1. Introduction
Radiation therapy and most traditional anti-cancer chemotherapeutic agents exert their anti-cancer effect through DNA damage. The development of inhibitors of DNA repair enzymes and factors is one of the promising areas of modern pharmacology and is one of the ways to create effective cancer therapy, especially to combat drug-resistant tumors. Inhibition of key DNA repair enzymes in model systems significantly enhances the effectiveness of traditional methods of treatment [
1,
2].
Topoisomerases inhibitors such as topotecan, irinotecan, etoposide, and doxorubicin are commonly used anticancer drugs [
3,
4]. To control DNA topology, topoisomerases 1 and 2 (Top1 and Top2) generate breaks in the DNA backbone by forming transient enzyme-DNA cleavage complexes (TOPcc) with phosphotyrosyl bonds between the DNA end and the topoisomerase catalytic tyrosine residue. These transient complexes can be stabilized with the drugs listed above, resulting in disruption of genomic stability and cell death if such "stuck" complexes are not repaired [
5]. The repair of such damages is carried out by tyrosyl-DNA phosphodiesterases 1 and 2 (Tdp 1/2), which eliminate blocking damage from the 3′- (Top1) and 5′-ends (Top2) of DNA [
6].
Tdp1 belongs to the phospholipases superfamily that catalyzes the hydrolysis of DNA phosphodiester bond [
7]. Tdp1 plays an important role in removing DNA damages created by Top1, its inhibitor camptothecin, and other anticancer drugs. Tdp1 cleaves the 3′ phosphodiester bond between covalent adducts of different origin and the 3′ end of DNA [
8,
9,
10,
11]. Such adducts are formed, among other ways, under the action of radiotherapy and anticancer drugs [
10,
11]. Such drugs, in particular, are the anticancer drugs topotecan and irinotecan, which fix short-lived TOPcc and, thereby, lead to cell death. Thus, by eliminating these complexes, Tdp1 reduces the effectiveness of therapy. The idea that Tdp1 is responsible for resistance to these drugs is supported by a large number of studies, both
in vitro and
in vivo. Cells or organisms deficient in Tdp1 activity are more sensitive to the effects of Top1 poisons [
12,
13,
14,
15,
16]. Conversely, tumor cells with overexpression of Tdp1 are less sensitive to these drugs [
17,
18,
19,
20,
21].
The human DNA repair Tdp2 belongs to the family of metal-dependent phosphodiesterases and hydrolyzes 5′-phosphotyrosine, the product of covalent attachment of Top2 to DNA [
22,
23]. Top2 inhibitors (etoposide, doxorubicin) stabilize the Top2-DNA covalent complex and induce cell death [
24]. Top2 inhibitors are widely used in clinical practice as anticancer drugs. Tdp2 activity reduces the efficacy of these drugs, and vice versa, Tdp2 deficiency leads to a significant increase in sensitivity to Top2 inhibitors [
22,
25]. Thus, Tdp2 inhibitors can increase the effectiveness of chemotherapy through synergy with Top2 inhibitors. Such sensitization of the antitumor effect was successfully implemented for inhibitors of another similar pair of enzymes, Tdp1/Top1 [
26,
27,
28]. Top1, like Top2, forms a covalent complex with DNA, but from the 3′-end.
Of note is the recently discovered ability of Tdp1 and Tdp2 to take over each other's functions, albeit with lower efficiency [
29,
30,
31]. This makes highly promising either the combined use of selective inhibitors of these two enzymes, or the creation of agents capable to simultaneously inhibitTdp1 and Tdp2.
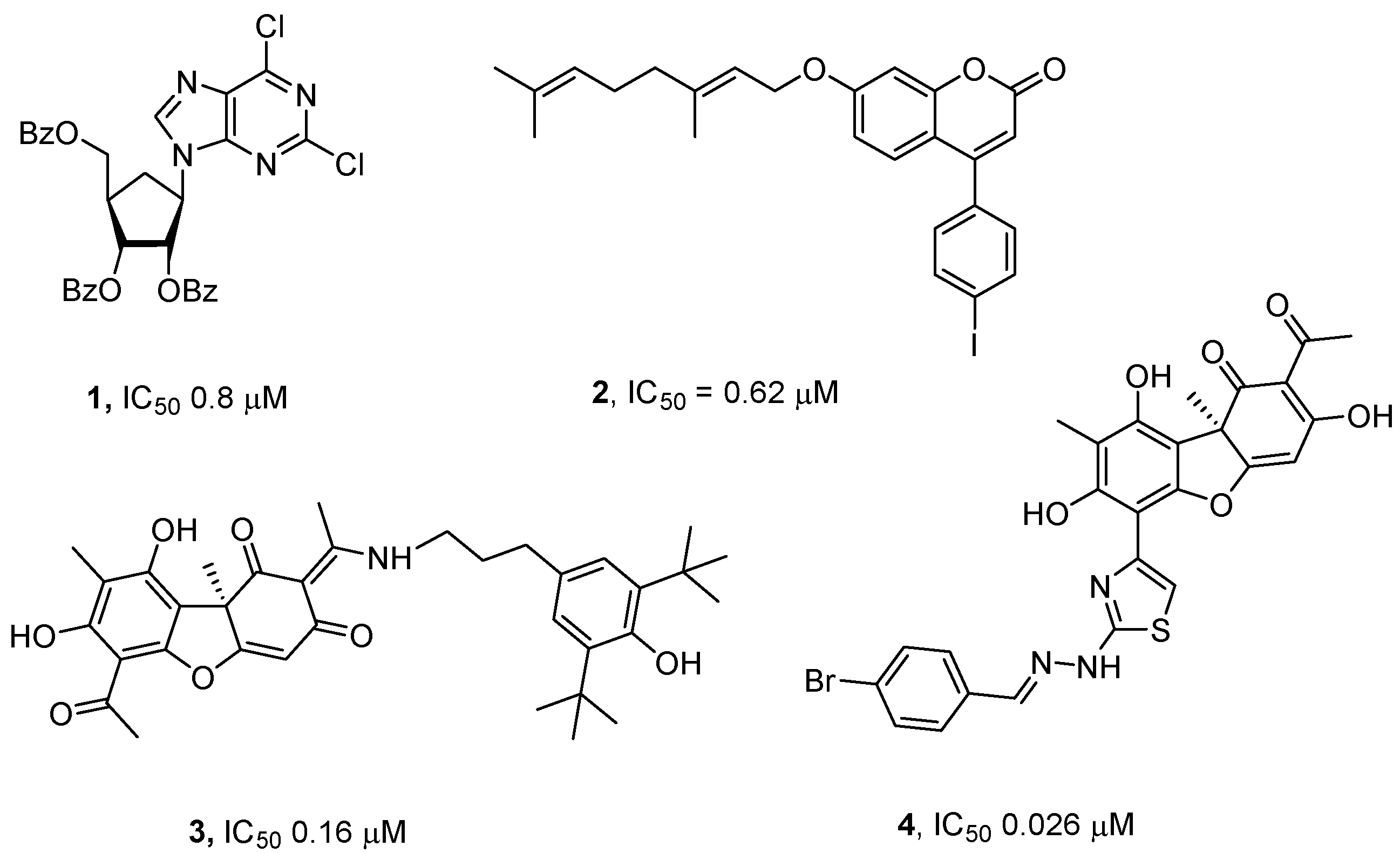
The list of Tdp1 inhibitors is quite extensive and includes both natural substances and their derivatives (detailed in the review [
28]) and synthetic compounds [
26] (
Figure 1). We have shown that the applying of Tdp1 inhibitors (compounds
1-4,
Figure 1) in combination with topotecan increases the antitumor and antimetastatic efficacy of the latter against tumors in mice
in vivo [
32,
33,
34,
35]. The most effective Tdp1 inhibitors known to date are usnic acid derivatives (similar to compound
4 in
Figure 1), acting in the nanomolar concentration range and capable of potentiating the antitumor and antimetastatic effects of topotecan
in vitro and
in vivo [
28].
The list of known Tdp2 inhibitors is significantly limited, mainly represented by deazaflavins [
36,
37] and quinoline derivatives [
38,
39] (
Figure 2). Our team found several mild Tdp2 inhibitors among derivatives of usnic acid and deoxycholic acid (compounds
7 and
8,
Figure 2) [
40,
41].
