1. Introduction
Breast cancer (BC), constituting 31% of female cancers, is the most commonly diagnosed malignancy and the second leading cause of morbidity among women worldwide [
1]. Especially, breast cancer in young individual has worse histological grading, higher ki-67, and more lymphocyte nodes infiltration, leading to more relapse and poorer survival outcomes [
2,
3]. Recent years have witnessed a gradual rise in the incidence of breast cancer among young individuals, necessitating focused attention in both research endeavors and clinical practice [
4].
Notably, previous studies have consistently demonstrated that young patients present with more severe clinicopathological features and unfavorable prognosis [
5]. However, the underlying biological mechanisms driving these disparities have remained elusive. Tumors, including breast cancer, are widely acknowledged to exhibit significant intertumoral and intratumoral heterogeneity [
6]. By leveraging transcriptional and genomic profiling, we sought to elucidate the heterogeneity between young and elder BC patients and gain a deeper understanding of the biological basis of the observed differences between these two patient groups. It’s widely recognized that tumors consist of a heterogeneous mix of cells, and the application of single-cell RNA-sequencing (scRNA-seq) on isolated tumor tissue allows for the characterization of heterogeneous tumor cells [
7,
8]. Notably, scRNA-seq enables the molecular distinction of identical cell types within a complex population mix, thereby revealing the distinct characteristics of specific cell types.
The mounting evidence highlights the involvement of metabolic regulation in cancer progression, metastasis, and therapy resistance [
9,
10]. Therefore, understanding metabolic heterogeneity among patients becomes crucial in predicting diverse survival outcomes. Our previous study has successfully validated the metabolic heterogeneity in triple negative breast cancer (TNBC) patients and proposed distinct clinical characteristics and potential therapeutic approaches for each subtype [
11,
12,
13]. Building upon these discoveries, the recent study aims to investigate the metabolic difference between different age groups, thereby elucidating the underlying factors contributing to the diverse prognosis observed.
Stemness is characteristics and properties exhibited by stem cell, which is associated with tumor initiation, progression, metastasis, therapy resistance, and relapse [
14,
15,
16,
17,
18]. The expression of breast cancer stem cell (CSC) markers, including CD44, CD29, and CD49f is correlated with aggressive tumor features and increased risk of recurrence.
Interestingly, our work revealed upregulation of galactose metabolism pathway specifically in young patients through the bulk RNA-seq data, and further uncovered a correlation between galactose metabolism and stemness through scRNA-seq data. Moreover, we experimentally validate this correlation in cell lines. Our work provides a novel perspective to explore the underlying biological basis of poor prognosis in young BC patients.
2. Material and Methods
2.1. Study cohorts and statistical analyses
Both clinical, transcriptional, and genomic data from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort were sourced from the cBio-Portal for Cancer Genomics [
19]. A total of 1980 patients were encompassed. The clinical data of 12790 operable BC patients were obtained from the Fudan University Shanghai Cancer Center (FUSCC), specific transcriptional and genomic data were obtained from 466 patients [
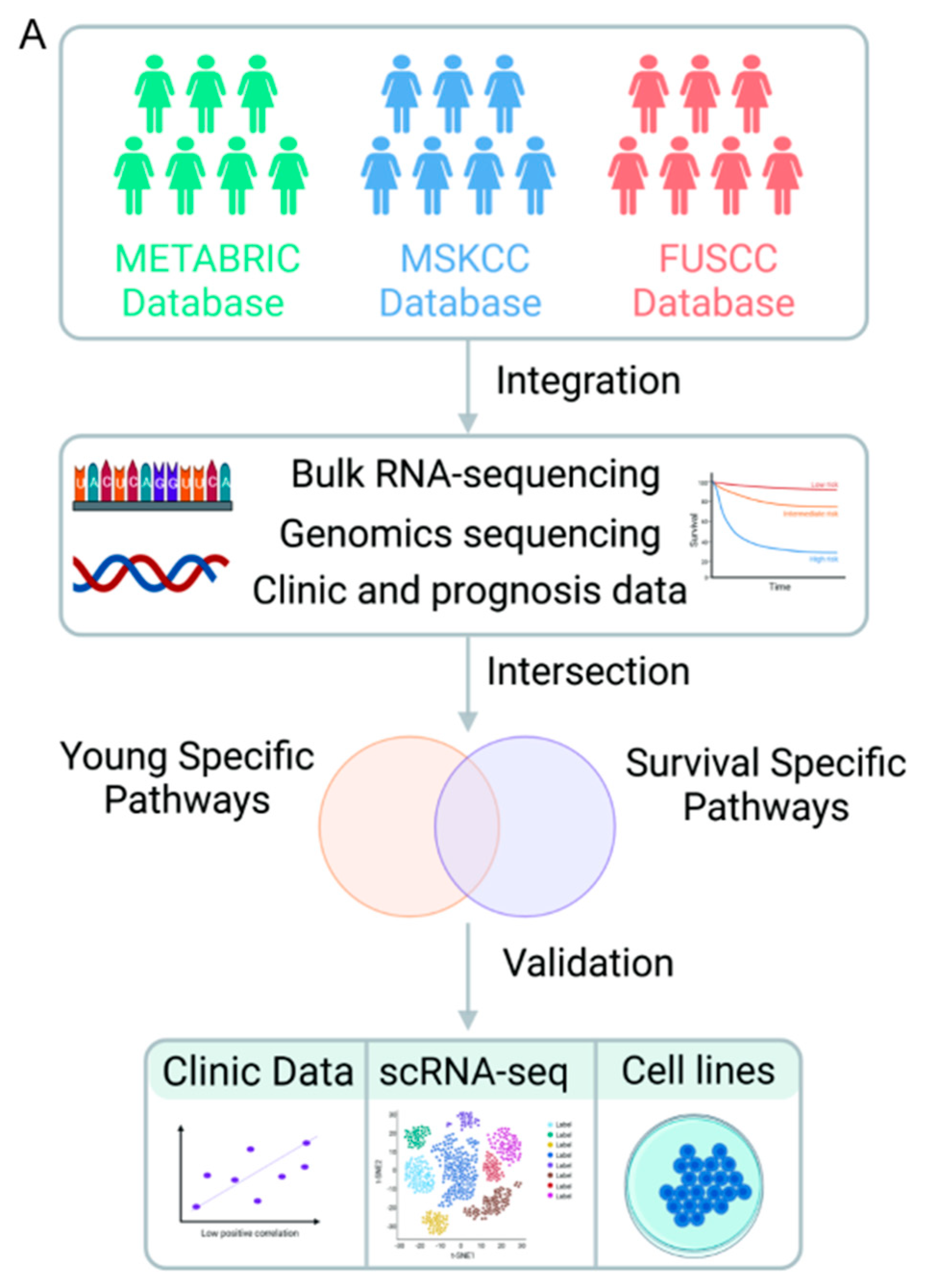
11]. Additionally, the genomic data from the Memorial Sloan Kettering Cancer Center (MSKCC) cohort included 1865 patients. The entire workflow of the study is visually presented in
Figure 1.
