1. Introduction
Endometrial cancer (EC) is the most common gynecological cancer in developed countries, with 417,367 new cases and 97,370 deaths worldwide in 2020, as reported via GLOBOCAN data [
1,
2]. In Korea, the rate of EC has seen a rapid increase recently (annual percent changes, 8.74%; 95% CI=4.99, 12.62) [
3], and in 2018, a total of 3,182 women were newly diagnosed with uterine corpus cancer [
4]. Patients diagnosed with localized EC (confined to the uterus) exhibit a favorable prognosis, with a five-year relative survival rate exceeding 95%, in contrast to 68% and 17% for patients with local and distant metastases, respectively [
5]. More than 50% of ECs are diagnosed at an early stage and low-risk disease and are managed with surgical treatment alone. In early-stage EC cases, adjuvant therapy is only recommended for patients at high risk of recurrence after risk stratification.
With the introduction of the Cancer Genome Atlas (TCGA) molecular classification [
6,
7], a clinically applicable surrogate-marker-based molecular classification system for EC was developed. It categorizes ECs into four molecular subtypes: POLE mutation (POLEmut), mismatch repair deficiency (MMR-D), p53 mutation (p53abn), and non-specific molecular profile (NSMP), using POLE gene sequencing, mismatch repair (MMR) protein immunohistochemistry (IHC) or MSI tests, and p53 IHC [
8,
9,
10,
11]. The POLE mutation subtype is characterized by mutations in the exonuclease domain (EDM) of polymerase-ε (POLE) and exhibits a favorable prognosis [
12,
13,
14]. The MMR-D subtype is a microsatellite instable group, with mutations in the MMR genes resulting in hypermutated tumors [
15,
16]. The p53 mutation subtype is characterized by p53 altered expression and high copy number alterations [
9]. The NSMP subtype represents cases with a non-specific molecular profile.
The L1 cell adhesion molecule (L1CAM; CD171) has been reported as a poor prognostic marker in various solid tumors, including EC [
17,
18,
19,
20], colorectal cancer [
21], gastric cancer [
22,
23], ovarian cancer [
24], and breast cancer [
25]. This poor prognosis is attributed to the ability of L1CAM to enhance cell motility, promoting tumor cell invasion and migration.
In early-stage EC, precise risk assessment is crucial for providing more tailored treatment recommendations, therapy avoiding overtreatment, and ultimately improving patient outcomes. Clinicopathologic factors for determining risk groups include histologic type, histologic grade, International Federation of Gynecology and Obstetrics (FIGO) stage, myometrial invasion, age, and lymphovascular space invasion (LVSI) [
26]. Recently, molecular classification has been integrated into the European Society of Gynaecological Oncology (ESGO)/European Society for Radiotherapy and Oncology (ESTRO)/European Society of Pathology (ESP) guidelines in 2021 [
27]. In 2023, FIGO staging was updated to encompass the latest advancements in pathology and molecular findings, clinical trial results, prognosis, and survival data. The updated 2023 FIGO staging introduced new subclassifications to incorporate molecular and histological staging systems. Specifically, when molecular classification indicates p53abn or POLEmut status in stages I and II, this leads to the disease being upstaged or downstaged (IICmp53abn or IAmPOLEmut)[
28].
In our study, on 247 early-stage EC cases, we performed molecular classification using clinically applicable surrogate markers and L1CAM immunohistochemical staining to gather additional prognostic information. Furthermore, we collected comprehensive clinicopathological information including the updated 2023 FIGO stage to explore its clinical significance, particularly in the context of risk stratification.
2. Materials and Methods
2.1. Patients
From January 2013 to December 2018, 247 patients with early-stage EC, who underwent surgical treatment at Seoul St. Mary’s Hospital were analyzed. All patients received close follow-up, radiotherapy, chemotherapy, or sequential chemotherapy and radiotherapy based on their risk stratification. Pathological slides of these patients were reviewed by two senior pathologists (Ahwon Lee & Misun Lee) to confirm the pathological parameters, including histologic type and histologic grade based on the 2020 WHO classification system [
29], and tumor staging based on the 2009 FIGO staging system [
30] and updated 2023 FIGO staging system [
28]. During the pathological review, represented tumor areas for tissue microarray (TMA) and molecular tests were selected, and cases with insufficient remaining tumor tissue or without formalin-fixed, paraffin-embedded (FFPE) blocks were excluded. This study was approved by the Institutional Review Board of Seoul St. Mary’s Hospital (KC20SISI0979). Investigations were carried out following the rules of the Declaration of Helsinki of 1975, revised in 2013.
2.2. Immunohistochemistry for p53, MMR proteins, and L1CAM
Immunohistochemical staining was conducted using TMA. Two cores of 2 mm diameter tissue were punched out from each representative tumor specimen and arrayed into a recipient block using a manual microarrayer (Quick-Ray set, Unitma, Seoul, Korea). Each TMA block contained 40 cores of EC.