Usnic acid is a natural compound of the dibenzofuran class, a major metabolite of a wide variety of lichens. This metabolite exists in two enantiomeric forms, the biological activity of which can significantly differ. As a rule, only one of the enantiomers is contained in various types of lichens. The content of (+)-usnic acid is quite high, which, together with the simple procedure of isolation and high optical purity, makes it commercially available. The availability of (+)-usnic acid, its structure and the presence of functional groups available for chemical modification made it attractive object for the synthesis of various derivatives, often having a more pronounced biological activity than the native compound itself. (-)-Usnic acid is less common and less commercially available. The biological activity of usnic acid derivatives can also vary depending on the structure of the chiral centers of both the usnic acid core and substituents [
42,
43].
The aim of this work was to study the inhibitory activity of usnic acid derivatives, which are effective inhibitors of Tdp1, against Tdp2 in order to identify possible dual Tdp1/Tdp2 inhibitors. Inhibition of both enzymes should lead to an increase in the efficiency of sensitization and also to expand the boundaries of the use of sensitizers in anticancer therapy, including therapy with both Top1 poisons (camptothecin derivatives), and with Top2 inhibitors (etoposide and doxorubicin).
In this work, we have shown that some highly effective inhibitors of Tdp1 that we have previously discovered [
44] are also capable to inhibit Tdp2. If Tdp1 was inhibited with both usnic acid enantiomers with approximately the same efficiency, then only (-)-enantiomers inhibited Tdp2.
2. Materials and Methods
2.1. Gel-Based Tdp2 Activity Assay
Oligonucleotide 5′- tyrosine -AAC GTC AGG GTC TTC C- FAM -3′ was synthesized in the Laboratory of Nucleic Acid Chemistry at the Institute of Chemical Biology and Fundamental Medicine (Novosibirsk, Russia) and used for the detection of Tdp2 enzyme activity in polyacrylamide gel. Human Tdp2 was expressed and isolated as described in [
40]. Tdp2 gel-based assays are also described there [
40]. Briefly, samples with a volume of 20 µl contained 100 nM substrate and 200 nM recombinant human Tdp2 in the absence or presence of an inhibitor in a buffer containing 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 7 mM β-mercaptoethanol, and BSA. Samples were incubated for 10 min at 37°C, then reactions were terminated by the addition of a gel loading buffer (TBE, 10% formamide, 7 M carbamide, 0.1% xylene cyanol, and 0.1% bromophenol blue, and 20 mM EDTA). The samples were heated before loading at 90°C for 7 min. The reaction products were separated by electrophoresis in a 20% denaturing PAGE with 7 M carbamide at a ratio of acrylamide to bisacrylamide of 19:1. A Typhoon FLA 9500 phosphorimager (GE Healthcare, Boston, MA, USA) was used for gel scanning and imaging, and the data were analyzed with QuantityOne 4.6.7 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
2.2. Cytotoxicity Assay
Analysis of the intrinsic toxicity of the compounds was examined against human cell line HEK293FT (human embryonic kidney) using a standard MTT test [
45] by colorimetric measurement of the amount of formazan converted from 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide in cells exposed to the compounds. The cells were grown in DMEM-F12 medium, with 50 IU/mL penicillin, and 50 µg/mL streptomycin (Thermo Fisher Scientific, Waltham, MA, USA), and in the presence of 10% fetal bovine serum (Biolot, Saint-Petersburg, Russia) in 5% CO
2 atmosphere. After formation of a 30-50% monolayer, tested compounds were added to the medium (the volume of added reagents was 1/100 of the total volume of the culture medium, the amount of DMSO was 1% of the final volume), and the cell culture was monitored for 3 days. Control cells were grown in the presence of 1% DMSO. The measurements were carried out in three parallel experiments.
Different concentrations of etoposide and fixed concentrations of usnic acid derivatives 10 μM were used to evaluate the effect of the compounds on the cytotoxic effect of etoposide.
2.3. Tdp2 Knockout HEK293FT Cell Clones
Selection of protospacers for human
Tdp2 gene knockout was performed using the Benchling CRISPR tool (
https://www.benchling. com/) (accessed date 13 February 2020). Two 20-nt protospacer sequences (PAM sequences in brackets) were selected for deletion of exon 6 of the human Tdp2 gene (NM_016614.3): TDP2-gRNA1 ATGCCTTTGGTGTATTGAGG(AGG) and TDP2-gRNA2 CAGAAATTAGCCGGGCATTG(TGG). Scheme of protospacers in
Tdp2 gene is shown on
Figure S9A in the Supplementary. Oligonucleotides were cloned in plasmid pSpCas9(BB)-2A-GFP (PX458) that was a gift from Feng Zhang (Addgene plasmid #48138;
http://n2t.net/addgene:48138; RRID:Addgene_48138) as previously described [
46]. Transfection-grade plasmid DNA was isolated using the Plasmid Plus Midi Kit (QIAGEN, Hilden, Germany).
HEK293FT cells were transfected with the constructed plasmids pX458-TDP2-gRNA1 and pX458-TDP2-gRNA2 (0.25 µg of each) using Lipofectamine 3000 Reagent (Thermofisher Scientific, Waltham, MA USA). Growth medium contained DMEM/F12 (Thermofisher Scientific, Waltham, MA, USA) 1:1, 10% fetal bovine serum (FBS) (Thermofisher Scientific, Waltham, MA USA), 100 U/mL penicillin-streptomycin (Thermofisher Scientific, Waltham, MA USA), and 1x GlutaMAX (Thermofisher Scientific, Waltham, MA USA). Forty-eight hours after transfection, the GFP-positive cell population was enriched by cell sorting using BD FACSAria III Cell Sorter (BD Biosciences, East Rutherford NJ, USA). Transfected GFP-positive cells were plated onto a 96-well plate, one cell per well. Single cell clones grew for nearly two weeks before they were replicated to another 96-well plate, so we obtained two equal 96-well plates with cell clones: one plate was used for PCR analysis of the deletion in the Tdp2 gene while the other plate was used for the mutant cell clone multiplication.
Genome DNA was analyzed by PCR amplification of the target region with primers to detect the presence of the CRISPR/Cas9-mediated deletions (TDP2-Del-F 5′-GCCAGGCCCTTAATAATACAGC-3′ and TDP2-Del-R 5′-TGTTTGCTCATTCACACTCCAG-3′) and wild-type alleles (TDP2-In-F 5′-CGTGTCAGGAAATGAGCTTTG-3′ and TDP2-In-R 5′-GGAACAATTATCACGGTCCAATC-3′). Scheme of primers location in TDP2 gene is shown in
Figure S9A in the Supplementary. Both reactions were run on an S1000 Thermal Cycler (Bio-Rad, Singapore) using BioMaster HS-Taq PCR-Color (2×) (Biolabmix, Novosibirsk, Russia) with the following program: 95 °C for 3 min; 35 cycles: 95 °C for 30 s; 65 °C for 30 s; 72 °C for 30 s; and 72 °C for 3 min. Products of the reactions were resolved in 1.5% agarose gel stained with ethidium bromide. As a result, two HEK293FT cell clones were found, which contained only alleles with deletions (
Figure S9B in Supplementary).
Homozygous mutation in target gene was confirmed with western blotting analysis. The whole cell extracts (HEK293FT WT and two knockout cell clones) were separated by Laemmli electrophoresis in 10% SDS-PAAG and were transferred on a nitrocellulose membrane (TransBlot Turbo, BIO-RAD, Hercules, CA, USA) using the semidry western blotting technique, and were probed with rabbit antibody to Tdp2 (99279 Rabbit Abcam) and beta-tubulin (Abcam6046). Blots were then probed with horseradish peroxidase-coupled goat anti-rabbit antibody (1:15,000, made by L. Matveev, Biotechnological Laboratory, ICBFM SB RAS, Novosibirsk, Russia), and immunoreactivity was detected by chemiluminescence (Pierce ECL Western Blotting Substrate, Thermo Scientific, Waltham, MA, USA). The western blotting analysis showed the absence of Tdp2 in the Tdp2-knockout cells.