To analyze the somatic mutation profiles of different patient groups, the maftools package (v2.6.05) was employed. The primary endpoints of interest were overall survival (OS) and disease-free survival (DFS). Kaplan-Meier plots were used to estimate survival rates among different age groups, and the log-rank test was utilized to identify metabolic pathways associated with DFS. All bioinformatics analyses were performed using R version 4.2.0 and Python version 3.9.
2.2. scRNA-seq cohorts and data processing
Quality-controlled scRNA-seq data of BC patients are deposited at Gene Expression Omnibus (GEO accession numbers: GSE176078) and 36 treatment-naïve patients were enrolled for subsequent analysis [
20]. Briefly, data normalization, batch correction, dimensionality reduction, and cell clustering were performed via Seurat4 package (v4.1.0). Later, cell clusters were annotated using the Garnett package (v0.2.18) and inferCNV package (v1.6.0). The normalized enrichment score (NES) of the metabolic pathways for each cell was calculated using the ssMWW-GST function [
21]. Ultimately, the cytoTRACE score for individual cell was calculated via the CYTOTRACE package (v0.3.3) [
22].
2.3. Cell lines and D-galactose treatment
Human BC cell lines, Hs578t, MDA-MB-231, MDA-MB-468, BT549, HCC1806, BT20, and HCC1937 cell lines, were purchased from the American Type Culture Collection (ATCC) and confirmed without mycoplasma contamination. Hs578t and MDA-MB-231 cells were cultured in high-glucose DMEM (BasalMedia, L110) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (BasalMedia, S110B)) at 37 °C with 5% CO2 incubator.
For the D-galactose treatment, cells were seeded in six-well plates and allowed to incubate overnight. Subsequently, 25mM D-galactose (Sigma-Aldrich, G5388) was supplemented to the growth medium [
23]. Cells were cultured for 24 hours for RNA-extraction experiments and 120 hours for the mammosphere formation assay.
2.4. RNA extraction, RT, and real-time qPCR
Total RNA was extracted using the RNA-Quick Purification Kit (ES science, RN001), and reverse-transcribed (RT) into complementary DNA (cDNA) using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech, R211). Real-time quantitative Polymerase Chain Reaction (qPCR) was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Q711). All primers for real-time qPCR were provided in supplementary table 1.
2.5. Mammosphere formation assay
The mammosphere culture was conducted following the procedures described in a prior study [
17]. MDA-MB-231 and Hs578t cells were seeded in 24-well ultra-low attachment plates (Corning, 3473) at a density of 2x10
3 cells per well. Cells were cultured in DMEM/F12 reduced serum median (Gibco, 12364010) supplemented with B27 (ThermoFisher, 17504044), 20 ng/ml epidermal growth factor (EGF, Peprotech, 100-47), 20 ng/ml basic fibroblast growth factor (bFGF, ThermoFisher, 13256029), 0.4% albumin from bovine serum (BSA, BasalMedia, S476T7), 2 μg/ml heparin (STEMCELL, 07980), and insulin-transferrin-selenium (ITS-G, BasalMedia, S450J7). Following a 5-day incubation, the quantification of mammospheres (diameter > 50 μm) was performed, and representative images were captured.
3. Results
3.1. Clinicopathological and survival features of BC in young patients
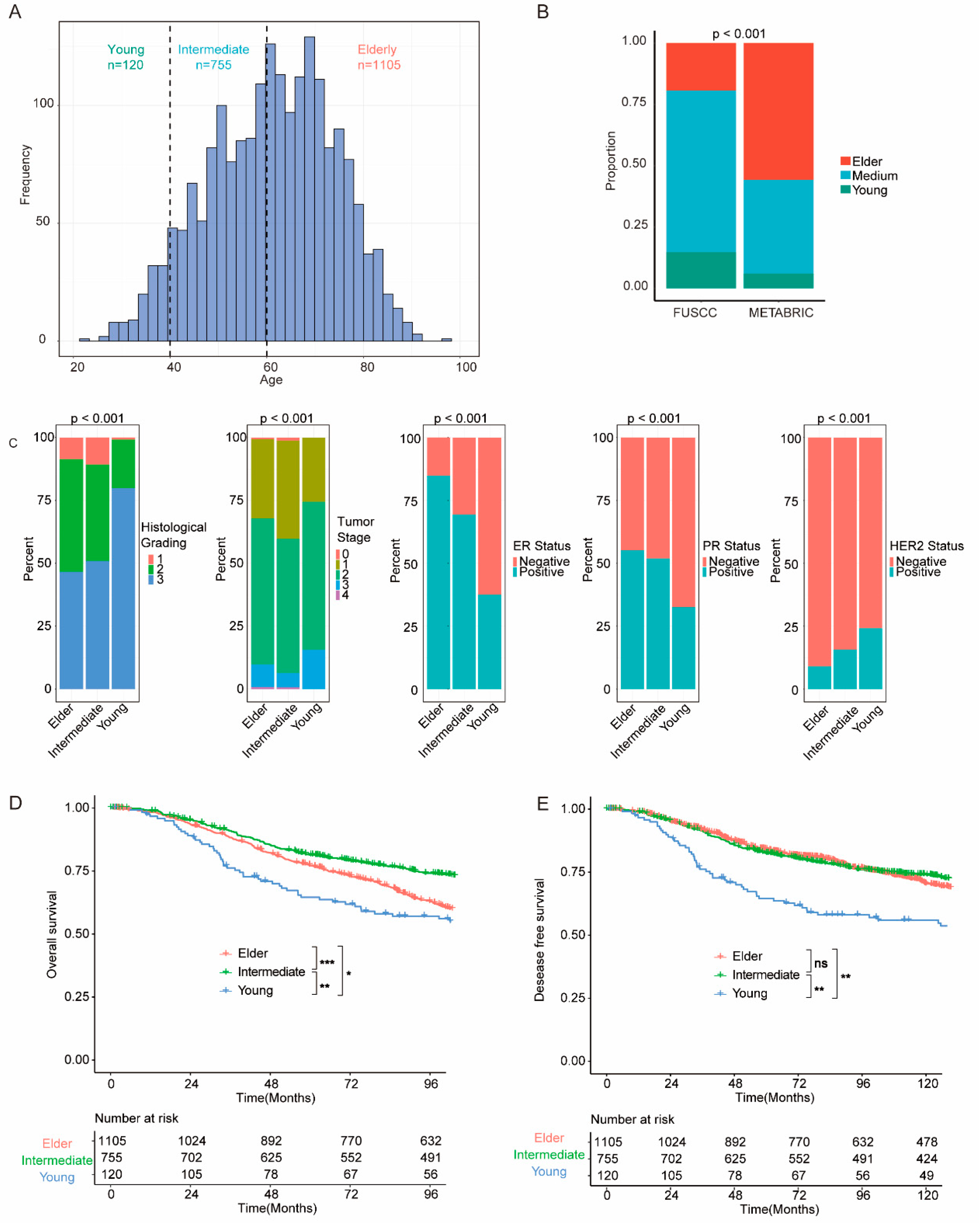
From the METABRIC cohort, 1980 patients with BC were eligible for further analyses. These patients were categorized into different age groups: the young group comprising 120 patients (aged ≤39 years), the intermediate group consisting of 755 patients (aged 40 to 59 years), and the elderly group comprising 1105 patients (aged ≥60 years), as depicted in
Figure 2a. Otherwise, the FUSCC cohort exhibited a larger proportion of patients in the young and intermediate groups compared with the METABRIC cohort (
Figure 2b and supplementary Figure S1a). Clinicopathological features of individual groups were analyzed, and significantly different features (P<0.001) between groups are illustrated in
Figure 2c. In the METABRIC cohort, young patients displayed higher histological grading and worse tumor stage. Additionally, young patients are more likely to be negative for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2).
We further investigated the survival outcomes of patients within different groups. The young group showed worse OS (overall survival) and DFS (disease-free survival) compared to both the intermediate group (OS P=0.050; DFS, P=0.002) and the elder group (OS P=0.003; DFS, P=0.008) (
Figure 1d,e). Likewise, the young group demonstrated worse OS in the FUSCC cohort (Supplementary Figure S1b).
3.2. Genomics features of BC in young patients
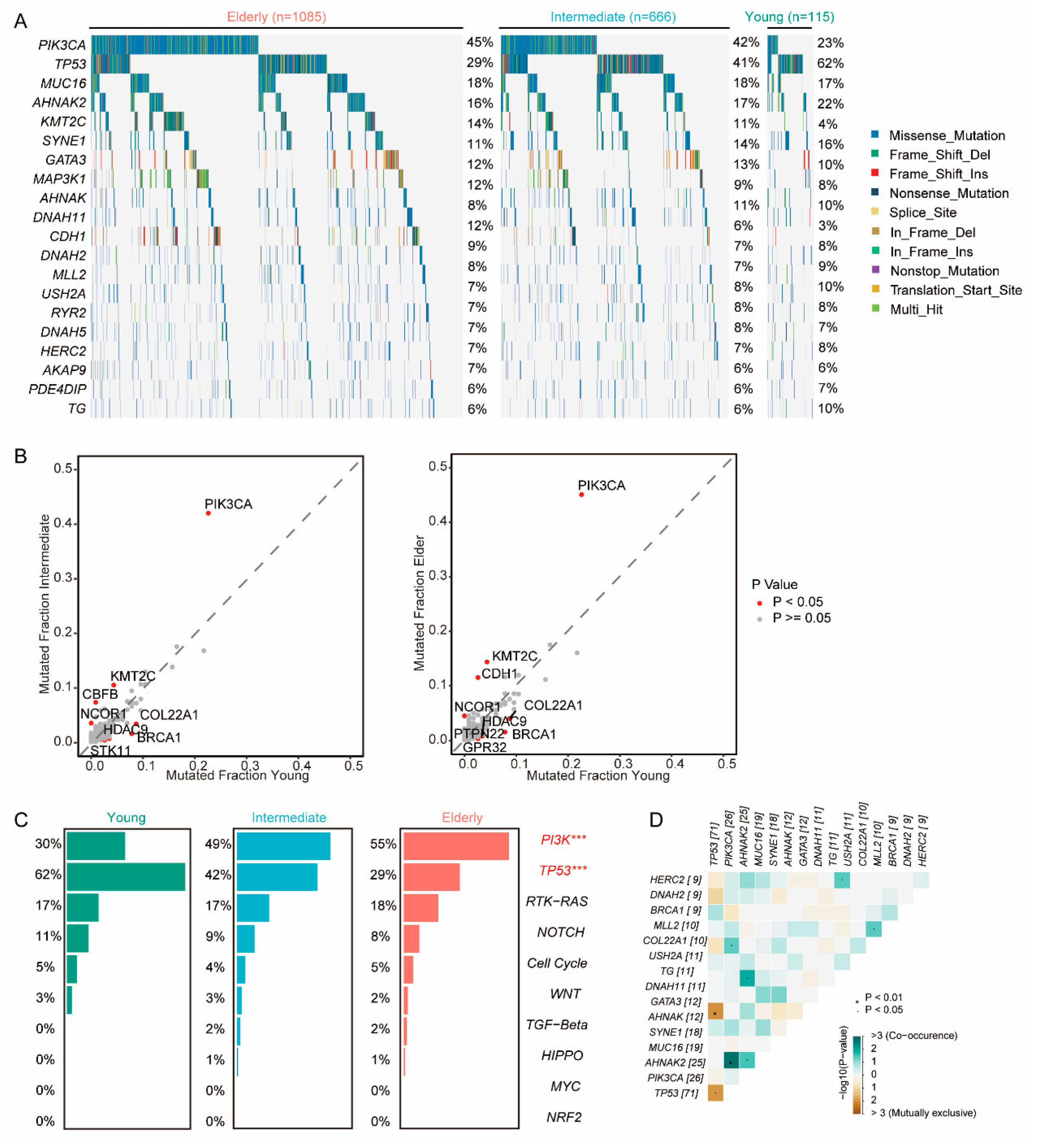
Given that mutation of oncogenes and tumor suppressor genes play a crucial role in tumor progression, we explored the genomic heterogeneity of patients across different age groups. Young group exhibited a higher frequency of mutations in TP53 (young vs intermediate vs elder: 61.7% vs 40.8% vs 29.0%, P <0.001), BRCA1(7.8% vs 1.6% vs 1.6%, P<0.001), and COL22A1(8.7% vs 3.5% vs 4.1%, P=0.019), and HDAC9(3.4% vs 0.8% vs 0.8%, P=0.031); a lower frequency in PIK3CA (22.6% vs 42.0% vs 45.1%, P <0.001) and KMT2C (4.3% vs 10.5% vs 14.4% P=0.001) (
Figure 3a,b). Furthermore, young patients had a higher prevalence of somatic mutations in TP53 signaling family (62% vs 42% vs 29%, P<0.001) and a lower frequency in PI3K signaling family (30% vs 49% vs 55%, P<0.001) (
Figure 3c). To further explore the genomic features of the young patients, we found co-occurrent mutations in PIK3CA and AHNAK2 and mutually exclusive mutations in TP53 and ANNAK (
Figure 3d). Remarkably, similar trends were observed in the MSKCC cohorts (Supplementary Figure S2).
3.3. Transcriptomic features of BC in young patients
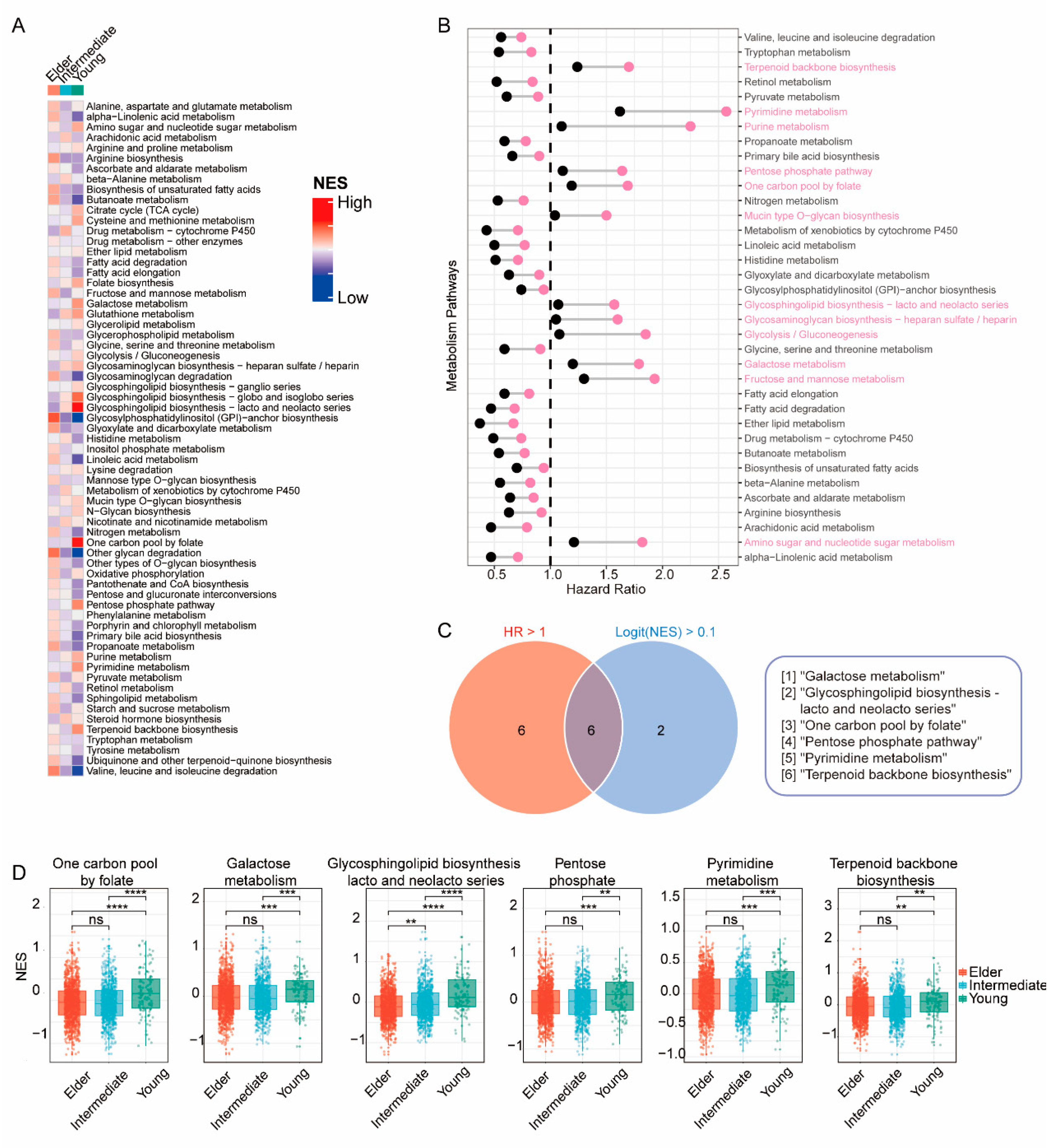
To investigate the potential mechanisms contributing to poorer survival outcomes in the young group, we performed the Mann-Whitney-Wilcoxon gene set test (MWW-GST) and calculated the enrichment normalized score (NES) of 65 metabolic pathways in individual bulk RNA-seq samples from the METABRIC cohort. Notably, the young group exhibited significant upregulation of 8 pathways (logFC (NES) > 0.1 & qValue < 0.01) compared with the intermediate and elder groups (
Figure 4a), including the glycosphingolipid biosynthesis pathway. Moreover, 12 pathways (HR >1 & P < 0.01) were identified as determinant prognostic features for worse DFS (
Figure 4b), such as the fructose and mannose metabolism pathway (HR 1.58, 95% CI 1.300-1.930, p<0.001, downregulation as reference). Among these metabolic pathways, 6 pathways demonstrated simultaneous upregulation in the young group and statistical significance for DFS via multivariate analysis (
Figure 4c,d), including the galactose metabolism pathway.
3.4. Galactose metabolic pathways are upregulated in less-differentiated cancer cells
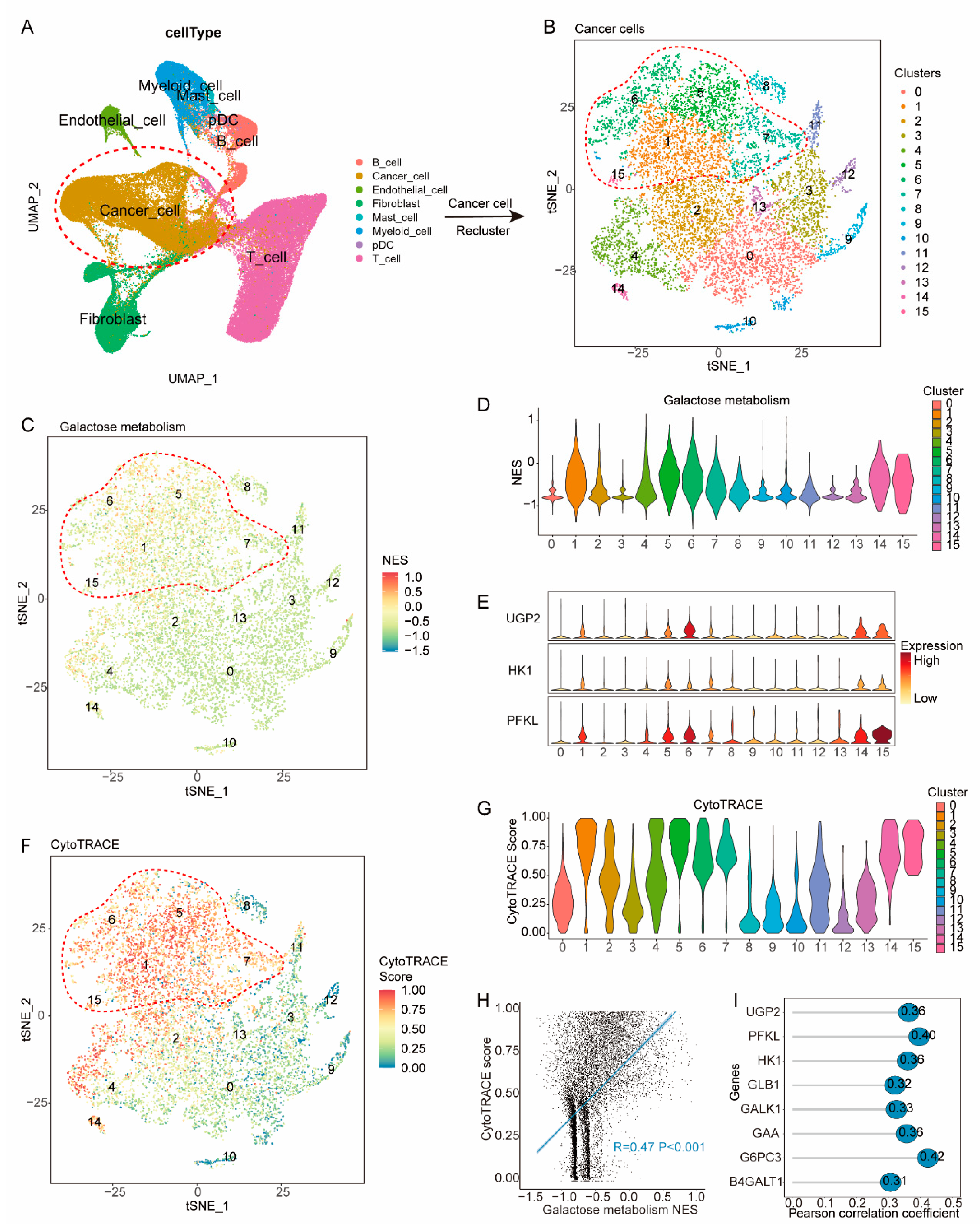
To gain insight into how galactose metabolism may contribute to worse DFS, we focused our analysis on cells from 35 treatment-naïve breast cancer samples in the published single-cell dataset (GSE176078) (
Figure 5a, Supplementary Figure S3). Following quality control, data integration, batch-effect removal, and cell annotation, 28,661 cancer cells were identified and re-clustered into 16 clusters (
Figure 5b). We then assessed the NES of galactose metabolism pathways in individual cancer cells, and cells in cluster1, 5, 6, 7, 14, and 15 showed upregulation of metabolism pathways and were annotated as HGM (Hyper-galactose metabolic) cells (
Figure 5c,d). Additionally, HGM cells significantly expressed UGP2, HK1, and PFKL, which are critical genes in galactose metabolism pathway (
Figure 5e).
To uncover the characteristics of HGM cells, we calculated the cytoTRACE score of individual cells, which is indicative of their differentiation states. Interestingly, HGM cells displayed significantly higher cytoTRACE scores compared to others (
Figure 5f,g), indicating their less-differentiate status. Previous studies have associated less-differentiated cells with relapse and metastasis. Among all cancer cells, the cytoTRACE score positively correlated with the NES of galactose metabolism (R=0.47 P<0.001) (
Figure 5h). Additionally, the cytoTRACE score exhibited significant correlations with the expression of critical genes in galactose metabolism pathway, including UGP2, HK1, PFKL, GLB1, GALK1, GAA, G6PC3 and B4GALT1 (R>0.3 P<0.001) (
Figure 5i).
3.5. Galactose metabolism regulates breast cancer stemness
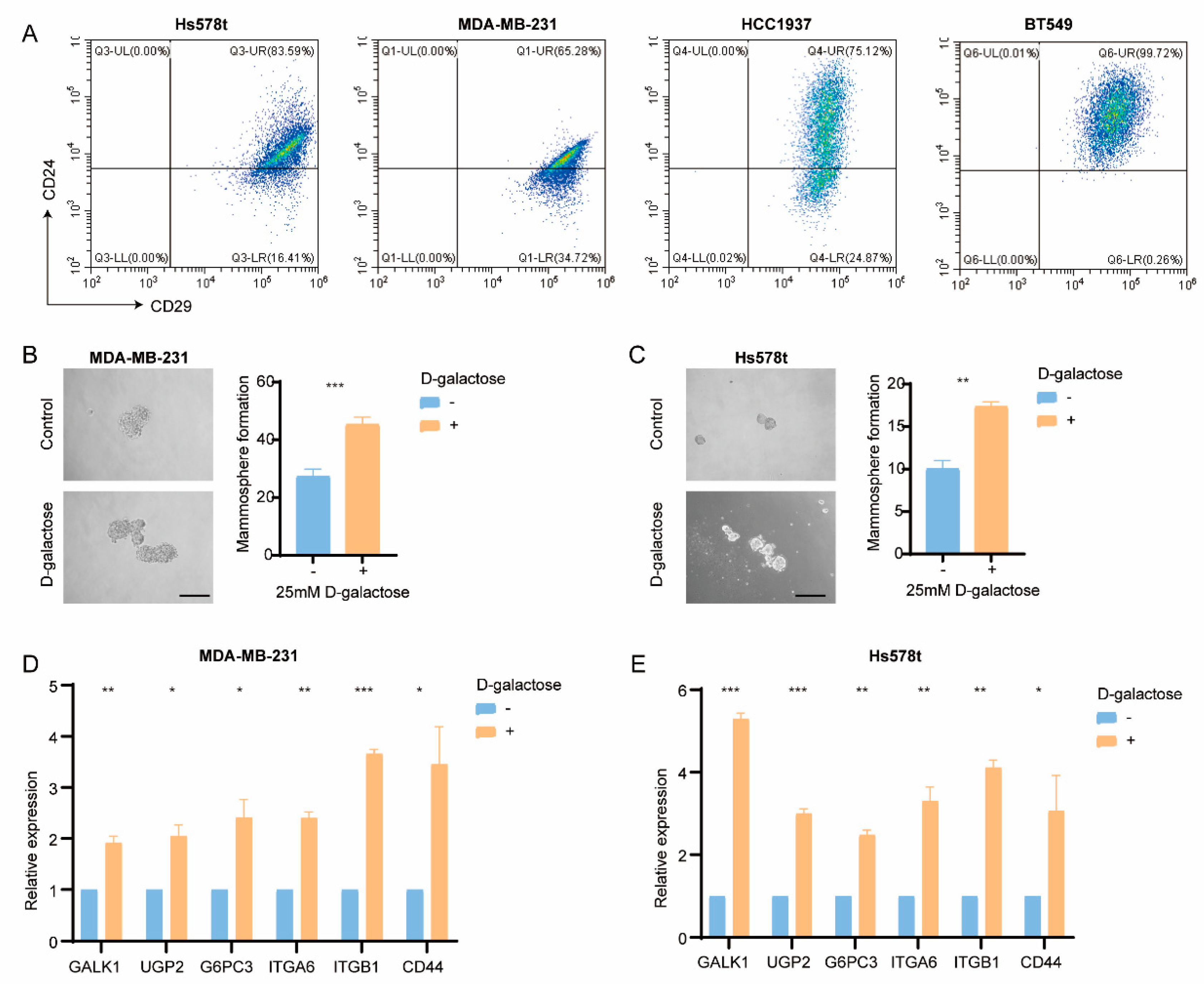
To validate the influence of galactose metabolism in regulating BC stemness in vitro, BC cell lines were utilized. According to the RNA-seq data from 19 cell lines, Hs578t, MDA-MB-231, MDA-MB-436, HCC1395, BT549, HCC1143, and HCC1937 cell lines showed higher enrichment scores of galactose metabolism and stemness signature. Moreover, the CD29
highCD24
+ (stem-like) subpopulation was found in these cell lines (
Figure 6a). Subsequently, we examined the impact of galactose metabolism on the tumor cells. Interestingly, D-galactose treatment significantly enhanced the mammosphere formation efficacy of MDA-MB-231 and Hs578t cells (
Figure 6b,c). Meanwhile, the real-time qPCR results showed the upregulation of GALK1, UGP2, G6PC3, ITGA6, ITGB1, and CD44 in cells treated with D-galactose (
Figure 6d,e), suggesting the potential role of galactose metabolism in regulating stemness.
4. Discussion
The result of our study confirmed differences in breast cancer among young patients compared to other age groups. In terms of clinicopathological features, breast cancer in young individuals was characterized by higher histological grading, greater likelihood of being ER/PR/HER2 negative, and worse DFS. Examining the multi-omics perspective, young patients harbored more somatic mutations in TP53, BRCA1, COL22A1, and HDAC9, but fewer mutations in PIK3CA and KMT2C; harbored more mutations in TP53 signaling family, but fewer in PI3K-AKT-mTOR signaling family. Additionally, our analysis revealed an upregulation of the galactose metabolism pathway in young patients, which emerged as a significant prognostic feature linked to worse DFS outcomes. Eventually, the scRNA-seq data from breast cancer patients and experimental results from cell lines confirmed that galactose metabolism showed the potential to modulate cancer stemness, ultimately contributing to the poorer DFS outcomes observed in young breast cancer patients.
We conducted an extensive genomic analysis to elucidate prognostic mutations and potential therapeutic targets. Notably, young patients displayed a higher incidence of mutations in BRCA1 and TP53 genes, which were also evident in the corresponding pathways. Emerging evidence indicates that TP53 mutation is inclined to reach a complete response (CR) after chemotherapy; however, it is also linked to worse OS and DFS in BC patients [
24,
25,
27]. Furthermore, TP53-mutated patients showed a higher risk of tumor recurrence following breast-conserving therapy (BCT) than wild-type patients, thereby warranting a recommendation of mastectomy for TP53-mutated individuals [
26]. In contrast, a lower mutation frequency of PIK3CA was observed in young patients, consistent with the findings in the PI3K pathway. A large pooled analysis found that PIK3CA mutation was associated with increasing age and improved DFS and OS in early-stage BC [
28]. Additionally, results from the SAFIR02 trial (NCT02299999) indicated that PIK3CA mutation was associated with a better OS in metastatic triple-negative BC [
29]. These genomic features may provide insight into the rationale behind the poorer survival outcomes observed in young BC patients.
From the transcriptional data, we observed significant upregulation of galactose metabolism pathways in young patient groups and identified the galactose metabolism pathway as a prognostic factor for worse survival outcomes. Notably, recent studies have highlighted the role of galactose metabolic enzymes in modulating the stemness of cancer stem cells (CSC) and influencing clinical outcomes. In the Leloir pathway, galactose is initially converted to galactose-1-phosphate by galactokinase (GALK1), which allows cancer cells to utilize galactose as an alternative fuel source instead of glucose in glioblastoma (GBM) [
30]. Also, the expression of GALK1 has been found to be correlated with poor clinical outcomes in GBM. Moreover, GALK1 has also been implicated in promoting high epithelial-mesenchymal transition (EMT) and wore OS in colorectal cancer (CRC) [
31]. Additionally, the conversion of glucose-1-phosphate to UDP-glucose, a critical step in galactose metabolism, is catalyzed by UDP-glucose pyrophosphorylase-2 (UGP2). UGP2 has been recognized as a crucial factor in cancer maintenance and has emerged as a potential therapeutic target in pancreatic ductal adenocarcinoma (PDAC) [
32]. UGP2 upregulation has been observed in leukemic stem cell-enriched factions and has been identified as a determinant prognostic factor in OS and DFS [
33]. Another enzyme, Beta-1,4-Galactosyltransferase 1 (B4GALT1), has been implicated in maintenance of the stemness in lung adenocarcinoma (LUAD) and CRC [
34,
35]. Our findings align well with previous studies, collectively suggesting that the galactose metabolism pathway may indeed play a pivotal role in modulating the stemness of BC cells. Therefore, targeting this pathway could hold promise as a potential therapeutic approach to improve survival outcomes in young breast cancer patients.
The current study has limitations. Firstly, the clinicopathological and multi-omics features identified in young breast cancer patients were based on retrospective cohorts, which may introduce inherent biases and limit the generalizability of our findings. Secondly, the scRNA-seq data available for young patients were insufficient in quantity, hindering a comprehensive analysis of significantly enriched cell clusters in this subgroup. Thirdly, while we have identified the galactose metabolism pathway as a potential prognostic factor in young BC patients, the underlying mechanism by which this pathway modulates stemness remains unexplored. Additionally, further investigation into potential therapeutic targets within this pathway is warranted to enhance its clinical relevance. Lastly, in order to translate our findings into clinical practice, thorough validation in prospective cohorts is essential and rigorous clinical validation is imperative to ascertain their efficacy and safety profiles.
5. Conclusion
Through the integration and comprehensive analysis of multi-omics data in breast cancer (BC), we have successfully identified distinct clinicopathological characteristics, specific genetic vulnerabilities, and metabolic features unique to young patient groups. Specifically, our investigation revealed a significant upregulation of the galactose metabolism pathway in these young patients. Furthermore, we unveiled its potential role in modulating stemness, ultimately contributing to the observed worse survival outcomes. These findings hold promising implications for the development of potential therapeutic strategies for young BC patients.
Author Contributions
XH: HL, and XCH designed this study. XCH and BYH collected data. XCH, HL, and BYH analyzed the statistics and wrote the manuscript. XCH, XH and HL revised the manuscript. All authors have approved the final manuscript.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81872137, 82072917 and 82272957) and a grant from the Shanghai Science and Technology Innovation Action Plan (22DZ2204400). The funders had no role in the study design, collection and analysis of the data, decision to publish, or manuscript preparation.
Conflicts of Interest
The authors have declared that no conflicts of interest exist in this work.
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Tichy, J.R.; Lim, E.; Anders, C.K. Breast Cancer in Adolescents and Young Adults: A Review with a Focus on Biology. J. Natl. Compr. Cancer Netw. 2013, 11, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Paluch-Shimon, S.; Cardoso, F.; Partridge, A.; Abulkhair, O.; Azim, H.; Bianchi-Micheli, G.; Cardoso, M.; Curigliano, G.; Gelmon, K.; Gentilini, O.; et al. ESO–ESMO fifth international consensus guidelines for breast cancer in young women (BCY5). Ann. Oncol. 2022, 33, 1097–1118. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chan, P.S.; Lok, V.; Chen, X.; Ding, H.; Jin, Y.; Yuan, J.; Lao, X.-Q.; Zheng, Z.-J.; Wong, M.C. Global incidence and mortality of breast cancer: A trend analysis. Aging 2021, 13, 5748–5803. [Google Scholar] [CrossRef] [PubMed]
- Eiriz, I.; Batista, M.V.; Tomás, T.C.; Neves, M.; Guerra-Pereira, N.; Braga, S. Breast cancer in very young women—A multicenter 10-year experience. ESMO Open 2021, 6, 100029. [Google Scholar] [CrossRef]
- Hu, X.; Myers, K.S.; Oluyemi, E.T.; Philip, M.; Azizi, A.; Ambinder, E.B. Presentation and characteristics of breast cancer in young women under age 40. Breast Cancer Res. Treat. 2021, 186, 209–217. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Liu, S.-Q.; Gao, Z.-J.; Wu, J.; Zheng, H.-M.; Li, B.; Sun, S.; Meng, X.-Y.; Wu, Q. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J. Hematol. Oncol. 2022, 15, 19. [Google Scholar] [CrossRef]
- Gong, Y.; Ji, P.; Yang, Y.-S.; Xie, S.; Yu, T.-J.; Xiao, Y.; Jin, M.-L.; Ma, D.; Guo, L.-W.; Pei, Y.-C.; et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021, 33, 51.e9–64e9. [Google Scholar] [CrossRef]
- Yu, T.-J.; Ma, D.; Liu, Y.-Y.; Xiao, Y.; Gong, Y.; Jiang, Y.-Z.; Shao, Z.-M.; Hu, X.; Di, G.-H. Bulk and single-cell transcriptome profiling reveal the metabolic heterogeneity in human breast cancers. Mol. Ther. 2021, 29, 2350–2365. [Google Scholar] [CrossRef]
- Jiang, Y.-Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.-D.; Liu, Y.-R.; Yu, Y.; et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019, 35, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, D.; Yang, Y.-S.; Yang, F.; Ding, J.-H.; Gong, Y.; Jiang, L.; Ge, L.-P.; Wu, S.-Y.; Yu, Q.; et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res. 2022, 32, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiao, Y.; Ding, J.-H.; Jin, X.; Ma, D.; Li, D.-Q.; Shi, J.-X.; Huang, W.; Wang, Y.-P.; Jiang, Y.-Z.; et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023, 35, 84–100. [Google Scholar] [CrossRef]
- Wang, D.; Hu, X.; Liu, C.; Jia, Y.; Bai, Y.; Cai, C.; Wang, J.; Bai, L.; Yang, R.; Lin, C.; et al. Protein C receptor is a therapeutic stem cell target in a distinct group of breast cancers. Cell Res. 2019, 29, 832–845. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.P.; Chan, T.E.; Lombardo, Y.; Corleone, G.; Rotmensz, N.; Bravaccini, S.; Rocca, A.; Pruneri, G.; McEwen, K.R.; Coombes, R.C.; et al. Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy. Nat. Commun. 2019, 10, 3840. [Google Scholar] [CrossRef] [PubMed]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Sun, Y.; Hou, Y.; Yang, L.; Wan, X.; Qin, Y.; Liu, Y.; Wang, R.; Zhu, P.; Teng, Y.; et al. A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J. Hematol. Oncol. 2021, 14, 178. [Google Scholar] [CrossRef]
- Wang, H.; Xiang, D.; Liu, B.; He, A.; Randle, H.J.; Zhang, K.X.; Dongre, A.; Sachs, N.; Clark, A.P.; Tao, L.; et al. Inadequate DNA Damage Repair Promotes Mammary Transdifferentiation, Leading to BRCA1 Breast Cancer. Cell 2019, 178, 135–e19. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Garofano, L.; Migliozzi, S.; Oh, Y.T.; D’Angelo, F.; Najac, R.D.; Ko, A.; Frangaj, B.; Caruso, F.P.; Yu, K.; Yuan, J.; et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Rev. Cancer 2021, 2, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Gulati, G.S.; Sikandar, S.S.; Wesche, D.J.; Manjunath, A.; Bharadwaj, A.; Berger, M.J.; Ilagan, F.; Kuo, A.H.; Hsieh, R.W.; Cai, S. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020, 367, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, P.S.; O’prey, J.; Cardaci, S.; Barthet, V.J.A.; Sakamaki, J.-I.; Beaumatin, F.; Roseweir, A.; Gay, D.M.; Mackay, G.; Malviya, G.; et al. Mannose impairs tumour growth and enhances chemotherapy. Nature 2018, 563, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Blondeaux, E.; Arecco, L.; Punie, K.; Graffeo, R.; Toss, A.; De Angelis, C.; Trevisan, L.; Buzzatti, G.; Linn, S.C.; Dubsky, P.; et al. Germline TP53 pathogenic variants and breast cancer: A narrative review. Cancer Treat. Rev. 2023, 114, 102522. [Google Scholar] [CrossRef] [PubMed]
- Shahbandi, A.; Nguyen, H.D.; Jackson, J.G. TP53 Mutations and Outcomes in Breast Cancer: Reading beyond the Headlines. Trends Cancer 2020, 6, 98–110. [Google Scholar] [CrossRef]
- Guo, Y.; Wan, Q.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Xie, Y. Risk of ipsilateral breast tumor recurrence and contralateral breast cancer in patients with and without TP53 variant in a large series of breast cancer patients. Breast 2022, 65, 55–60. [Google Scholar] [CrossRef]
- Tung, N.M.; Boughey, J.C.; Pierce, L.J.; Robson, M.E.; Bedrosian, I.; Dietz, J.R.; Dragun, A.; Gelpi, J.B.; Hofstatter, E.W.; Isaacs, C.J.; et al. Management of Hereditary Breast Cancer: American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology Guideline. J. Clin. Oncol. 2020, 38, 2080–2106. [Google Scholar] [CrossRef]
- Zardavas, D.; Marvelde, L.T.; Milne, R.L.; Fumagalli, D.; Fountzilas, G.; Kotoula, V.; Razis, E.; Papaxoinis, G.; Joensuu, H.; Moynahan, M.E.; et al. Tumor PIK3CA Genotype and Prognosis in Early-Stage Breast Cancer: A Pooled Analysis of Individual Patient Data. J. Clin. Oncol. 2018, 36, 981–990. [Google Scholar] [CrossRef]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Dien, A.T.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.-P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef]
- Sharpe, M.A.; Ijare, O.B.; Baskin, D.S.; Baskin, A.M.; Baskin, B.N.; Pichumani, K. The Leloir Cycle in Glioblastoma: Galactose Scavenging and Metabolic Remodeling. Cancers 2021, 13, 1815. [Google Scholar] [CrossRef]
- Liu, G.; Wu, X.; Chen, J. Identification and validation of a glycolysis-related gene signature for depicting clinical characteristics and its relationship with tumor immunity in patients with colon cancer. Aging 2022, 14, 8700–8718. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Zhou, Q.; Toska, E.; Galeas, J.; Ku, A.A.; Koche, R.P.; Bandyopadhyay, S.; Scaltriti, M.; Lebrilla, C.B.; McCormick, F.; et al. UDP-glucose pyrophosphorylase 2, a regulator of glycogen synthesis and glycosylation, is critical for pancreatic cancer growth. Proc. Natl. Acad. Sci. USA 2021, 118, e2103592118. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, H.J.M.; Woolthuis, C.M.; Vos, A.Z.; Mulder, A.; Berg, E.v.D.; Kluin, P.M.; van der Weide, K.; de Bont, E.S.J.M.; Huls, G.; Vellenga, E.; et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011, 25, 1825–1833. [Google Scholar] [CrossRef] [PubMed]
- De Vitis, C.; Corleone, G.; Salvati, V.; Ascenzi, F.; Pallocca, M.; De Nicola, F.; Fanciulli, M.; di Martino, S.; Bruschini, S.; Napoli, C.; et al. B4GALT1 Is a New Candidate to Maintain the Stemness of Lung Cancer Stem Cells. J. Clin. Med. 2019, 8, 1928. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, Z.; Guo, X.; Dong, H.; Zhou, K.; Huang, Z.; Xiao, Z. lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity. J. Cell. Physiol. 2019, 234, 18524–18534. [Google Scholar] [CrossRef]
Figure 1.
Graphical overview of the investigation.
Figure 1.
Graphical overview of the investigation.
Figure 2.
Clinicopathological and survival features of breast cancer patients across different age groups. (A) Distribution of age at diagnosis and the defined age groups in the study cohort. (B) Comparison of age group distributions between the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort and the Fudan University Shanghai Cancer Center (FUSCC) cohort. (C) Histological grading, tumor stage, estrogen receptor (ER) status, progesterone receptor (PR) status, and human epidermal growth factor receptor 2 (HER2) status in each age group. (D) Overall survival (OS) in the MTEABRIC cohort. (E) Disease-free survival (DFS) in the METABRIC cohort. Survival comparisons were performed, and significant differences were annotated as *P < .05; **P < .01; ***P < .001; or not significant (ns) (P > .05).
Figure 2.
Clinicopathological and survival features of breast cancer patients across different age groups. (A) Distribution of age at diagnosis and the defined age groups in the study cohort. (B) Comparison of age group distributions between the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort and the Fudan University Shanghai Cancer Center (FUSCC) cohort. (C) Histological grading, tumor stage, estrogen receptor (ER) status, progesterone receptor (PR) status, and human epidermal growth factor receptor 2 (HER2) status in each age group. (D) Overall survival (OS) in the MTEABRIC cohort. (E) Disease-free survival (DFS) in the METABRIC cohort. Survival comparisons were performed, and significant differences were annotated as *P < .05; **P < .01; ***P < .001; or not significant (ns) (P > .05).
Figure 3.
Genomics features of breast cancer patients in each age group. (A) Mutation profile of patients in elder, intermediate, and young groups. The top 20 genes were listed. (B) Comparison of mutations in young, intermediate (left), and elder (right) groups. (C) Mutations of oncogenic pathways in young, intermediate, and elder groups. (D) Co-occurrence and mutually exclusive mutations in the young group.
Figure 3.
Genomics features of breast cancer patients in each age group. (A) Mutation profile of patients in elder, intermediate, and young groups. The top 20 genes were listed. (B) Comparison of mutations in young, intermediate (left), and elder (right) groups. (C) Mutations of oncogenic pathways in young, intermediate, and elder groups. (D) Co-occurrence and mutually exclusive mutations in the young group.
Figure 4.
Metabolic heterogeneity of breast cancer patients in different age groups. (A) Heatmap displaying the normalized enrichment score (NES) of 65 metabolic pathways in each age group. (B) Forest plot illustrating the hazard ratio (HR) and 95% confidence interval (CI) of each metabolic pathway in the METABRIC population. (C) Venn diagram of the intersection between metabolic pathways upregulated in the young group (logit (NES) > 0.1) and pathways associated with worse disease-free survival (HR > 1). (D) Box plots presenting NES values of intersected metabolic pathways in each age group; p-values: Kruskal-Wallis test for multiple comparisons.
Figure 4.
Metabolic heterogeneity of breast cancer patients in different age groups. (A) Heatmap displaying the normalized enrichment score (NES) of 65 metabolic pathways in each age group. (B) Forest plot illustrating the hazard ratio (HR) and 95% confidence interval (CI) of each metabolic pathway in the METABRIC population. (C) Venn diagram of the intersection between metabolic pathways upregulated in the young group (logit (NES) > 0.1) and pathways associated with worse disease-free survival (HR > 1). (D) Box plots presenting NES values of intersected metabolic pathways in each age group; p-values: Kruskal-Wallis test for multiple comparisons.
Figure 5.
Correlation of the galactose metabolic pathway with less-differentiated cancer cells. (A) tSNE plot of 84,854 cells (GSE176078) color-coded to indicate cell type. (B) Sub-clustering cancer cells into 16 clusters, as indicated by the color-coded legend. (C, D) tSNE and violin (D) plots showing the NES of the galactose metabolism pathway for each cluster. (E) Violin plot showing the expression of UGP2, HK1, and FPKL for each cluster. (F, G) tSNE (F) and violin (G) plot showing the CytoTRACE score for each cluster. (H) Dot plot showing the NES of galactose metabolism pathway positively correlated with the CytoTRACE score. (I) Lollipop plot showing the expression of critical genes in the galactose metabolism pathway positively correlated with the CytoTRACE score.
Figure 5.
Correlation of the galactose metabolic pathway with less-differentiated cancer cells. (A) tSNE plot of 84,854 cells (GSE176078) color-coded to indicate cell type. (B) Sub-clustering cancer cells into 16 clusters, as indicated by the color-coded legend. (C, D) tSNE and violin (D) plots showing the NES of the galactose metabolism pathway for each cluster. (E) Violin plot showing the expression of UGP2, HK1, and FPKL for each cluster. (F, G) tSNE (F) and violin (G) plot showing the CytoTRACE score for each cluster. (H) Dot plot showing the NES of galactose metabolism pathway positively correlated with the CytoTRACE score. (I) Lollipop plot showing the expression of critical genes in the galactose metabolism pathway positively correlated with the CytoTRACE score.
Figure 6.
Correlation of the galactose metabolic pathway with stemness in breast cancer cell lines. (A) Representative FACS plots of Hs578t, MDA-MB-231, HCC1937, and BT549 cells using the cancer stem cell (CSC) markers CD29 and CDC4. (B-C) Representative images (left) and mammosphere formation counting (right) of mammospheres formed from MDA-MB-231 (B) and Hs578t (C) tcells reated with vehicle and D-galactose. (D-E) qRT-PCR analysis of GALK1, UGP2, G6PC3, ITGA6, ITGB1, and CD44 expression in MDA-MB-231 (D) and Hs578t (E) cells treated with vehicle and D-galactose.
Figure 6.
Correlation of the galactose metabolic pathway with stemness in breast cancer cell lines. (A) Representative FACS plots of Hs578t, MDA-MB-231, HCC1937, and BT549 cells using the cancer stem cell (CSC) markers CD29 and CDC4. (B-C) Representative images (left) and mammosphere formation counting (right) of mammospheres formed from MDA-MB-231 (B) and Hs578t (C) tcells reated with vehicle and D-galactose. (D-E) qRT-PCR analysis of GALK1, UGP2, G6PC3, ITGA6, ITGB1, and CD44 expression in MDA-MB-231 (D) and Hs578t (E) cells treated with vehicle and D-galactose.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).