IHC for p53 and MMR proteins (MLH1, MSH2, MSH6, and PMS2) was performed using primary antibodies for p53 (clone DO7, Ventana Medical Systems, Tucson, USA), MLH1 (clone M1; Ventana), MSH2 (clone G219-1129; Ventana), MSH6 (clone SP93; Ventana), and PMS2 (clone A16-4; Ventana) in a fully automated manner on a Ventana BenchMark ULTRA platform with an iVIEW DAB detection system (Roche, Mannheim, Germany). The results of p53 IHC were interpreted as altered expression (mutated type) staining, characterized by either the strong nuclear expression of tumor nuclei (>80%) or the complete absence of the expression of tumor nuclei (0%) [
29]. If there was no nuclear expression observed in the tumor cells of the TMA, they were re-stained with the whole slide to confirm the complete absence of the expression of tumor nuclei with retained internal control. The results of MMR protein staining were interpreted as MMR protein loss if any of the following criteria were met: the complete loss of the nuclear expression of both MLH1 and PMS, the loss of both MSH2 and MSH6, the loss of MSH6, or the loss of PMS2. Strong nuclear staining of normal endometrial glands, stromal cells, and lymphoid cells adjacent to the tumor served as the internal positive control. In cases where the MMR protein staining results were inconsistent with the MSI-PCR result, the whole slide of the corresponding section was re-stained to verify the result.
IHC for L1CAM was performed in line with the manufacturer’s protocol. For L1CAM staining, antigen retrieval was carried out in a pressure cooker (Electric Pressure Cooker CPC-600; Cuisinart, East Windsor, USA) using 1× citrate buffer (pH 6.0).
Sections were incubated with the primary antibody L1CAM (clone 14.10; diluted 1:50; Biolegend, San Diego, USA) at room temperature (22–25°C) in a humidified chamber.
Sections were subsequently incubated with secondary antibodies (EnVision+System-HRP labeled polymer anti-mouse, K4001, DAKO) at room temperature. The signal of immunoreaction was amplified and revealed using a liquid DAB+ Substrate kit (GBI, Bothell, USA). Subsequently, these slides were counterstained with Harris's hematoxylin (YD Diagnostics, Yongin, Korea). If L1CAM immunostaining showed positivity in 10% or more of the tumor cells, it was considered L1CAM-positive. The 10% cut-off was determined based on previous studies [
31,
32]
2.3. POLE Sanger Sequencing
Total genomic DNA was extracted from manually dissected whole tissue sections of FFPE blocks using a QIAamp DNA FFPE tissue kit (QIAGEN, Venlo, Netherlands). PCR reaction was performed using POLE exon 9-specific primer, POLE exon 13-specific primer, and POLE exon 14-specific primer (
Table S1) [
33]. Bidirectional sequencing was performed using the same primers and BigDye Terminator v3.1Cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) on a 3730xl DNA analyzer (Applied Biosystems), as previously described [
34].
2.4. Droplet digital PCR reaction assay to detect POLE mutation
The Droplet digital PCR (ddPCR) was performed using a ddPCR system (QX200 Droplet Digital PCR System; Bio-Rad, Hercules, CA, USA) and a commercial kit (Droplex POLE Mutation Test, Gencurix, Seoul, South Korea) as per the manufacturer’s recommended protocol [
34]. In brief, after thermal cycling, the plate was loaded to measure the endpoint fluorescence signal from each droplet using a Qx200 Droplet Reader (Bio-Rad). The droplet reader was connected to a laptop computer running data analysis QuantaSoft software (v1.6.6.0320; Bio-Rad) and run data analysis was carried out as follows: Each individual droplet was defined on the basis of the fluorescent amplitude as being either positive or negative. The threshold was determined manually based on the amplitude of positive control wells containing wild-type genomic DNA and standard positive DNA. The numbers of positive and negative droplets were distinguished by the threshold, and the given numbers were used for calculating the concentration target in terms of copies/µL. The threshold values of P286R, S297F, V411L, A456P, and S459F were (3000), (3500), (2800), (3200), and (3400), respectively, which were adjusted according to each test condition. According to the fluorescent signal of the negative control, the cut-off for POLE mutation was ≥6 copies/20 µL or a mutation index (MI, %, mutant copies for each mutation/the mutant copies of the internal control) ≥0.3%.
2.5. Microsatellite instability test using PNA probe-mediated real-time PCR
We performed a microsatellite instability (MSI) test with a U-TOP MSI Detection Kit Plus (Seasun Biomaterials, Daejeon, Korea), a peptide nucleotide acid (PNA) probe-mediated real-time PCR-based MSI test, as previously described [
15]. The U-TOP MSI Detection Kit Plus detects the MSI status by using amplicon melting analysis of five quasi-monomorphic mononucleotide repeat markers (NR21, NR24, NR27, BAT25, and BAT26) and an internal control. Samples with alterations in more than one MSI marker were determined as MSI-H, whereas samples with an alteration in a single MSI marker or no alteration were determined as MSI-L or MSS, respectively. MSI-L and MSS were grouped together for statistical analysis based on a previous report of no significant clinicopathological or molecular differences between MSI-L and MSS colorectal cancer [
15].
2.6. Statistical Analysis
All of the statistical analyses were performed using SPSS software (version 25.0; IBM Corp., Armonk, NY, USA). The relationship between EC molecular classification and clinicopathological features was analyzed using the chi-square test. The Kaplan–Meier method was used to estimate the recurrence-free survival (RFS) or overall survival (OS), and the differences were compared using the log-rank test. The OS was determined as from the pathologic diagnostic date to the last follow-up or the date of the patient’s death, while RFS was defined as the period from the date of primary surgery to the date of cancer recurrence. The prognostic factors were analyzed using Cox’s proportional hazard regression model, including those that were statistically significant in the univariate analysis. For statistical significance, a P-value < 0.05 was considered significant.