2.4. Molecular modeling
Crystallographic data of the Tdp2 (PDB [
47] code 4GYZ [
48], chain B) were downloaded from noncommercial Protein data bank. The geometrical parameters of the protein were prepared for the calculation appropriately: hydrogen atoms were added and minimized, bond multiplicities and side chains of amino acids were restored, water molecules and excess low molecular weight compounds were removed [
49]. Molecular docking was performed using an induced fit docking (or flexible) protocol under the following conditions: flexible protein and ligand, grid size 15Å, a.a. within a radius of 5Å additionally optimized considering the effect of the ligand on the side chains. Ten docking positions were set. Docking results were ranked based on the evaluation of the following scoring functions: docking score is the main score of docking results, consisting of various energy terms, taking into account clash interaction penalties, e-model is the energy of ligand clustering in the binding site, and IFD score is the energy of the ligand-protein complex taking into account the term of the Coulomb interaction.
3. Results
3.1. Chemistry
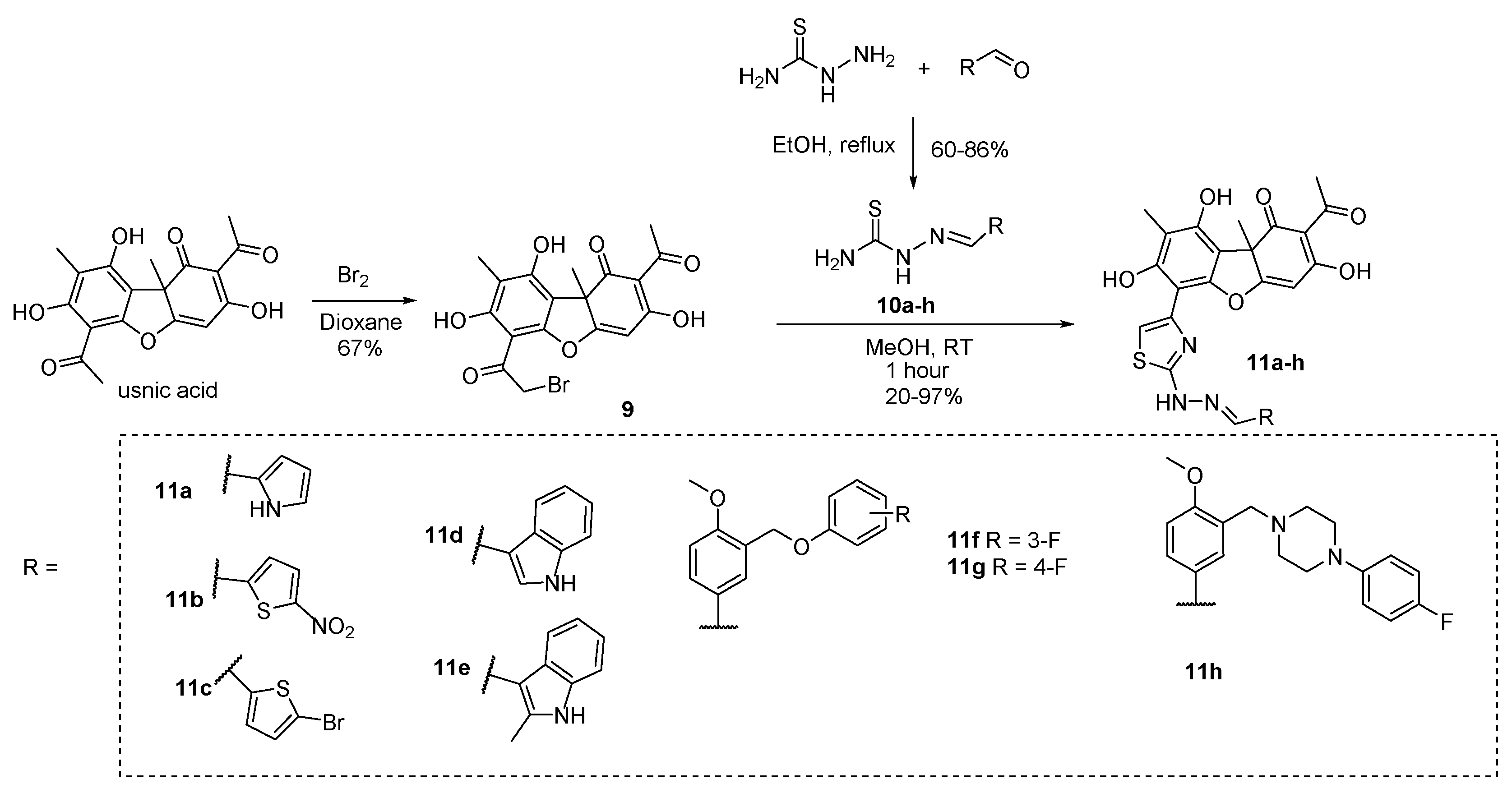
The compounds
11a-h were synthesized by a technique described previously [
44]. Thiosemicarbazones
10a-h were obtained using reaction of aldehydes with thiosemicarbazide in ethanol (
Scheme 1). The precipitate formed was filtered off, washed with water, and then air dried. Thiosemicarbazones
10a-h were obtained with the yields from 60% to 86%.
R-(+)-Usnic acid and
S-(-)-usnic acid were obtained by extraction from a mixture of the lichen genus
Usnea and
Cladonia stellaris and used as starting material [
50]. The synthesis of the bromo-substituted derivatives (+)-
9 and
(-)-9 was performed by reaction of corresponding enantiomer of usnic acid with bromine in dioxane [
51]. Hydrazonothiazoles (+)-
11a-h and (-)-
11a-h were synthesized by the heating of the bromo-substituted derivatives
9 with thiosemicarbazones
10a-h in methanol [
44]. The precipitate was filtered off, dissolved in methylene chloride, washed with sodium bicarbonate solution, and the organic phase was evaporated (
Scheme 1). Thus, usnic acid derivatives
(+)-11a-h and
(-)-11a-h were obtained with the yields from 20% to 97%.
3.2. Effect of compounds on the activity of purified Tdp2.
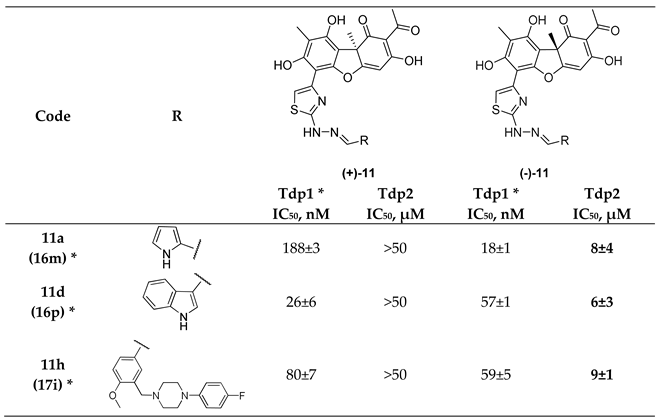
To evaluate the effect of the synthesized compounds on Tdp2 activity, we carried out the reaction of tyrosine cleavage from the 5′-end of the oligonucleotide. A fluorescent label at the 3′-end makes it possible to monitor the mobility of the initial oligonucleotide and the cleavage products in the acrylamide gel. Eight enantiomer pairs were tested, five of them had mild effect on Tdp2 activity at a concentration of 50 μM (the residual activity of the enzyme was not less than 60%). Among the three pairs remaining,
11a,d,h, (-)-enantiomers inhibited Tdp2 in the micromolar concentration range, while their (+)-counterparts at 50 μM concentration had little effect on enzyme activity. IC
50 values (the concentration of a compound that gives half-maximal inhibition) for active (-)-derivatives are shown in
Table 1.
* Data and compound numbers from [
44].
3.3. Cytotoxicity assay
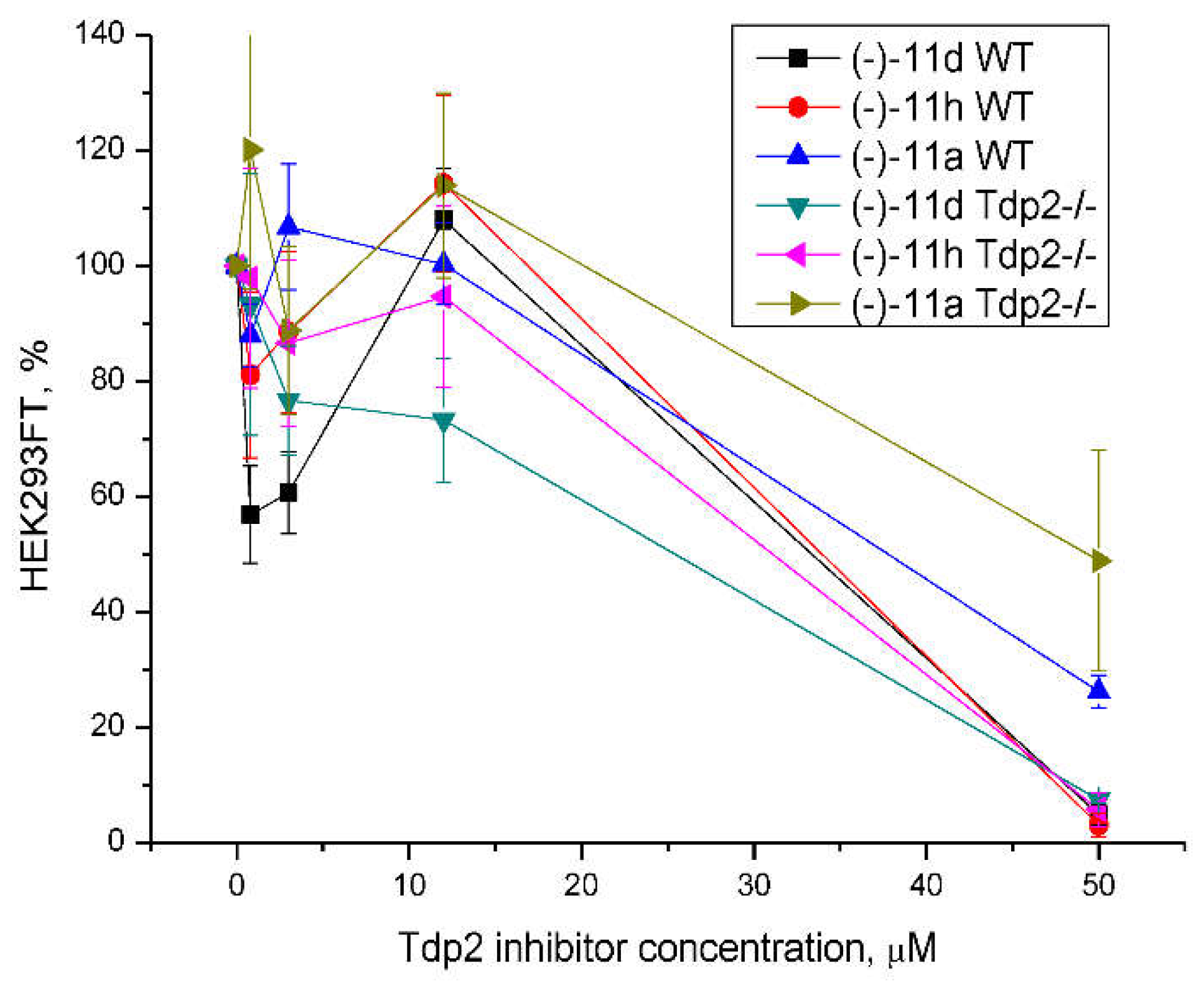
Since Tdp inhibitors are intended to be used in therapeutic cocktails, non-toxic concentrations of the compounds are required so as not to exacerbate the already serious side effects of therapy. We studied the intrinsic cytotoxicity of the compounds (-)-
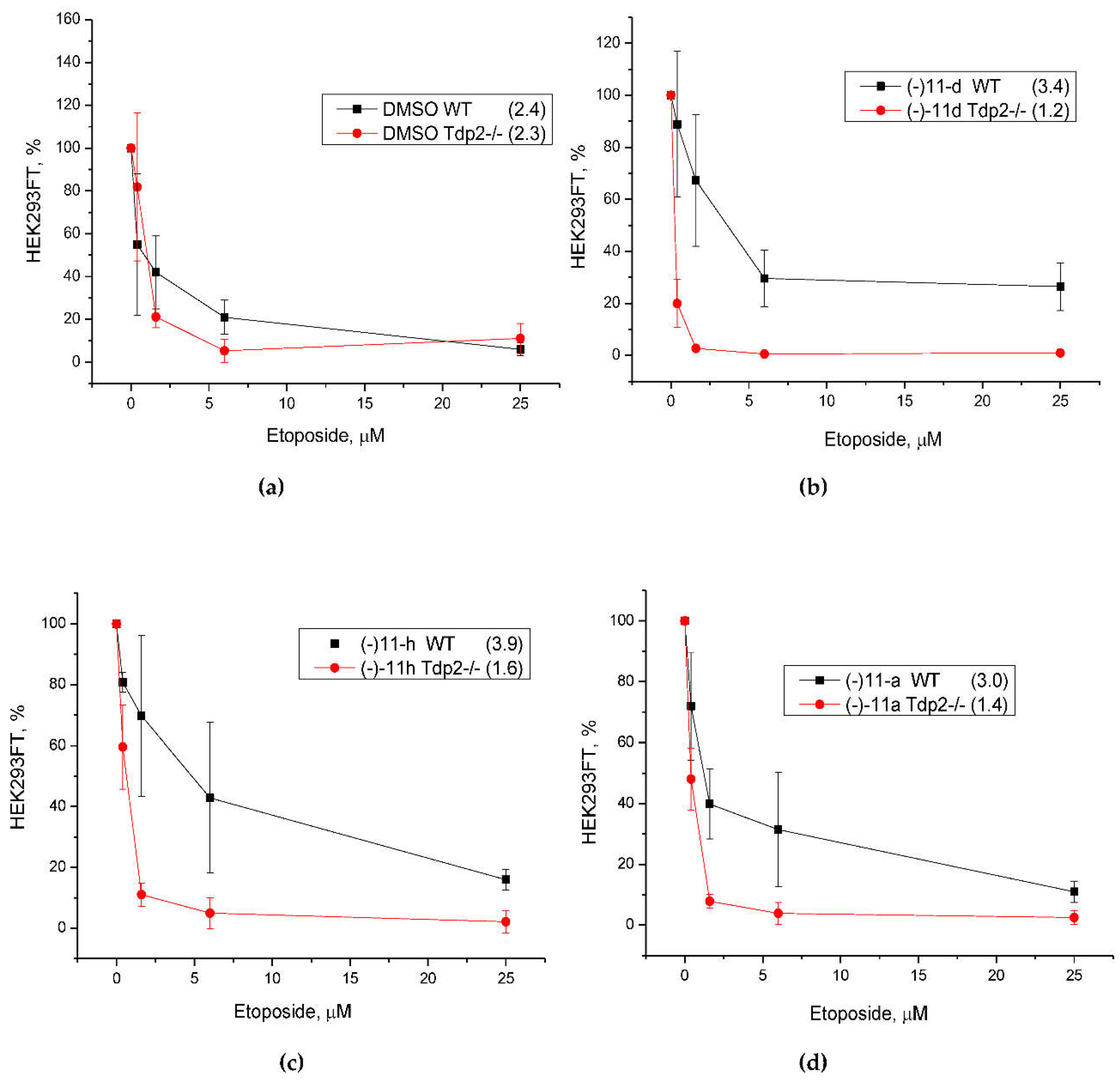
11a,d,h that inhibited Tdp2 on HEK293FT wild-type cells and Tdp2 knockout (Tdp2-/-) cells (
Figure 3). Tdp2 knockout HEK293FT cells were obtained using the CRISPR/Cas9 method. At 10 μM concentration, the cell survival was not less than 80% for both types of cells. This concentration was chosen for further studies of the ability of the compounds to sensitize the effect of Top2 inhibitor etoposide.
We studied the effect of Tdp2 inhibitors on the cytotoxic effect of etoposide. The CC
50 for etoposide was measured in the presence and absence of compounds. The sensitivity to etoposide in Tdp2 knockout cells, contrary to expectations, did not change compared to wild-type cells (
Figure 4a). The cytotoxicity of etoposide increased with the addition of compounds - inhibitors of Tdp2 (
Figure 4b-d).
3.3. Molecular Modeling
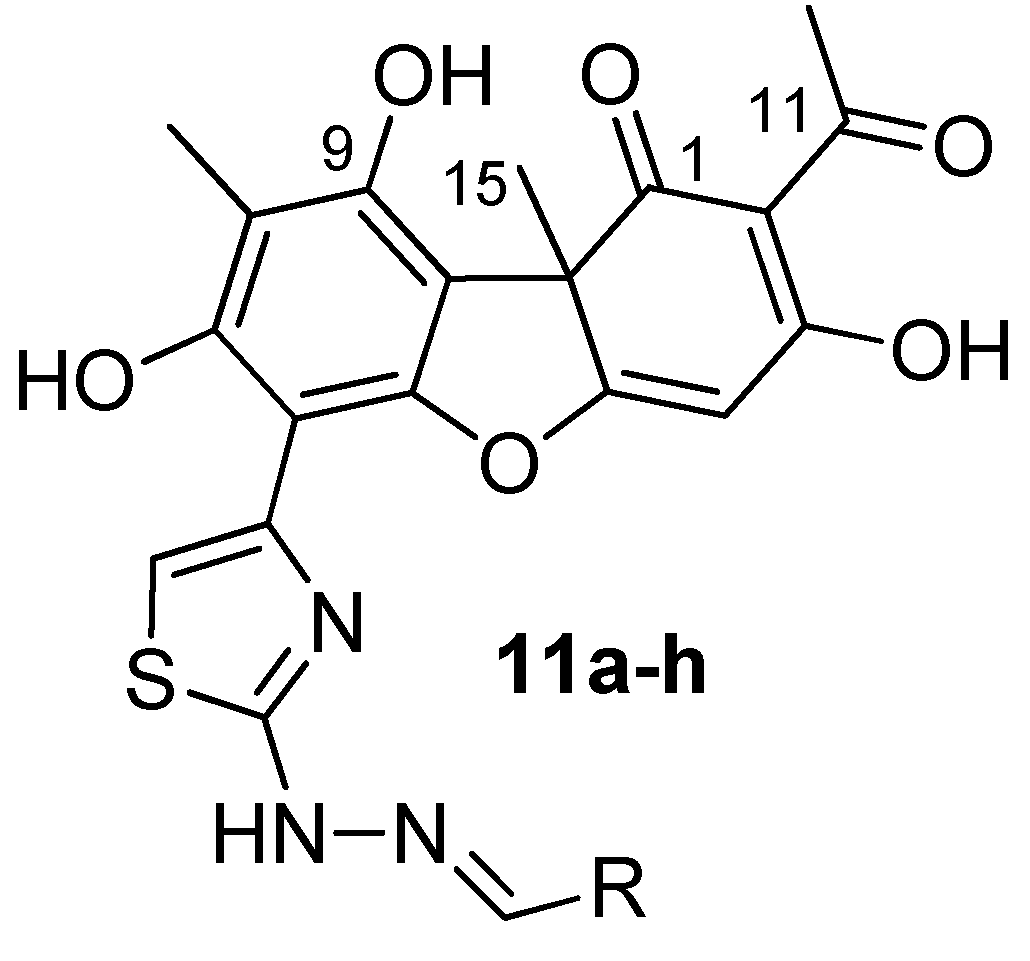
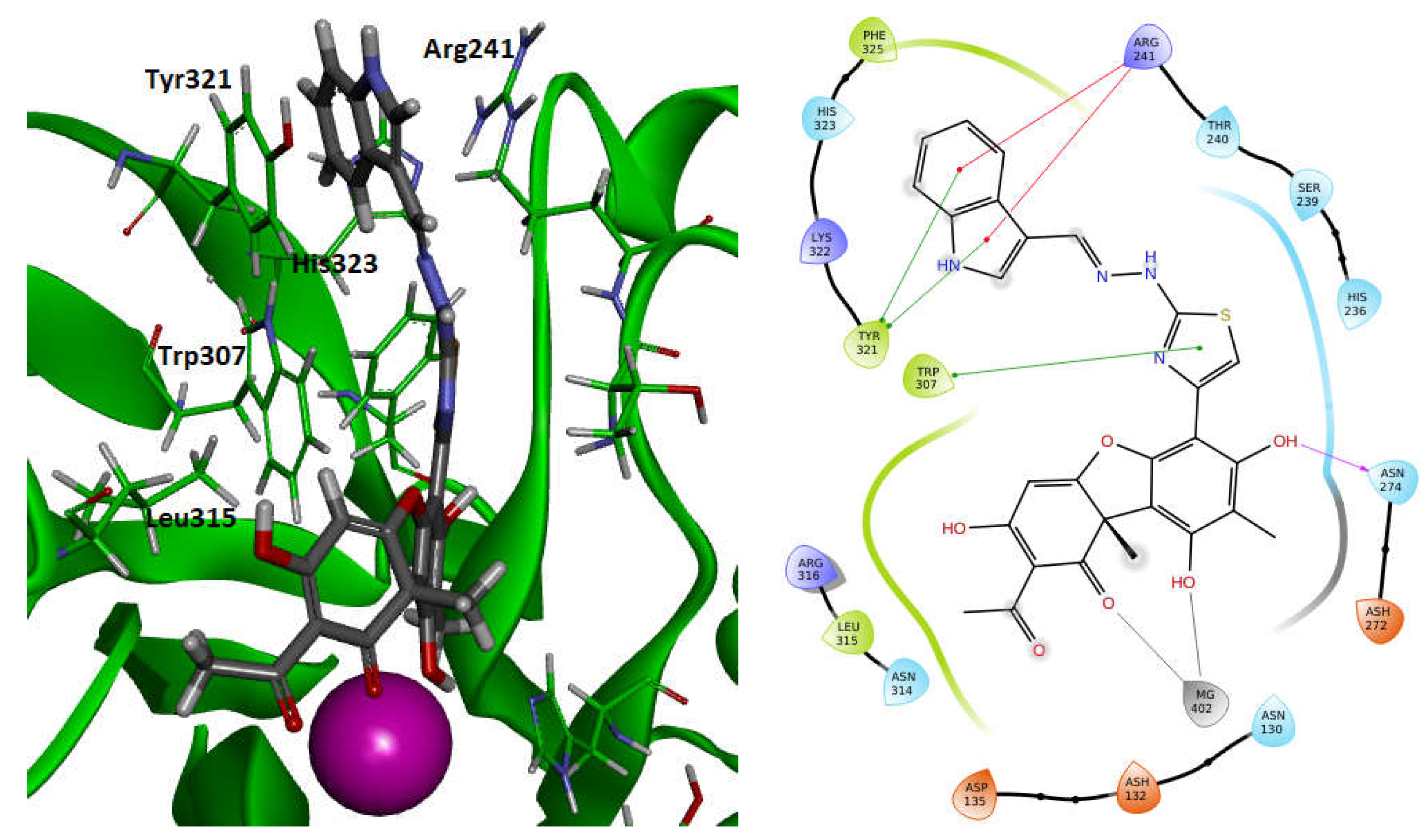
For the influence elucidation of the stereoisomeric usnic acid fragments on the Tdp2 inhibitory activity, the derivatives
(+)-11a-h and
(-)-11a-h were docked into the binding site. Due to the significant size of the molecules as well as their conformational flexibility, a number of the significant differently docking poses were obtained. Nevertheless, some poses with the specific binding pattern were found only for the (-)-enantiomers. Thus, the usnic acid fragment seemed to be a bidentate ligand of the magnesium ion located into the active site and playing a key role in the Tdp2 activity [
52]. The Mg
2+ binding could be realized by C
1=O and either C
9-O or C
11=O oxygen atoms of the usnic acid fragment (see the atomic numeration on
Figure 5). At the same time usnic acid core interacts with the lipophilic flat surface formed by Ley-315 and Trp-307. The angular C-15 methyl group could be oriented to the solvent or, to the small lipophilic cavity formed by Ley-134, Tyr-188, Met-214 and His-236. That orientation seems to be more preferable for both (-)- and (+)- stereoisomers. The extended substituent, containing thiazole fragment located in the corresponding elongate cavity, formed by Trp-307, Tyr-321, Arg-241, His-323. A number of stacking interactions could be formed, i.e. between Trp-307, Tyr-321 and thiazole or usnic acid fragments, or with aromatic systems of the substituents R in the hydrazone moiety (
Figure 6). It should be noted, that Trp-307 participate in the 5’-nucleoside binding during the catalysis by Tdp2 [
53].
For the inactive derivatives of (-)-usnic acid, in particular compounds
(-)-11b-g, the less efficient binding with the extended cavity was found. Here we considered only poses with the same pattern, that explained the difference in activity for enantiomeric (-)- and (+)-usnic acid derivatives. In particular, only poses with the usnic acid core binding magnesium ion and the heterocycle tail binding the cavity Arg-241, Trp-307, Tyr-321, His-323 were taken into account. In this context, the inactive compounds
(-)-11b-g demonstrated in general higher docking scores being up to -8.0 kcal/mol, while the active compounds
(-)-11a,d,h were found to bind the protein with the better docking scores being about -8.5 kcal/mol (
Table S1 in supplementary). For the inactive thiophene derivatives
(-)-11b,c the usnic acid fragment bind the magnesium ion as well, but 2-substituted thiophene fragments interacted with the protein less effective than pyrrole fragment of
(-)-11a. In particular, for
(-)-11b the only one corresponding pose was obtained. Considering the diphenyl- derivatives
(-)-11f,g the substituent seems to be too extended with a bad orientation of aromatic fragments. Thus, no corresponding poses were found at all for the
(-)-11f, and the only one was obtained for the
(-)-11g. Comparing indole derivatives
(-)-11d and
(-)-11e, for the inactive compound
(-)-11e the same poses were found as for the active compound
(-)-11d, but the
(-)-11e was docked with the worse docking score being -7.524 comparing to -8.613 for the
(-)-11d that could be an evidence of the worth binding resulting in the loss of the activity.
4. Discussion
To date hydrazinothiazoles based on usnic acid are the most effective known inhibitors of Tdp1, and this enzyme is not sensitive to the position of the angular methyl group of usnic acid core, the IC
50 values for both enantiomers are within the same order of magnitude [
44,
54,
55].
In this work, we have tested as Tdp2 inhibitors eight pairs of enantiomers of hydrazinothiazoles of usnic acid, among them 13 compounds had low inhibitory effect on Tdp2 activity at a concentration of 50 μM. And three derivatives of (-)-usnic acid inhibit the enzyme in the micromolar concentration range: (-)-11a,d,h.
As a rule, the cytotoxic effect of usnic acid and its derivatives does not depend on the structure of the asymmetric center if the cytotoxicity is associated with the triketone fragment of the C ring of usnic acid [
42], and therefore, in those processes where this mechanism is the main one, the difference in the action of the usnic acid enantiomers and its derivatives is insignificant. A significant influence of the position of the angular methyl group is observed in cases where specific binding of usnic acid to a substrate occurs, for example, proteins, enzymes. This is noted in publications where usnic acid-enzyme interaction was observed [
42], as well as in works on the antiviral and sensitizing effects of usnic acid and its derivatives [
43]. However, with Tdp1, the difference in the action of enantiomeric pairs of the compounds studied in this work was not significant. Previously we showed [
55] that the methyl group attached to an asymmetric carbon atom of usnic moiety in both isomers forms hydrophobic contacts with the same amino acid residues of Tdp1 (the side chains of the Tyr204 and the deprotonated His263 residues), and the inhibitor molecules themselves are oriented in the active center specularly relative to each other.
However, pairs of (+)- and (-)-usnic acid derivatives 11a-h showed a difference in Tdp2 inhibition: (+)-isomers did not affect the activity of the enzyme at concentrations up to 50 μM, while the IC50 values for some (-)-isomers were 6-9 μM. Interestingly, that according to docking experiment the location of the heterocyclic fragment in that cavity does not let the (+)-usnic acid to be oriented similar to its (-)-stereoisomer. From the other hand, the location of the (+)-usnic fragments in a similar way to the (-)-isomer does not let the extended heterocyclic chain to bind the cavity. It seems that proper binding of the extended substituent and the preferable location of the usnic acid into the active cite with simultaneous magnesium ion binding are significant for the demonstrated activity.
Thus, we have shown that (+)-isomers are selective inhibitors of Tdp1, while (-)-usnic acid derivatives
(-)-11a,d,h suppress the activity of both enzymes. Such inhibitors are relevant due to the recently discovered ability of Tdp1 and Tdp2 to take on each other's functions [
23,
29,
30,
31]. Dual Tdp1/2 inhibitors can be used to increase the efficacy of a large range of clinically important anticancer drugs (Top1/2 inhibitors) and to overcome resistance to them in various types of tumors. It should be noted that significant differences in the structure of the active centers of the Tdp1 and Tdp2 enzymes make this task extremely nontrivial.
Triple inhibitors of Tdp1/2, which also suppress the activity of Top1, were created by the group of Yves Pommier [
56,
57]. These inhibitors are not highly effective against purified Tdp1 and Tdp2 enzymes (ED
50 for Tdp1/2 ranges from 1 to 40 μM), but suppress the growth of tumor cells in the nanomolar concentration range. Activity against Top1 was comparable to that of camptothecin. The authors explain this by the presence of a synergistic effect when two or three targets are suppressed at once. But the
in vivo experiments with these compounds were not conducted, there are no data on their toxicity to animals.
Our team previously discovered dual inhibitors of Tdp1/2 among deoxycholic acid derivatives [
41] and thioether derivatives of usnic acid [
40]. These compounds were non-toxic or slightly toxic to the cells, in contrast to the isoquinolines of the Pommier’s group, but have a mild inhibitory effect against Tdp2 (in submillimolar concentrations) and were quite effective against Tdp1 (in micromolar and submicromolar concentrations). The (-)-usnic acid derivatives
(-)-11a,d,h studied in the present work inhibit both enzymes much more efficiently (in nanomolar range against Tdp1 and micromolar range against Tdp2) and can be used as a platform for designing dual Tdp1/2 inhibitors.
We obtained completely unexpected results when studying the effect of compounds on the cytotoxic effect of etoposide. We expected that the compounds would have a sensitizing effect on wild-type cells, while they would not have any effect on Tdp2 knockout cells due to the lack of a target. Or we were ready to see no effect on HEK293FT cells, as was the case with combinations of topotecan and Tdp1 inhibitors on related HEK293A cells [
58,
59]. For wild-type cells, we saw some protective effect of the compounds: an increase in CC
50 values from 2.4 µM to 3 µM and higher (compare the black plots in
Figure 4a and
Figure 4b-d). As for Tdp2-/- cells, we observed a two-fold decrease in CC
50 values for etoposide in the presence of usnic acid derivatives
(-)-11a,d,h (red graphs in
Figure 4). At the same time, in the presence of compounds, the cytotoxicity of etoposide significantly increased for Tdp2-/- cells compared to HEK293FT WT (compare black and red plots in
Figure 4b-d). We hypothesize that the additional toxicity of etoposide to Tdp2 -/- cells in the presence of the compounds is due to the effective suppression of Tdp1 in them in addition to the absent Tdp2. As mentioned above, usnic acid hydrazinothiazole derivatives are the most effective inhibitors of Tdp1 known to date and act in the nanomolar concentration range.
5. Conclusions
In the present work, we have shown that hydrazonothiazole derivatives of usnic acid with previously discovered inhibitory properties against Tdp1 are also capable of inhibiting Tdp2. At the same time, both (+)- and (-)-enantiomers of compounds act equally effectively against Tdp1, and only some (-)-enantiomers are active against Tdp2. Surprisingly, the compounds protect HEK293FT WT cells from the cytotoxic effect of etoposide but potentiate it against Tdp2-/- cells. Presumably, the sensitizing effect of the compounds in the absence of Tdp2 is associated with effective inhibition of Tdp1, which could take over the functions of Tdp2.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figures S1–S8: The 1H spectra of compounds 11a-h; Figure S9: Scheme of protospacers (TDP2-gRNA1 and TDP2-gRNA1) and primers location in TDP2 gene (A) and PCR analysis of CRISPR/Cas9-induced deletion (B); Table S1: Molecular docking results.
Author Contributions
Conceptualization, A.L.Z. and O.A.L.; methodology, A.A.M., N.S.D. and A.S.F.; software, A.S.F. and E.S.M.; validation, A.L.Z., O.A.L., A.A.M., and N.S.D.; formal analysis, S.P.M.; investigation, A.S.F., N.S.D., A.A.M., and A.L.Z.; resources, N.S.D., O.A.L., S.M.Z., and N.F.S.; data curation, A.L.Z., N.S.D., and S.P.M.; writing—original draft preparation, A.L.Z., O.A.L., and N.S.D.; writing—review and editing, S.M.Z., N.F.S., and O.I.L.; visualization, O.A.L., A.L.Z., and N.S.D.; supervision, S.M.Z., N.F.S., and O.I.L.; funding acquisition, A.L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation (Moscow, Russia) grant number 21-14-00105
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Data Availability Statement
Data is contained within the article or supplementary material
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kelley, M.R.; Logsdon, D.; Fishel, M.L. Targeting DNA repair pathways for cancer treatment: what's new? Future Oncol. 2014, 10, 1215–1237. [Google Scholar] [CrossRef]
- Curtin, N.J. Inhibiting the DNA damage response as a therapeutic manoeuvre in cancer. Br J Pharmacol. 2013, 169, 1745–1765. [Google Scholar] [CrossRef]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin Jr, A.B.; Stewart, L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 15387–15392. [Google Scholar] [CrossRef]
- Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009, 9, 338–350. [Google Scholar] [CrossRef]
- Pommier, Y. Drugging topoisomerases: lessons and challenges. ACS Chem Biol. 2013, 8, 82–95. [Google Scholar] [CrossRef]
- Pommier, Y.; Huang, S.Y.; Gao, R.; Das, B.B.; Murai, J.; Marchand, C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst). 2014, 19, 114–129. [Google Scholar] [CrossRef]
- Interthal, H.; Pouliott, J.J.; Champoux, J.J. The Tyrosyl-DNA Phosphodiesterase Tdp1 Is a Member of the Phospholipase D Superfamily. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 12009–12014. [Google Scholar] [CrossRef]
- Ben Hassine, S.; Arcangioli, B. Tdp1 protects against oxidative DNA damage in non-dividing fission yeast. EMBO J. 2009, 28, 632–640. [Google Scholar] [CrossRef]
- Povirk, L.F. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012, 2012, 345805. [Google Scholar] [CrossRef]
- Comeaux, E.Q.; Van Waardenburg, R.C.A.M. Tyrosyl-DNA Phosphodiesterase I Resolves Both Naturally and Chemically Induced DNA Adducts and Its Potential as a Therapeutic Target. Drug Metab. Rev. 2014, 46, 494–507. [Google Scholar] [CrossRef]
- Brettrager, E.J.; van Waardenburg, R.C.A.M. Targeting Tyrosyl-DNA phosphodiesterase I to enhance toxicity of phosphodiester linked DNA-adducts. Cancer Drug Resist. 2019, 2, 1153–1163. [Google Scholar] [CrossRef]
- Hirano, R.; Interthal, H.; Huang, C.; Nakamura, T.; Deguchi, K.; Choi, K.; Bhattacharjee, M.; Arimura, K.; Umehara, F.; Izumo, S.; et al. Spinocerebellar ataxia with axonal neuropathy: Consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007, 26, 4732–4743. [Google Scholar] [CrossRef]
- He, X.; van Waardenburg, R.C.A.M.; Babaoglu, K.; Price, A.C.; Nitiss, K.C.; Nitiss, J.L.; Bjornsti, M.-A.; White, S.W. Mutation of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison. J. Mol. Biol. 2007, 372, 1070–1081. [Google Scholar] [CrossRef]
- Interthal, H.; Chen, H.J.; Kehl-Fie, T.E.; Zotzmann, J.; Leppard, J.B.; Champoux, J.J. SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005, 24, 2224–2233. [Google Scholar] [CrossRef]
- Katyal, S.; Khamisy, S.F.; Russell, H.R.; Li, Y.; Ju, L.; Caldecott, K.W.; McKinnon, P.J. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007, 26, 4720–4731. [Google Scholar] [CrossRef]
- El-Khamisy, S.F.; Katyal, S.; Patel, P.; Ju, L.; McKinnon, P.J.; Caldecott, K.W. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair. 2009, 8, 760–766. [Google Scholar] [CrossRef]
- Barthelmes, H.U.; Habermeyer, M.; Christensen, M.O.; Mielke, C.; Interthal, H.; Pouliot, J.J.; Boege, F.; Marko, D. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 2004, 279, 55618–55625. [Google Scholar] [CrossRef]
- Nivens, M.C.; Felder, T.; Galloway, A.H.; Pena, M.M.; Pouliot, J.J.; Spencer, H.T. Engineered resistance to camptothecin and antifolates by retroviral coexpression of tyrosyl DNA phosphodiesterase-I and thymidylate synthase. Cancer Chemother. Pharmacol. 2004, 53, 107–115. [Google Scholar] [CrossRef]
- Alagoz, M.; Gilbert, D.C.; El-Khamisy, S.; Chalmers, A.J. DNA repair and resistance to topoisomerase I inhibitors: Mechanisms, biomarkers and therapeutic targets. Curr. Med. Chem. 2012, 19, 3874–3885. [Google Scholar] [CrossRef]
- Perego, P.; Cossa, G.; Tinelli, S.; Corna, E.; Carenini, N.; Gatti, L.; De Cesare, M.; Ciusani, E.; Zunino, F.; Luison, E.; Canevari, S.; Zaffaroni, N.; Beretta, G.L. Role of tyrosyl-DNA phosphodiesterase 1 and inter-players in regulation of tumor cell sensitivity to topoisomerase I inhibition. Biochem. Pharmacol. 2012, 83, 27–36. [Google Scholar] [CrossRef]
- Meisenberg, C.; Ward, S.E.; Schmid, P.; El-Khamisy, S.F. TDP1/TOP1 ratio as a promising indicator for the response of small cell lung cancer to topotecan. J. Cancer Sci. Ther. 2014, 6, 258–267. [Google Scholar] [CrossRef]
- Zeng, Z.; Cortés-Ledesma, F.; El Khamisy, S.F.; Caldecott, K.W. TDP2/TTRAP is the major 5'-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J Biol Chem. 2011, 286, 403–409. [Google Scholar] [CrossRef]
- Ledesma, F.C.; El Khamisy, S.F.; Zuma, M.C.; Osborn, K.; Caldecott, K.W. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 2009, 461, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Herreros, F.; Romero-Granados, R.; Zeng, Z.; Alvarez-Quilón, A.; Quintero, C.; Ju, L.; Umans, L.; Vermeire, L.; Huylebroeck, D.; Caldecott, K.W.; Cortés-Ledesma, F. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013, 9, e1003226. [Google Scholar] [CrossRef]
- Zakharenko, A.; Dyrkheeva, N.; Lavrik, O. Dual DNA topoisomerase 1 and tyrosyl-DNA phosphodiesterase 1 inhibition for improved anticancer activity. Med Res Rev. 2019, 39, 1427–1441. [Google Scholar] [CrossRef]
- Kawale, A.S.; Povirk, L.F. Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation. Nucleic Acids Res. 2018, 46, 520–537. [Google Scholar] [CrossRef]
- Zakharenko, A.L.; Luzina, O.A.; Chepanova, A.A.; Dyrkheeva, N.S.; Salakhutdinov, N.F.; Lavrik, O.I. Natural Products and Their Derivatives as Inhibitors of the DNA Repair Enzyme Tyrosyl-DNA Phosphodiesterase 1. Int J Mol Sci. 2023, 24, 5781. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Das, B.B.; Dexheimer, T.S.; Takeda, S.; Pommier, Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J. Biol. Chem. 2012, 287, 12848–12857. [Google Scholar] [CrossRef]
- Zeng, Z.; Sharma, A.; Ju, L.; Murai, J.; Umans, L.; Vermeire, L.; Pommier, Y.; Takeda, S.; Huylebroeck, D.; Caldecott, K.W.; El-Khamisy, S.F. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 2012, 40, 8371–8380. [Google Scholar] [CrossRef]
- Maede, Y.; Shimizu, H.; Fukushima, T.; Kogame, T.; Nakamura, T.; Miki, T.; Takeda, S.; Pommier, Y.; Murai, J. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol. Cancer Ther. 2014, 13, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Zakharenko, A.L.; Luzina, O.A.; Sokolov, D.N.; Kaledin, V.I.; Nikolin, V.P.; Popova, N.A.; Patel, J.; Zakharova, O.D.; Chepanova, A.A.; Zafar, A.; et al. Novel tyrosyl-DNA phosphodiesterase 1 inhibitors enhance the therapeutic impact of topotecan on in vivo tumor models. Eur. J. Med. Chem. 2019, 161, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Khomenko, T.M.; Zakharenko, A.L.; Chepanova, A.A.; Ilina, E.S.; Zakharova, O.D.; Kaledin, V.I.; Nikolin, V.P.; Popova, N.A.; Korchagina, D.V.; Reynisson, J.; et al. Promising New Inhibitors of Tyrosyl-DNA Phosphodiesterase I (Tdp 1) Combining 4-Arylcoumarin and Monoterpenoid Moieties as Components of Complex Antitumor Therapy. Int. J. Mol. Sci. 2019, 21, 126. [Google Scholar] [CrossRef]
- Nikolin, V.P.; Popova, N.A.; Kaledin, V.I.; Luzina, O.A.; Zakharenko, A.L.; Salakhutdinov, N.F.; Lavrik, O.I. The influence of an enamine usnic acid derivative (a tyrosyl-DNA phosphodiesterase 1 inhibitor) on the therapeutic effect of topotecan against transplanted tumors in vivo. Clin. Exp. Metastasis 2021, 38, 431–440. [Google Scholar] [CrossRef]
- Chernyshova, I.A.; Zakharenko, A.L.; Kurochkin, N.N.; Dyrkheeva, N.S.; Kornienko, T.E.; Popova, N.A.; Nikolin, V.P.; Ilina, E.S.; Zharkov, T.D.; Kupryushkin, M.S.; et al. Lipophilic Purine Nucleoside-Tdp1 Inhibitor-Enhances DNA Damage Induced by Topotecan In Vitro and Potentiates the Antitumor Effect of Topotecan In Vivo. Molecules 2022, 28, 323. [Google Scholar] [CrossRef]
- Marchand, C.; Abdelmalak, M.; Kankanala, J.; Huang, S.Y.; Kiselev, E.; Fesen, K.; Kurahashi, K.; Sasanuma, H.; Takeda, S.; Aihara, H.; Wang, Z.; Pommier, Y. Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chemical Biology. 2016, 11, 1925–1933. [Google Scholar] [CrossRef] [PubMed]
- Kankanala, J.; Ribeiro, C.J.A.; Kiselev, E.; Ravji, A.; Williams, J.; Xie, J.; Aihara, H.; Pommier, Y.; Wang, Z. Novel Deazaflavin Analogues Potently Inhibited Tyrosyl DNA Phosphodiesterase 2 (TDP2) and Strongly Sensitized Cancer Cells toward Treatment with Topoisomerase II (TOP2) Poison Etoposide. J. Med. Chem. 2019, 9, 4669–4682. [Google Scholar] [CrossRef]
- Yu, L.M.; Hu, Z.; Chen, Y.; Ravji, A.; Lopez, S.; Plescia, C.B.; Yu, Q.; Yang, H.; Abdelmalak, M.; Saha, S.; Agama, K.; Kiselev, E.; Marchand, C.; Pommier, Y.; An, L.K. Synthesis and structure-activity relationship of furoquinolinediones as inhibitors of Tyrosyl-DNA phosphodiesterase 2 (TDP2). Eur. J. Med. Chem. 2018, 10, 777–796. [Google Scholar] [CrossRef]
- Kont, Y.S.; Dutta, A.; Mallisetty, A.; Mathew, J.; Minas, T.; Kraus, C.; Dhopeshwarkar, P.; Kallakury, B.; Mitra, S.; Üren, A.; Adhikari, S. Depletion of tyrosyl DNA phosphodiesterase 2 activity enhances etoposide-mediated double-strand break formation and cell killing. DNA Repair. 2016, 43, 38–47. [Google Scholar] [CrossRef]
- Dyrkheeva, N.S.; Filimonov, A.S.; Luzina, O.A.; Orlova, K.A.; Chernyshova, I.A.; Kornienko, T.E.; Malakhova, A.A.; Medvedev, S.P.; Zakharenko, A.L.; Ilina, E.S.; et al. New Hybrid Compounds Combining Fragments of Usnic Acid and Thioether Are Inhibitors of Human Enzymes TDP1, TDP2 and PARP1. Int. J. Mol. Sci. 2021, 22, 11336. [Google Scholar] [CrossRef]
- Salomatina, O.V.; Dyrkheeva, N.S.; Popadyuk, I.I.; Zakharenko, A.L.; Ilina, E.S.; Komarova, N.I.; Reynisson, J.; Salakhutdinov, N.F.; Lavrik, O.I.; Volcho, K.P. New Deoxycholic Acid Derived Tyrosyl-DNA Phosphodiesterase 1 Inhibitors Also Inhibit Tyrosyl-DNA Phosphodiesterase 2. Molecules 2021, 27, 72. [Google Scholar] [CrossRef]
- Luzina, O.A.; Salakhutdinov, N.F. Biological activity of usnic acid and its derivatives: Part 1. Activity against unicellular organisms. Russ. J. Bioorg. Chem. 2016, 42, 115–132. [Google Scholar] [CrossRef]
- Luzina, O.A.; Salakhutdinov, N.F. Biological activity of usnic acid and its derivatives: Part 2. effects on higher organisms. Molecular and physicochemical aspects. Russ. J. Bioorg. Chem. 2016, 42, 249–268. [Google Scholar] [CrossRef]
- Filimonov, A.S.; Chepanova, A.A.; Luzina, O.A.; Zakharenko, A.L.; Zakharova, O.D.; Ilina, E.S.; Dyrkheeva, N.S.; Kuprushkin, M.S.; Kolotaev, A.V.; Khachatryan, D.S.; et al. New Hydrazinothiazole Derivatives of Usnic Acid as Potent Tdp1 Inhibitors. Molecules 2019, 24, 3711. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.J. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Immunol Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, M.J.; Appel, C.D.; Adhikari, S.; Robertson, P.D.; Ramsden, D.A.; Williams, S. Mechanism of repair of 5′-topoisomerase II–DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat. Struct. Mol. Biol. 2012, 19, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef]
- Polovinka, M.P.; Salakhutdinov, N.F.; Panchenko, M.Y. Method for preparing usninic acid. Patent RU231 7076, 2008. [Google Scholar]
- Luzina, O.A.; Sokolov, D.N.; Shernyukov, A.V.; Salakhutdinov, N.F. Synthesis of aurones based on usninic acid. Chem. Nat. Compd. 2012, 48(3), 385–391. [Google Scholar] [CrossRef]
- Riccio, A.A.; Schellenberg, M.J.; Williams, R.S. ; Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution. Cell Mol. Life Sci. 2020, 77, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kawale, A.S.; Povirk, L.F. Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation. Nucleic Acids Res. 2018, 46, 520–537. [Google Scholar] [CrossRef] [PubMed]
- Zakharenko, A.; Luzina, O.; Koval, O.; Nilov, D.; Gushchina, I.; Dyrkheeva, N.; Švedas, V.; Salakhutdinov, N.; Lavrik, O. Tyrosyl-DNA Phosphodiesterase 1 Inhibitors: Usnic Acid Enamines Enhance the Cytotoxic Effect of Camptothecin. J. Nat. Prod. 2016, 79, 2961–2967. [Google Scholar] [CrossRef] [PubMed]
- Dyrkheeva, N.; Luzina, O.; Filimonov, A.; Zakharova, O.; Ilina, E.; Zakharenko, A.; Kuprushkin, M.; Nilov, D.; Gushchina, I.; Švedas, V.; Salakhutdinov, N.; Lavrik, O. Inhibitory Effect of New Semisynthetic Usnic Acid Derivatives on Human Tyrosyl-DNA Phosphodiesterase 1. Planta Med. 2019, 85, 103–111. [Google Scholar] [CrossRef]
- Wang, P.; