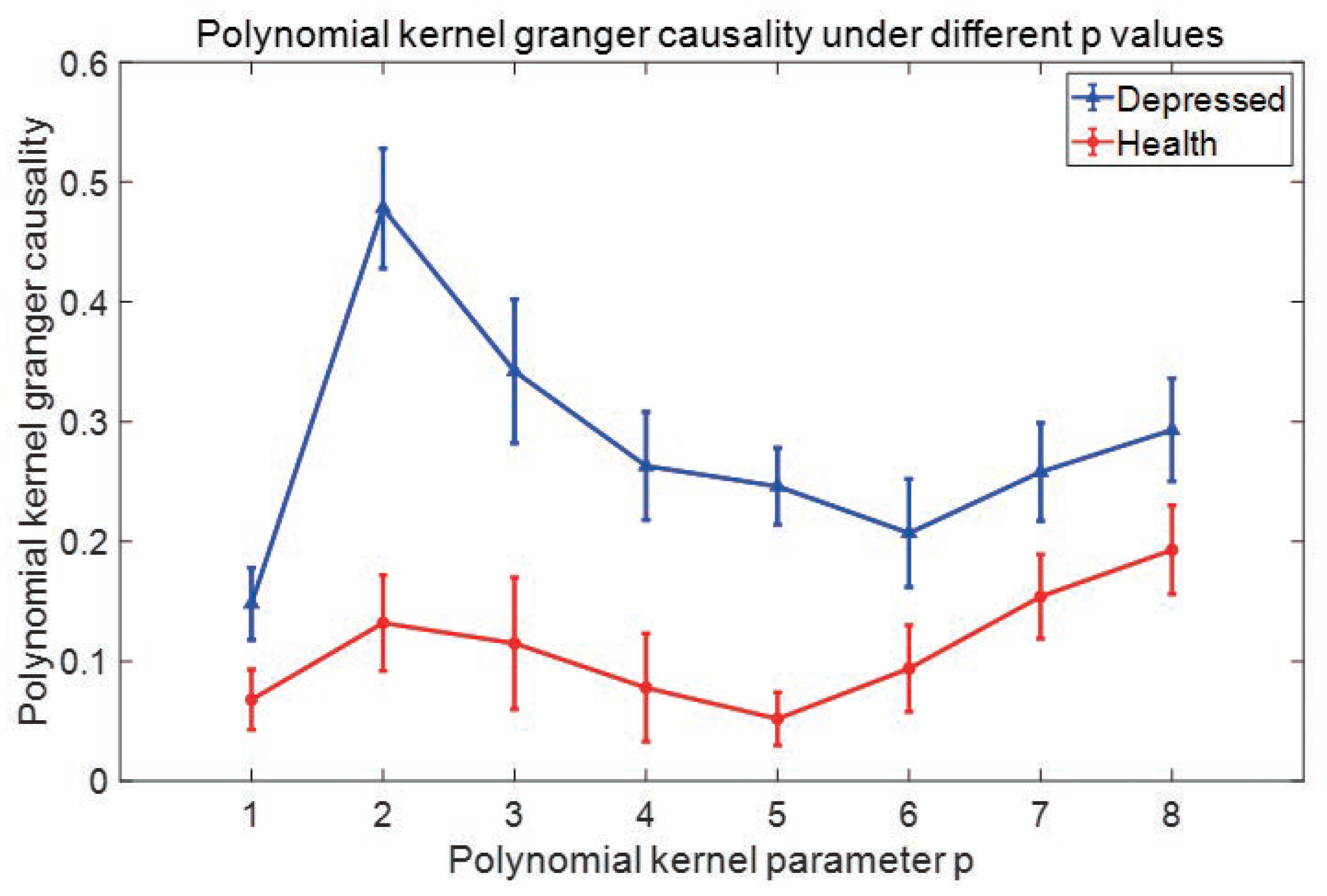
2.2. Granger causality based on polynomial Kernel
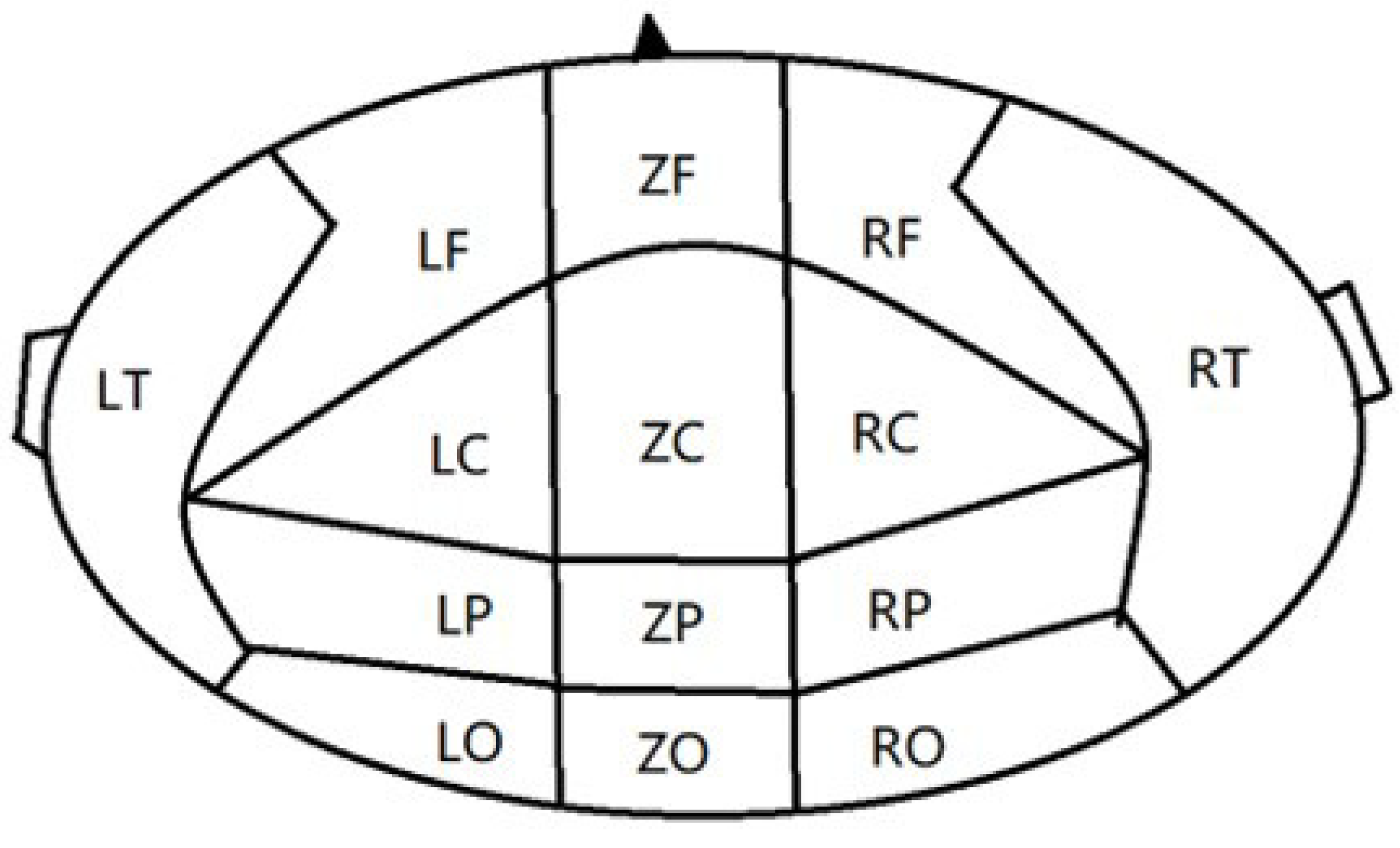
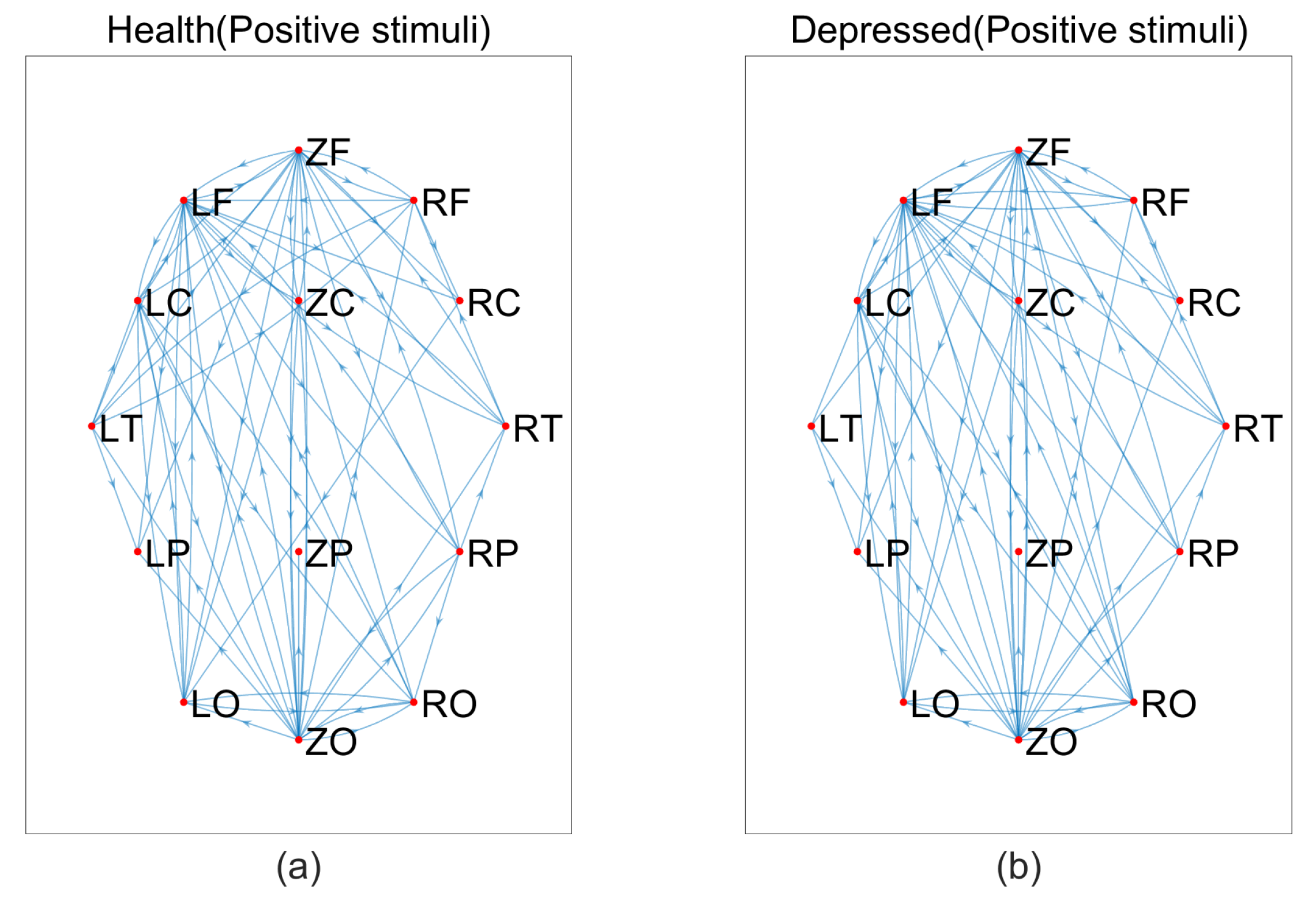
2.3. Introduction to topology Properties of brain network
-
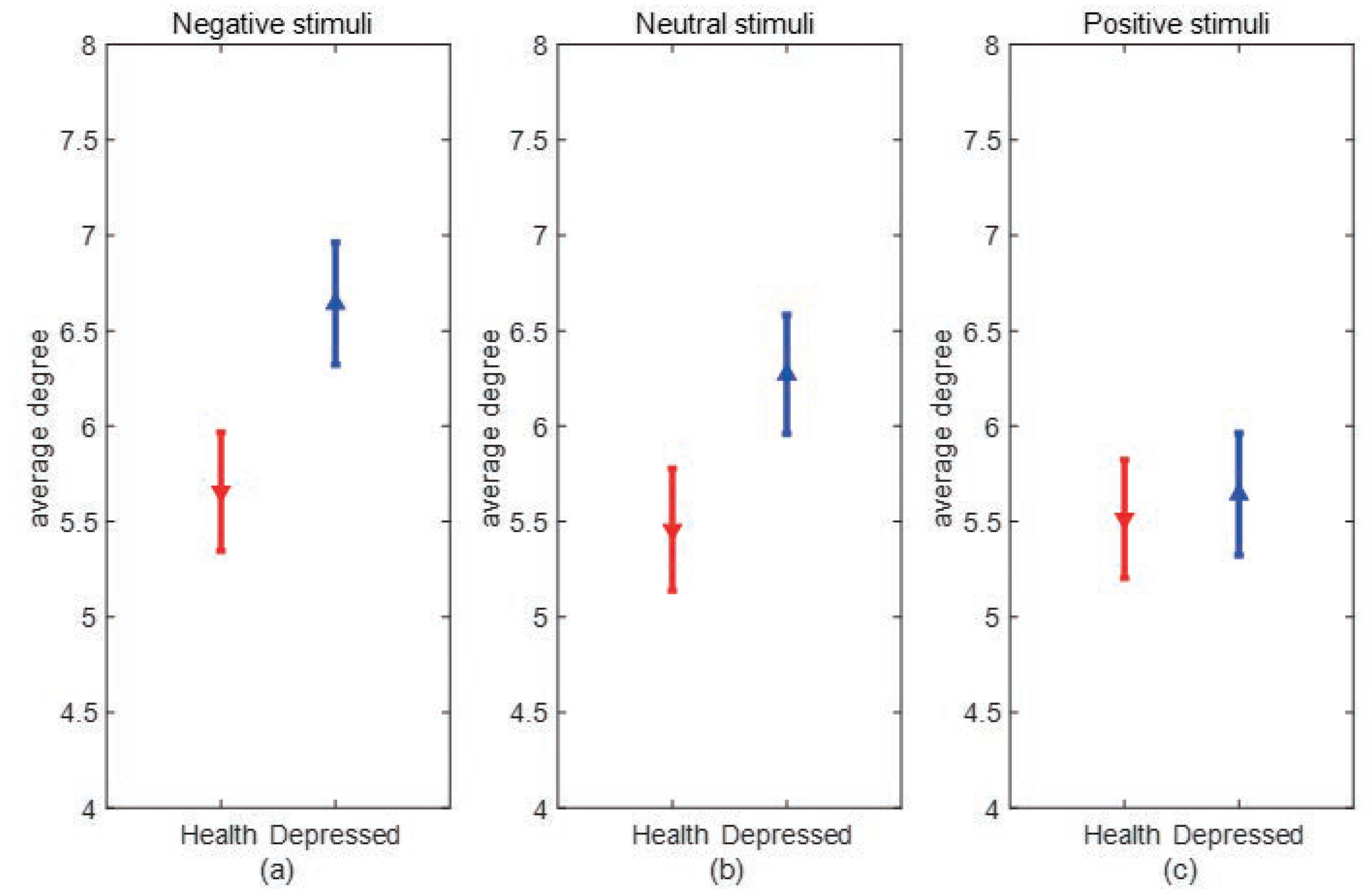
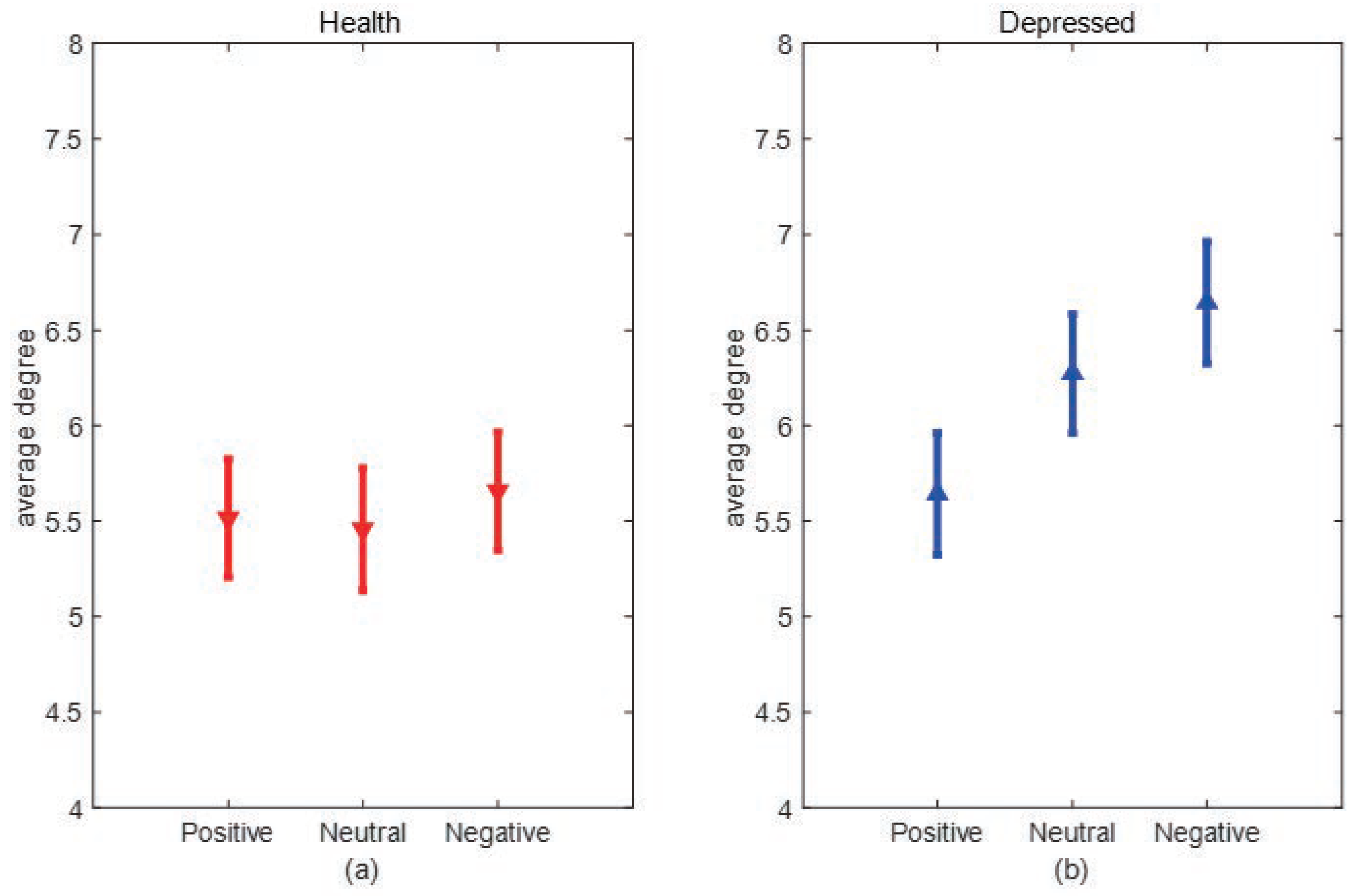
Degree and Degree distribution:The degree of a node is defined as the number of edges connected to node i in the network, and the degree of the I node can be expressed as:where is the connection state between nodes i and j, when there is a connection between i and j, =1, when there is no connection between i and j, =0. When the value of i increased, the more connected edges of the node, representing the node is more important. The average degree of all nodes in the network is the average degree of the network, which can directly reflect the connection level between the nodes in the network and measure the complexity of the network.
-
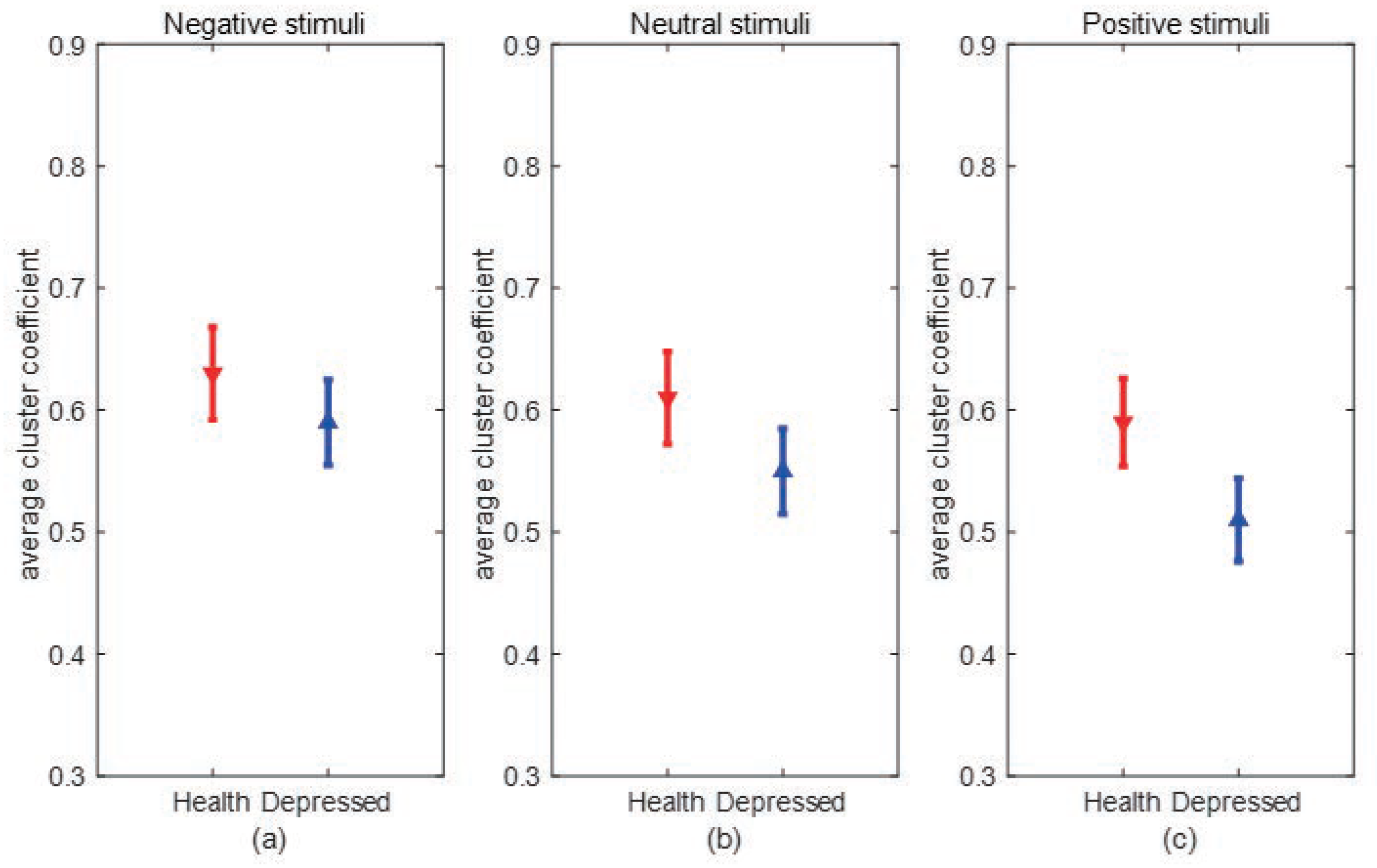
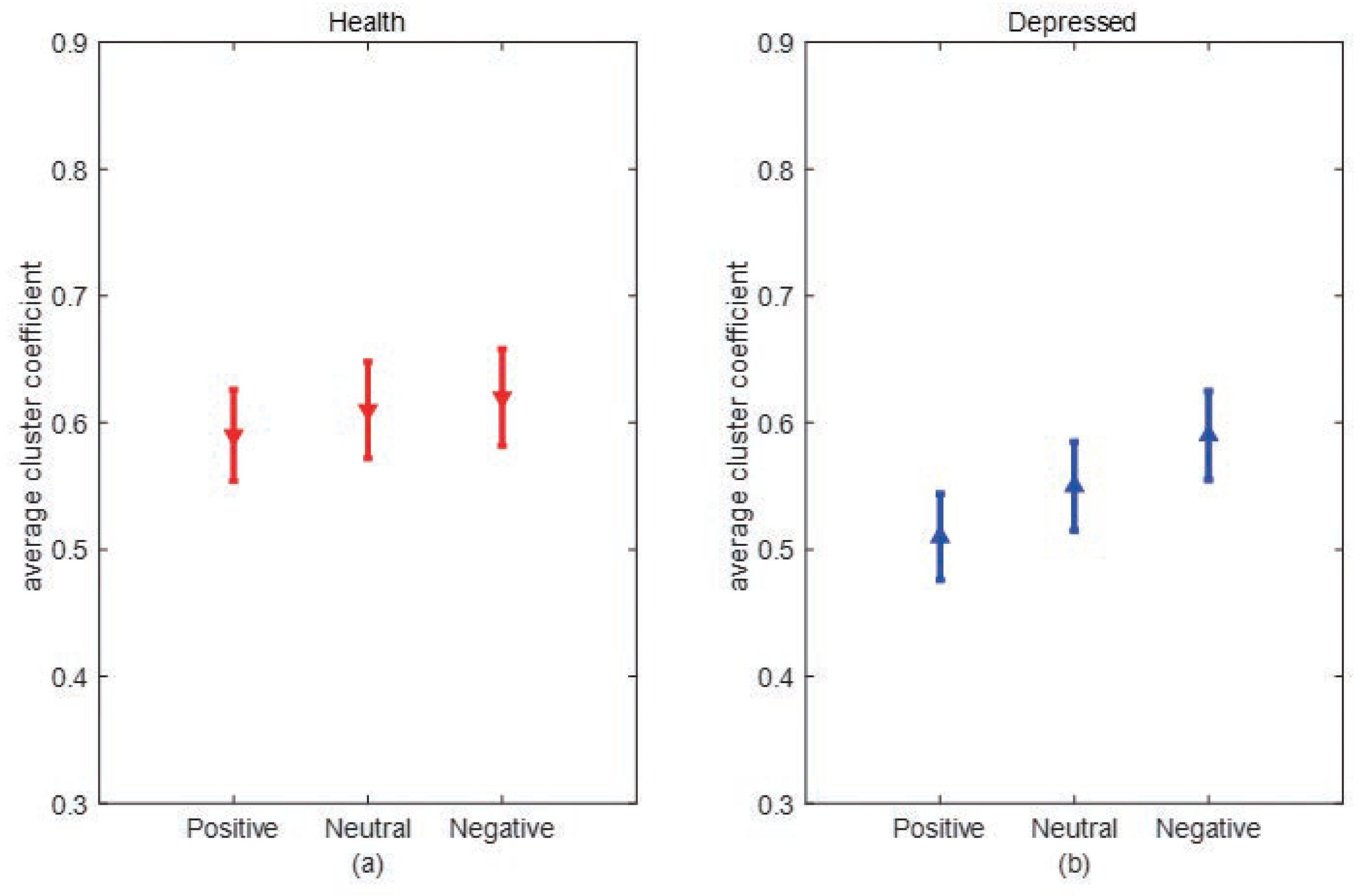
Clustering coefficient:The clustering coefficient C represents the clustering situation of nodes in the network. Generally speaking, C represents the probability that the neighbors of nodes are also neighbors to each other. Cluster coefficient of node i can be defined as:where represents the number of connections between nodes connected to node i, represents the maximum number of connected edges between nodes connected to node i. The average clustering coefficient of a network is the average clustering coefficient of all nodes in the network, which can be expressed as:N is the number of nodes in the network, and the value of is between 0 and 1. When there is a connection between two nodes in the network, , and when all nodes in the network do not have a connection, . C can reflect the degree of collectivization of the network, which is the tightness of connections between nodes.
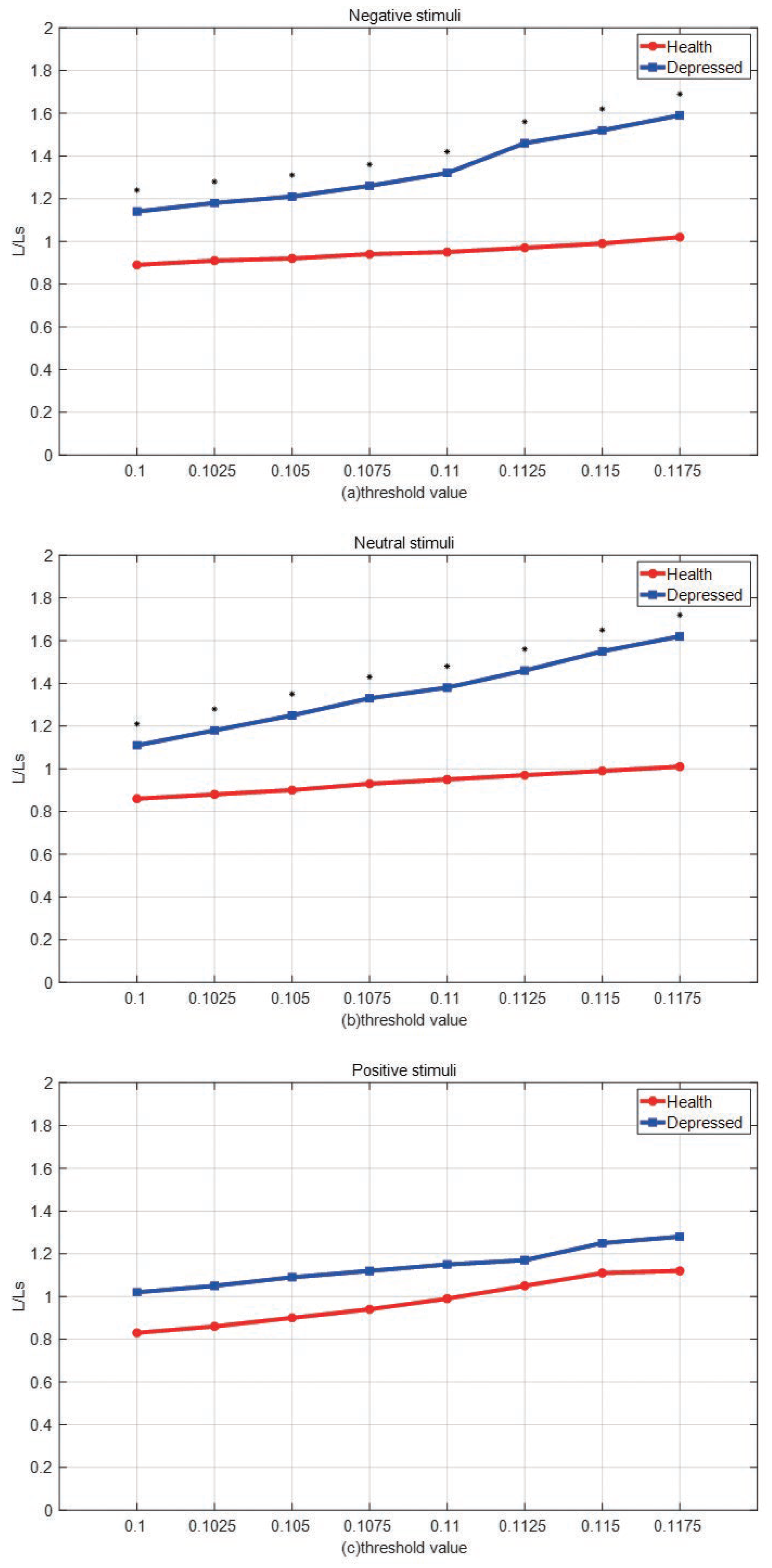
- Average path length The calculation formula for the average path length L of the network is:where N is the number of nodes in the network, is the shortest distance between node i and node j, representing the minimum number of connection edges required for connectivity between node i and node j. In small-scale networks, we usually use the Floyd algorithm to calculate . The average path length is an important indicator to measure the transmission efficiency of a network. Random networks typically have shorter average lengths, while regular networks typically have longer average path lengths.
3. Experimental results and analysis
3.1. Experimental data
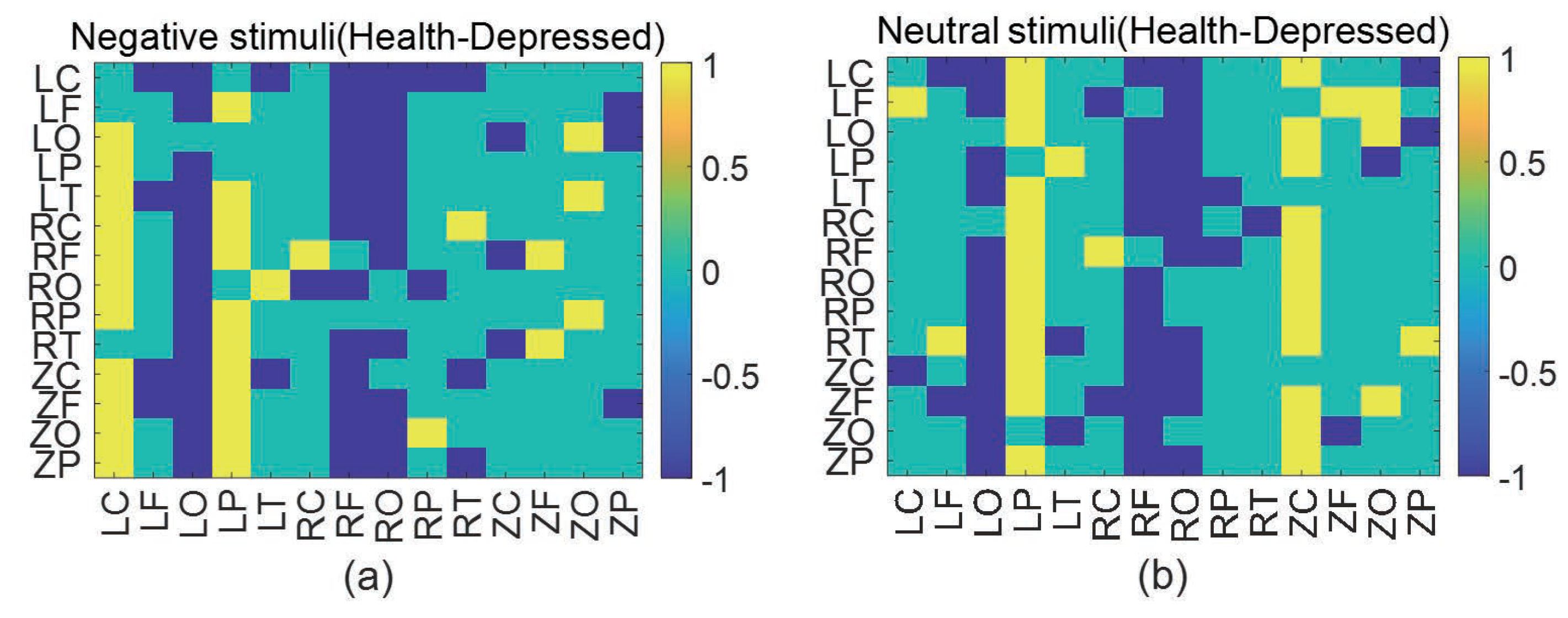
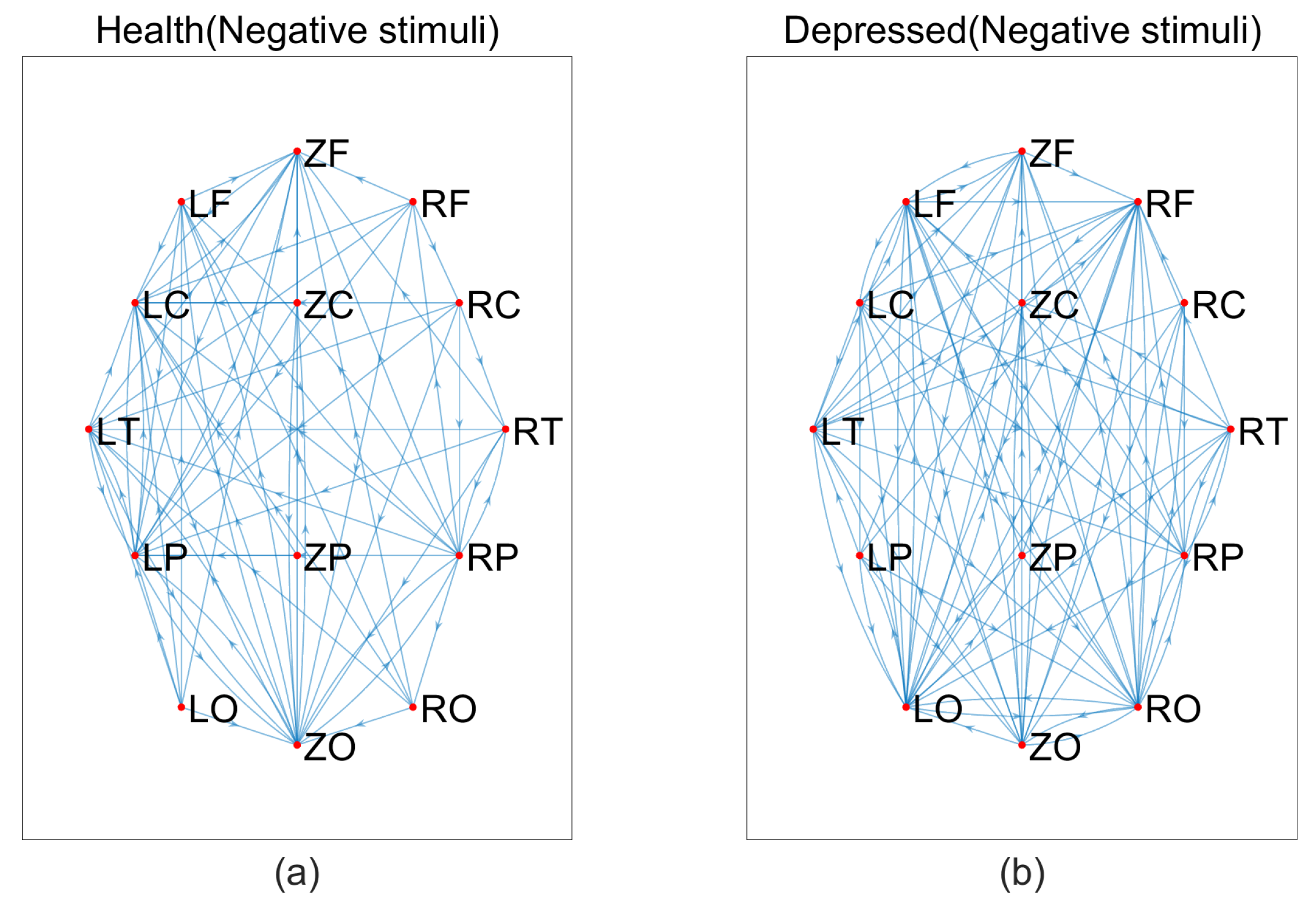
3.2. Experimental process and steps
| Depression | Normal | t | p | |
|---|---|---|---|---|
| ]1*Positive stimulus | 0.1155±0.0041 | 0.1158±0.0041 | 0.040 | 0.968 |
| ]1*Neutral stimulus | 0.1198±0.0041 | 0.1154±0.0041 | 20.158 | 0.000 |
| ]1*Negative stimulus | 0.1211±0.0041 | 0.1157±0.0041 | 21.573 | 0.000 |
4. Summary
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| MEG | Magnetoencephalogram |
References
- Ducasse D, Loas G, Dassa D, E.F. Anhedonia is associated with suicidal ideation independently of depressio n: A meta-analysis. Depression and anxiety 2018, 35, 382–392. [CrossRef] [PubMed]
- Lemoult J, Gotlib I H. Depression: A cognitive perspective. Clinical Psychology Review 2019, 69, 51–66. [CrossRef]
- Leonard B, E. Inflammation and depression: a causal or coincidental link to the pathophysiology? Acta Ne uropsychiatrica 2018, 30, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kalin N, H. The critical relationship between anxiety and depression. American Journal of Psychiatry 2020, 177, 365–367. [Google Scholar] [CrossRef]
- Tuckwiller B, Dardick W R. The critical relationship between anxiety and depression. Journal of Interdisciplinary Studies in Education 2018, 6, 32.
- Mišić B, Sporns O. From regions to connections and networks: new bridges between brain and behavior. Current opinion in neurobiology 2016, 40, 1–7. [CrossRef]
- Shao X, Sun S, Li J. E.F. Analysis of functional brain network in MDD based on improved empirical mode decomposition with resting state EEG data. IEEE Transactions on Neural Systems and Rehabilitation Engineering 2021, 29, 1546–1556. [CrossRef]
- Wang X, Ren Y, Zhang W. Depression disorder classification of fMRI data using sparse low-rank functional brain network and graph-based features. Computational and mathematical methods in medicine 2017, 2017, 3609821.
- Van Mierlo P, Höller Y, Focke N K. E.F. Network perspectives on epilepsy using EEG/MEG source connectivity. Frontiers in neurology 2019, 10, 721. [CrossRef]
- Zhang F F, Peng W, Sweeney J A. E.F. Brain structure alterations in depression: Psychoradiological evidence. CNS neuroscience and therapeutics 2018, 24, 994–1003. [CrossRef]
- Chiou-Wei S Z , Chen C F , Zhu Z. Economic growth and energy consumption revisited - Evidence from linear and nonlinear Granger causality. Energy Economics 2008, 30, 2.
- Previti E , Salinari S , Bertuzzi A. E.F. Glycemic control after metabolic surgery: a Granger causality and graph analysis. American Journal of Physiology 2017, 5, 313.
- Ding M , Chen Y , Bressler S L. Granger Causality: Basic Theory and Application to Neuroscience. John Wiley and Sons 2006, 17.
- Lionel B , Barrett A B , Seth A K. Solved problems for Granger causality in neuroscience: A response to Stokes and Purdon. Neuroimage 20186, 5, 67.
- Bilgi, Mustafa M, Ozalay, E.F. Small frontal gray matter volume in first-episode depression patients. Turkish journal of psychiatry 2010, 21, 185–194.
- Granger, C. Investigating causal relations by econometric models and cross-spectral methods. Harvard University Press 2001, 37, 424–438. [Google Scholar]
- Moguilner, S., García, A. M., Perl, Y. S.. Dynamic brain fluctuationsoutperform connectivity measures and mirror pathophysiological profiles across dementia subtypes: A multicenter study. NeuroImage 2021, 225, 117522. [CrossRef]
- LUO Lin-kai, YE Ling-jun, ZHOU Qi-feng. Geometric Measures and Properties of Commonly Used Kernel Functions. Journal of Xiamen University.Natural Science 2009, 48, 804–807.
- Güntekin B, Basar E. Emotional face expressions are differentiated with brain oscillations. International Jo urnal of Psychophysiology 2007, 64, 91–100. [CrossRef]
- Güntekin B, Başar E. Event-related beta oscillations are affected by emotional eliciting stimuli. Neuroscien ce letters 2010, 483, 173–178. [CrossRef]
- Akaike, H. Information theory and an extension of the maximum likelihood principle. Selected papers of hirotugu akaike 1998, 199–213. [Google Scholar]
- Yang C F, Le B J R, Bellanger J ,E.F. A new strategy for model order identification and its application to transfer entropy for EEG signals analysis. IEEE Trans Biomed Eng 2013, 60, 1318–1327. [CrossRef] [PubMed]
- Liu Y, Liang M, Zhou Y,E.F. Disrupted small-world networks in schizophrenia. Brain 2008, 131, 945–961. [CrossRef] [PubMed]
- Goldman R I, Stern J M, Engel Jr J,E.F. Simultaneous EEG and fMRI of the alpha rhythm. Neuroreport 2002, 13, 2487. [CrossRef]
- Garrett A, Kelly R, Gomez R,E.F. Aberrant brain activation during a working memory task in psychotic major depression. American Journal of Psychiatry 2011, 168, 173–182. [CrossRef] [PubMed]
- Tan Y L, Zou Y Z, Qu Y. Frontal Lobe Executive Function of Patients with Major Depression and OCD. Chinese Mental Health Journal 2003, 17, 617–619.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