3. Results
3.1. Patient characteristics
Table 1 shows the clinicopathological characteristics of the 247 patients. The mean age at diagnosis was 55.73 years (range: 30–83 years). The histological subtypes consist of 222 endometrioid carcinoma (89.9%) and 25 non-endometrioid carcinoma (9.7%; 9 serous, 1 clear cell, 2 mixed, 4 dedifferentiated, 2 mucinous, and 7 carcinosarcoma). The 2009 FIGO stages at diagnosis were stage IA (174/247 patients, 70.4%), stage IB (54/247 patients, 21.9%), stage II (17/247 patients, 6.9%), and stage III (3/247 patients, 0.8%). The updated 2023 FIGO stages at diagnosis were stage I (179/247 patients, 72.5%) and stage II (68/247 patients, 27.5%).
Patients were risk stratified according to the European Society for Medical Oncology (ESMO), ESTRO, and ESGO consensus guidelines [
26]. The mean follow-up period was 70.2 months (range: 0.3–126.1 months). During the follow-up, 14.2% of the patients experienced disease recurrence and 8.9% of patients died because of their disease. Kaplan–Meier survival analysis demonstrated reliable differences between clinical risk groups(
Figure S1).
3.2. Molecular classification using surrogate markers and its clinical significance
Among a cohort of 247 patients with early-stage EC, POLE EDM hotspot mutations were found in 32 patients (13.0%). They consisted of P286 (c.857 C>G, exon9) (n=16), V411L (c.1231 G>T/C, exon13) (n=12), A456P (c.1356 G>C, exon14) (n=1), and S459F (c.1376 C>T, exon14) (n=3). MMR protein loss, determined through MMR protein IHC, was found in 67 patients (27.1%). The MSI test revealed 51 cases of MSI-H (20.6%), 10 cases of MSI-L (4.0%), 185 cases of MSS (74.9%), and 1 case where the test failed. Discrepant cases between the MMR protein IHC and MSI test were a total of 28 cases (IHC negative and MSI-H, 6 cases; IHC positive and MSI-L or MSS, 22 cases). We classified cases with MMR protein loss detected by MMR protein IHC, as well as those with MSI-H identified by an MSI test, as MMR-D. The p53 IHC was positive in 22 cases (8.9%). Four cases in which POLE mutation and MMR protein loss by IHC or MSI-H using an MSI test were simultaneously observed were classified as the POLEmut subtype [
35]. One case in which MMR protein loss by IHC, MSI-H using an MSI test, and p53 altered expression were simultaneously observed was classified as an MMR-D subtype [
35]. The 274 ECs were classified into one of the four molecular subgroups; 32 (13.0%) were POLEmut, 69 (27.9%) were MMR-D, 21 (8.5%) were p53abn, and 125 (50.6%) were NSMP ECs.
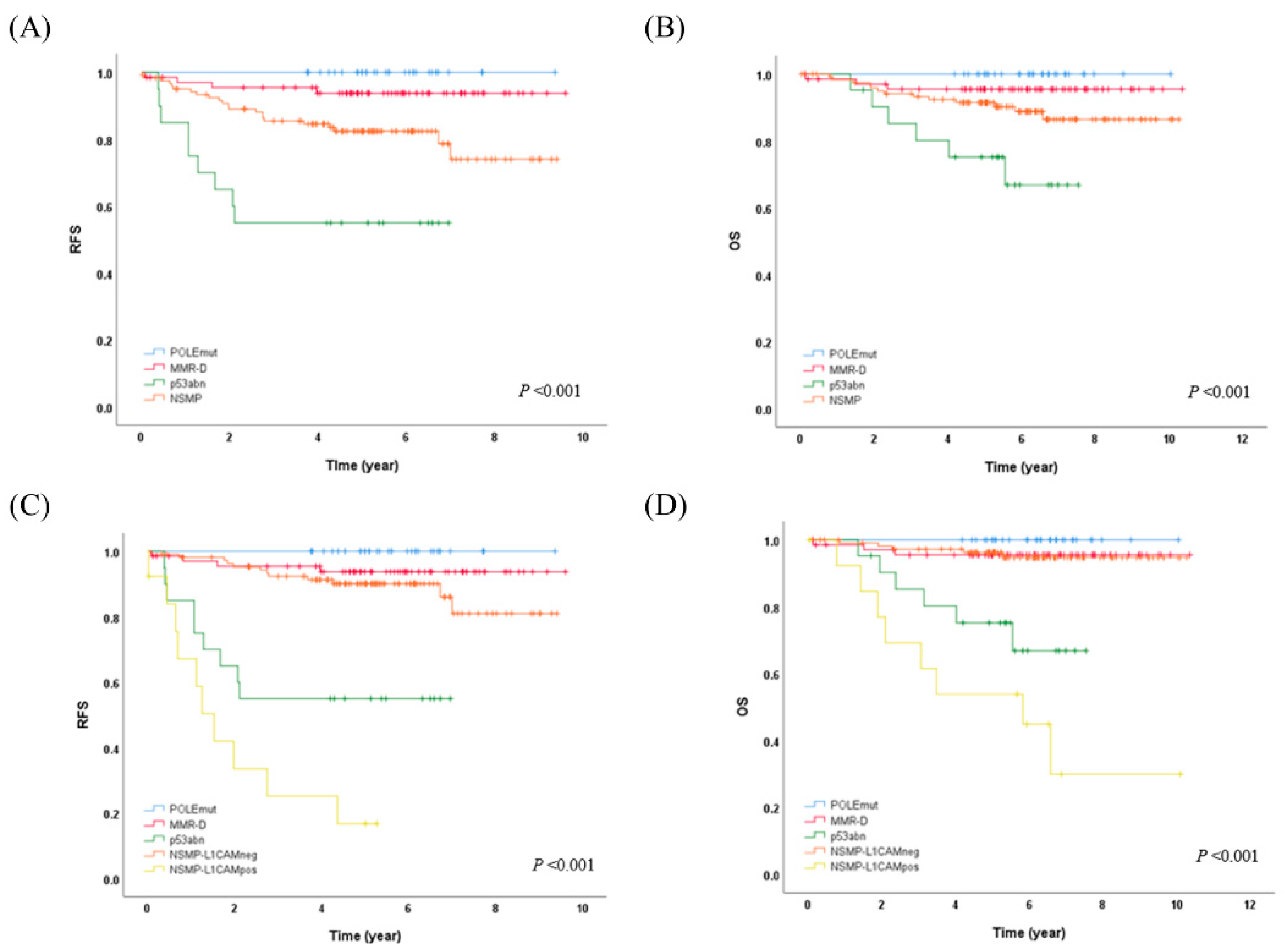
The POLEmut subtype showed a significant association with the endometrioid histologic subtype compared to non-endometrioid carcinoma (p=0.026). The MMR-D subtype was found to be associated with LVSI (p=0.033). Additionally, the p53abm subtype exhibited significant associations with the non-endometrioid histologic subtype, and histologically high-grade tumors (p<0.001, each). Kaplan–Meier analysis for RFS and OS demonstrated reliable differences between molecular subgroups (
Figure 1A,B, p=0.001, each). The five-year RFS and OS for patients with POLE mutation were 100% each. Notably, none of the patients with POLEmut experienced disease recurrence or succumbed to the disease. Mortality was observed in patients with MMR-D (4.3%, 3/69), p53abn (28.6%, 6/21), and NSMP (10.4%, 13/125). Patients with MMR-D, p53abn, and NSMP ECs showed a 5-year RFS of 93.7%, 55.0%, and 82.4%, respectively, and a 5-year OS of 95.5%, 75.2%, and 91.5%, respectively.
3.3. L1CAM expression and its impact on prognosis and molecular classification
L1CAM was positive in 26 patients (10.5%). According to molecular subgroups, L1CAM overexpression was noted in 3.1% (1/31), 1.44% (1/69), 47.6% (10/21), and 11.2% (14/125) in the POLEmut, MMR-D, p53abn, and NSMP subgroups, respectively. L1CAM expression was correlated with old age, the non-endometrioid histologic subtype, the histological high grade, and myometrial invasion (p=0.001, p<0.001, p<0.001, and p =0.022, respectively).
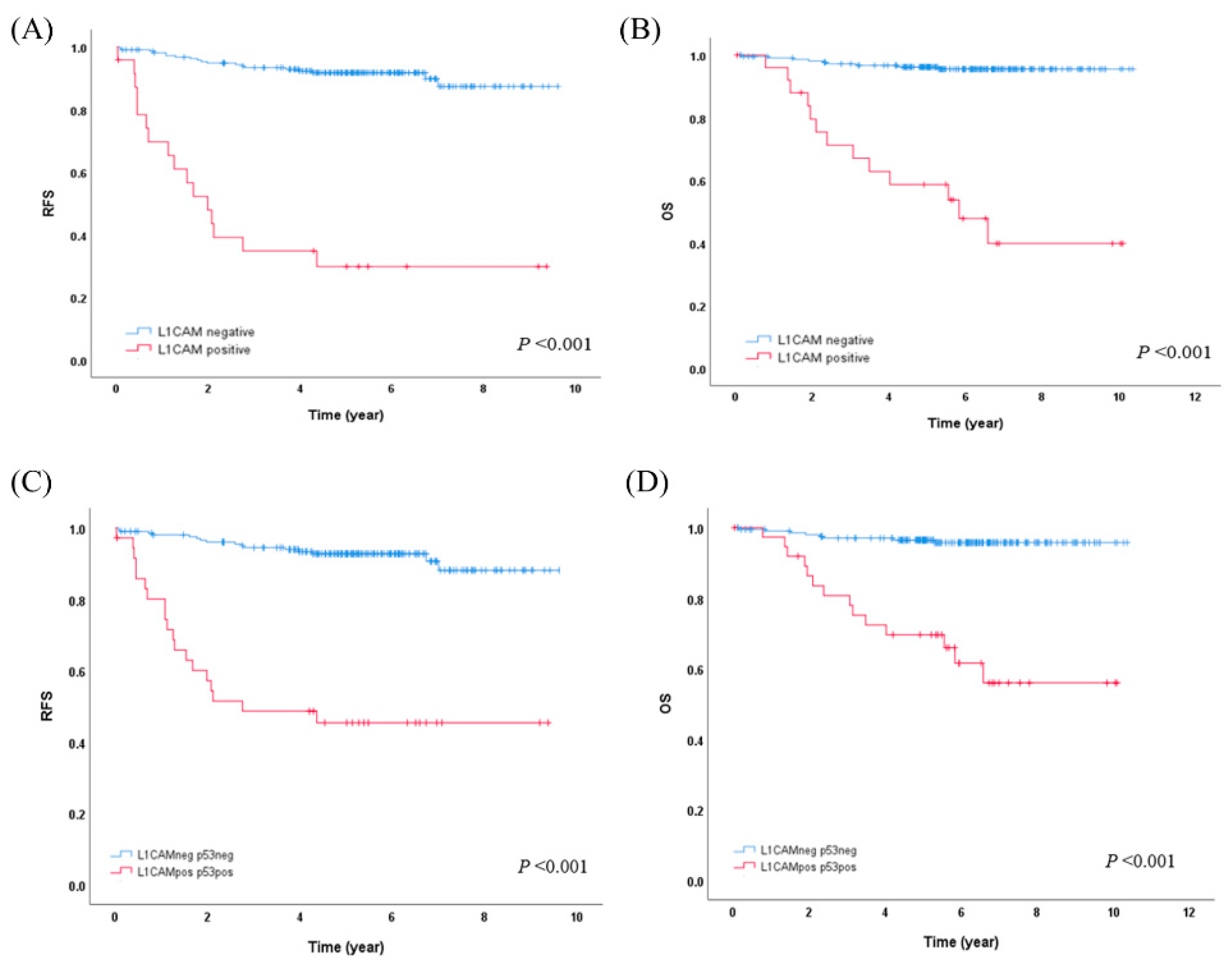
L1CAM overexpression showed a strong association with worse RFS (5-year RFS 29.9% vs. 91.8%, p<0.001) and OS (5-year OS 58.7% vs. 96.2%, p<0.001) (
Figure 2A,B). In the molecular subgroup analysis, L1CAM overexpression was statistically significantly associated with worse RFS and OS (5-year RFS 16.8% vs. 90.1%, p<0.001; 5-year OS 53.8% vs. 96.2%, p<0.001, respectively) only in the NSMP subgroup. In the p53abn subgroups, there were trends indicating that L1CAM overexpression might be associated with worse RFS and OS, but these trends did not reach statistical significance.
Building upon these findings and drawing from previous literature references [
18,
19,
20], we proceeded to subcategorize the NSMP subgroup into the NSMP-L1CAMneg and NSMP-L1CAMpos groups (from now on, we will call it molecular-L1CAM classification). Kaplan–Meier analysis for RFS and OS demonstrated reliable differences between molecular-L1CAM subgroups (
Figure 1C,D, p<0.001, each). In the molecular-L1CAM classification, patients with POLEmut, MMR-D, and NSMP-L1CAMneg (212/247, 85.8%) demonstrated favorable outcomes, in contrast to patients with p53abn and NSMP-L1CAMpos (35/247, 14.2%) who had less favorable outcomes (10-year OS 95.8% vs. 50.3%, p<0.001).
3.4. Enhanced Risk Stratification in Early-Stage EC by integrating Molecular-L1CAM Classification or L1CAM/p53 Categorization
A comprehensive analysis of the clinical characteristics of molecular-L1CAM classification was conducted (
Table 2). The results revealed significant associations with various factors, including histologic type (p<0.001), histologic grade (p<0.001), updated 2023 FIGO stage (p<0.001), and clinical risk group (p=0.001). Using multivariate analysis, the molecular-L1CAM classification was found to be an independent predictor of both RFS (p<0.001) and OS (p<0.001). Additionally, when the clinical risk groups were categorized as low, intermediate, and high, the intermediate-risk group was associated with worse RFS (p=0.009) compared to the low-risk group (
Table 3).
To simplify the risk stratification process for practical use, we opted to use only the p53 and L1CAM IHC results instead of the molecular-L1CAM classification, which involves the POLE mutation test. The patients were categorized as L1CAM/p53-negative if both L1CAM and p53 IHC were negative (209/247,84.6%), and as L1CAM/p53-positive if either L1CAM or p53 IHC were positive (38/247, 15.4%). The L1CAM/p53-positive group exhibited a strong association with worse RFS (5-year RFS 45.4% vs. 92.8%, p<0.001) and OS (5-year OS 69.6% vs. 96.5%; 10-year OS 56.0% vs. 95.8%, p<0.001) compared to L1CAM/p53-negative group (
Figure 2C,D). In the multivariate analysis, the L1CAM/p53 categorization remained an independent prognostic factor for both RFS (p<0.001) and OS (p=0.003). Additionally, when the clinical risk groups were categorized as low, intermediate, and high, the intermediate-risk group was associated with worse RFS (p=0.005) compared to the low-risk group, and similarly, high-risk group was associated with worse RFS (p=0.020) compared to the intermediate-risk group. A high histologic grade was associated with worse OS compared to a low histologic grade (p=0.017) (
Table 4).
This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
4. Discussion
While a significant number of ECs are successfully treated without recurrence, some cases of EC still result in death even at its early stages. In the recent ESGO/ESTRO/ESP guidelines, it is recommended to classify EC into four distinct molecular subgroups to tailor adjuvant therapy [
27]. In the updated 2023 FIGO staging system, the use of molecular classification is encouraged, and if the molecular subtype is known, "m" was added for molecular classification. Especially in early-stage EC, major changes were made including new substages that reflect the molecular alteration of POLE and p53 genes. On the other hand, it has been reported that positive L1CAM expression was associated with poor prognosis in all patients with EC [
18,
19,
20], in stage I EC[
32], and with p53 wild-type EC [
20]. Additionally, our study demonstrated positive L1CAM expression as a poor prognostic factor in the overall early-stage EC (5-year RFS 29.9% vs. 91.8%, p<0.001; 5-year OS 58.7% vs. 96.2%, p<0.001) (
Figure 2A,B) and specifically within the NSMP molecular subgroup of early-stage EC (5-year RFS 16.8% vs. 90.1%, p<0.001; 5-year OS 53.8% vs. 96.2%, p<0.001).
When we further categorized the NSMP group, which demonstrates an intermediate prognosis between the POLEmut/MMR-D group and the p53abn group, based on the L1CAM IHC result, we observed a distinct separation of a subgroup with a 10-year OS rate over 95%, suggesting a favorable prognosis. This subgroup comprises the POLEmut/MMR-D/NSMP-L1CAMneg groups (212/247, 85.8%). With this Molecular-L1CAM classification, we effectively differentiated good prognostic group by eliminating the intermediate prognostic group (
Figure 1).
In this study, we intended to detect the five most frequently occurring pathogenic variations within exons 9, 13, and 14 of the POLE EDM by Sanger sequencing or ddPCR. These mutations collectively account for 84% of the known POLE pathogenic mutations [
36]. To detect all reported pathogenic mutations, next-generation sequencing (NGS) testing, including the POLE EDM (exon 9-14), is currently available. However, caution should be exercised due to the possibility of false-positive interpretations. For the MMR-D subtype classification, we conducted both IHC testing for all four MMR proteins (MLH1, MSH2, MSH6, and PMS2) and MSI testing. A case was classified as MMR-D if either the MMR protein IHC or MSI testing came out positive. In the updated 2023 FIGO staging, MSH6 and PMS2 IHC are recommended as simplified surrogate markers for TCGA molecular classification [
28]. The primary use of IHC is recommended because it directly identifies the absent MMR protein(s), while PCR-based MSI tests are not thoroughly validated in non-colorectal cancer including endometrial cancer [
16]. In this study, we found four additional cases categorized into the MMR-D subgroup by additionally performing MSI tests. The differences and similarities of absent MMR proteins and MSI-H in EC need more evaluation. Regarding p53 IHC, cases with altered expression were classified as the p53abn subtype only if they did not belong to the POLEmut or MMR-D subtypes. This classification strategy for multiple classifiers was based on various factors, including morphologic features, the frequent subclonal overexpression of p53, hierarchical clustering study with TCGA data, and clinical outcomes [
35]. In our study, MMR proteins and p53 IHC were performed using TMA, which may not fully represent the entire tumor lesion. To address this concern, cases with p53 null expression or discrepancies between the MMR protein IHC and MSI test results were re-examined on whole-slide sections.
Upon classifying early-stage EC into molecular subtypes, the frequencies of the POLEmut, MMR-D, p53abn, and NSMP subtypes were found to be 13.0%, 27.9%, 8.5%, and 50.6%, respectively. In the PORTEC cohorts for early-stage EC, their frequencies were 6%, 26%, 9%, and 59%, respectively [
37]. In a PORTEC cohorts study, they only detected the POLE EDM mutation in exons 9 and 13. We additionally detected POLE A456P and S459F mutations in exon 14 (n=4).
In the univariate analysis, several factors were found to be statistically significant for worse RFS and OS. These factors include old age, non-endometrial histologic type, high histologic grade, higher prognostic risk group, higher FIGO stage, and the p53abn/NSMP-L1CAMpos subgroup of molecular-L1CAM classification (
Table 3). However, in the multivariate analysis, the molecular-L1CAM classification, and the intermediate prognostic risk group, when compared to the low prognostic risk group, were identified as independent prognostic factors for RFS, while molecular-L1CAM classification alone remained as an independent prognostic factor for OS.
In clinical practice, the application of molecular classification for all EC patients is challenging due to economic or medical insurance reasons. Specifically, the POLE mutation test poses additional challenges, as it requires repetitive sequencing or targeted NGS due to the dispersed hotspot regions encompassing exons 9-14. With this background, we developed a simplified classification method using only an IHC-omitting molecular test and still providing additional prognostic information to the traditional clinical risk group, which is known to have limitations due to subjectivity in histologic type and grade. In our study, we introduced the L1CAM/p53 categorization, which categorizes patients based on the combined results of p53 and L1CAM IHC. The L1CAM/p53-positive group exhibited a worse prognosis compared to the L1CAM/p53-negative group (10-year OS, 56.0% vs. 95.8%, p<0.001) (
Figure 2). Furthermore, when we included the L1CAM/p53 categorization instead of the molecular-L1CAM classification in the multivariable analysis, both RFS and OS were confirmed to be independent prognostic factors (
Table 4).
There are several limitations in this study, being that it was a retrospective study conducted in a single institution, with a relatively small sample size, particularly for cases of the NSMP-L1CAMpos subtype. Therefore, further validation through a large-scale, prospective multicenter study is necessary to confirm and generalize our findings.
5. Conclusion
In conclusion, our study aimed to enhance risk stratification in early-stage EC by integrating either the molecular-L1CAM classification or the simplified L1CAM/p53 categorization. Both the molecular-L1CAM classification and L1CAM/p53 categorization proved to be independent prognostic factors for both RFS and OS. Despite the limitations of being a single-institution retrospective study with a relatively small sample size, our findings indicate the potential benefit of integrating immunostaining-based L1CAM/p53 categorization into the current risk-stratification approach, particularly in cases where molecular test is unavailable. We acknowledge the need for further validation through large-scale, prospective multicenter studies to confirm the utility of our proposed classification method.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Survival analysis of four groups according to the clinical risk group of early-stage endometrial cancer. Kaplan-Meier curves are shown for (A) recurrence-free survival and (B) overall survival of 247 patients.; Table S1: Primer sequences for POLE Sanger sequencing.
Author Contributions
Conceptualization, A.L.; methodology, S.J. and M.L.; validation, S.J. and M.L.; formal analysis, M.L, J.K. and A.L.; investigation, S.J., M.L. and J.K.; resources, J.K., S.H., S.J.L. and K.H.L.; writing—original draft preparation, S.J. and A.L.; writing—review and editing, J.K., S.H., S.J.L., K.H.L and A.L.; visualization, S.J.; supervision, A.L.; funding acquisition, A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1F1A1070341).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Seoul Metropolitan Government-Seoul St. Mary’s Hospital (KC20SISI0979).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. This study did not collect or record any personally identifiable information. It has been confirmed as a minimal risk study, and has been exempted from obtaining informed consent by the Institutional Review Board of Seoul Metropolitan Government-Seoul St. Mary’s Hospital.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.I.; Chang, H.K.; Park, S.J.; Lim, J.; Won, Y.J.; Lim, M.C. The incidence and survival of cervical, ovarian, and endometrial cancer in Korea, 1999-2017: Korea Central Cancer Registry. Obstet Gynecol Sci 2021, 64, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Won, Y.J.; Lee, J.J.; Jung, K.W.; Kong, H.J.; Im, J.S.; Seo, H.G.; Community of Population-Based Regional Cancer, R. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2018. Cancer Res Treat 2021, 53, 301–315. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J Clin 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- McAlpine, J.; Leon-Castillo, A.; Bosse, T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J Pathol 2018, 244, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, S.; McConechy, M.K.; Kommoss, F.; Leung, S.; Bunz, A.; Magrill, J.; Britton, H.; Kommoss, F.; Grevenkamp, F.; Karnezis, A.; et al. Final validation of the ProMisE molecular classifier for endometrial carcinoma in a large population-based case series. Ann Oncol 2018, 29, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; Hoang, L.N.; McConechy, M.K.; Nakonechny, Q.; Leo, J.; Cheng, A.; Leung, S.; Yang, W.; Lum, A.; Kobel, M.; et al. Molecular classification of endometrial carcinoma on diagnostic specimens is highly concordant with final hysterectomy: Earlier prognostic information to guide treatment. Gynecol Oncol 2016, 143, 46–53. [Google Scholar] [CrossRef]
- Church, D.N.; Briggs, S.E.; Palles, C.; Domingo, E.; Kearsey, S.J.; Grimes, J.M.; Gorman, M.; Martin, L.; Howarth, K.M.; Hodgson, S.V.; et al. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet 2013, 22, 2820–2828. [Google Scholar] [CrossRef]
- Church, D.N.; Stelloo, E.; Nout, R.A.; Valtcheva, N.; Depreeuw, J.; ter Haar, N.; Noske, A.; Amant, F.; Tomlinson, I.P.; Wild, P.J.; et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst 2015, 107, 402. [Google Scholar] [CrossRef] [PubMed]
- McConechy, M.K.; Talhouk, A.; Leung, S.; Chiu, D.; Yang, W.; Senz, J.; Reha-Krantz, L.J.; Lee, C.H.; Huntsman, D.G.; Gilks, C.B.; McAlpine, J.N. Endometrial Carcinomas with POLE Exonuclease Domain Mutations Have a Favorable Prognosis. Clin Cancer Res 2016, 22, 2865–2873. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Kwon, Y.; Kim, H.; Kim, H.; Min, B.S.; Park, Y.; Kim, T.I.; Hong, S.P.; Kim, W.K. Microsatellite instability test using peptide nucleic acid probe-mediated melting point analysis: a comparison study. BMC Cancer 2018, 18, 1218. [Google Scholar] [CrossRef] [PubMed]
- Rüschoff, S.S.a.J. Mismatch Repair Deficiency and Microsatellite Instability. Encyclopedia 2022. [Google Scholar]
- Zeimet, A.G.; Reimer, D.; Huszar, M.; Winterhoff, B.; Puistola, U.; Azim, S.A.; Muller-Holzner, E.; Ben-Arie, A.; van Kempen, L.C.; Petru, E.; et al. L1CAM in early-stage type I endometrial cancer: results of a large multicenter evaluation. J Natl Cancer Inst 2013, 105, 1142–1150. [Google Scholar] [CrossRef]
- Asano, H.; Hatanaka, K.C.; Matsuoka, R.; Dong, P.; Mitamura, T.; Konno, Y.; Kato, T.; Kobayashi, N.; Ihira, K.; Nozaki, A.; et al. L1CAM Predicts Adverse Outcomes in Patients with Endometrial Cancer Undergoing Full Lymphadenectomy and Adjuvant Chemotherapy. Ann Surg Oncol 2020, 27, 2159–2168. [Google Scholar] [CrossRef]
- Van Gool, I.C.; Stelloo, E.; Nout, R.A.; Nijman, H.W.; Edmondson, R.J.; Church, D.N.; MacKay, H.J.; Leary, A.; Powell, M.E.; Mileshkin, L.; et al. Prognostic significance of L1CAM expression and its association with mutant p53 expression in high-risk endometrial cancer. Mod Pathol 2016, 29, 174–181. [Google Scholar] [CrossRef]
- Kim, J.; Kim, S.I.; Kim, N.R.; Kim, H.; Kim, H.S.; Chung, H.H.; Kim, J.W.; Lee, C.; Lee, M. Prognostic significance of L1CAM expression in addition to ProMisE in endometrial cancer. Gynecol Oncol 2023, 174, 231–238. [Google Scholar] [CrossRef]
- Kajiwara, Y.; Ueno, H.; Hashiguchi, Y.; Shinto, E.; Shimazaki, H.; Mochizuki, H.; Hase, K. Expression of l1 cell adhesion molecule and morphologic features at the invasive front of colorectal cancer. Am J Clin Pathol 2011, 136, 138–144. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Li, L.; Zhao, Z.S.; Wang, Y.X.; Ye, Z.Y.; Tao, H.Q. L1 and epithelial cell adhesion molecules associated with gastric cancer progression and prognosis in examination of specimens from 601 patients. J Exp Clin Cancer Res 2013, 32, 66. [Google Scholar] [CrossRef] [PubMed]
- Kodera, Y.; Nakanishi, H.; Ito, S.; Misawa, K.; Ito, Y.; Nakayama, G.; Koike, M.; Fujiwara, M.; Yamamura, Y.; Nakao, A. Expression of L1 cell adhesion molecule is a significant prognostic factor in pT3-stage gastric cancer. Anticancer Res 2009, 29, 4033–4039. [Google Scholar] [PubMed]
- Fogel, M.; Gutwein, P.; Mechtersheimer, S.; Riedle, S.; Stoeck, A.; Smirnov, A.; Edler, L.; Ben-Arie, A.; Huszar, M.; Altevogt, P. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 2003, 362, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Schroder, C.; Schumacher, U.; Fogel, M.; Feuerhake, F.; Muller, V.; Wirtz, R.M.; Altevogt, P.; Krenkel, S.; Janicke, F.; Milde-Langosch, K. Expression and prognostic value of L1-CAM in breast cancer. Oncol Rep 2009, 22, 1109–1117. [Google Scholar]
- Colombo, N.; Creutzberg, C.; Amant, F.; Bosse, T.; Gonzalez-Martin, A.; Ledermann, J.; Marth, C.; Nout, R.; Querleu, D.; Mirza, M.R.; et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: diagnosis, treatment and follow-up. Ann Oncol 2016, 27, 16–41. [Google Scholar] [CrossRef]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int J Gynecol Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef]
- Berek, J.S.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N.; Endometrial Cancer Staging Subcommittee, F.W.s.C.C. FIGO staging of endometrial cancer: 2023. Int J Gynaecol Obstet 2023, 162, 383–394. [Google Scholar] [CrossRef]
- Hohn, A.K.; Brambs, C.E.; Hiller, G.G.R.; May, D.; Schmoeckel, E.; Horn, L.C. 2020 WHO Classification of Female Genital Tumors. Geburtshilfe Frauenheilkd 2021, 81, 1145–1153. [Google Scholar] [CrossRef]
- Pecorelli, S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet 2009, 105, 103–104. [Google Scholar] [CrossRef]
- Kommoss, F.K.; Karnezis, A.N.; Kommoss, F.; Talhouk, A.; Taran, F.A.; Staebler, A.; Gilks, C.B.; Huntsman, D.G.; Kramer, B.; Brucker, S.Y.; et al. L1CAM further stratifies endometrial carcinoma patients with no specific molecular risk profile. Br J Cancer 2018, 119, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Bosse, T.; Nout, R.A.; Stelloo, E.; Dreef, E.; Nijman, H.W.; Jurgenliemk-Schulz, I.M.; Jobsen, J.J.; Creutzberg, C.L.; Smit, V.T. L1 cell adhesion molecule is a strong predictor for distant recurrence and overall survival in early stage endometrial cancer: pooled PORTEC trial results. Eur J Cancer 2014, 50, 2602–2610. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Shao, H.; Ban, X.; Zhang, H.; You, Y.; Zhou, N.; Mao, X.; Zhao, H.; Chen, J.; Lu, Z. Detection of POLE Subtypes in High-Grade Endometrioid Carcinoma by BaseScope-ISH Assay. Front Oncol 2019, 9, 831. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Lee, S.K.; Suh, D.H.; Kim, K.; No, J.H.; Kim, Y.B.; Kim, H. Clinical evaluation of a droplet digital PCR assay for detecting POLE mutations and molecular classification of endometrial cancer. J Gynecol Oncol 2022, 33, e15. [Google Scholar] [CrossRef] [PubMed]
- Leon-Castillo, A.; Gilvazquez, E.; Nout, R.; Smit, V.T.; McAlpine, J.N.; McConechy, M.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; et al. Clinicopathological and molecular characterisation of 'multiple-classifier' endometrial carcinomas. J Pathol 2020, 250, 312–322. [Google Scholar] [CrossRef]
- Leon-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; Rau, T.T.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol 2020, 250, 323–335. [Google Scholar] [CrossRef]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jurgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin Cancer Res 2016, 22, 4215–4224. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).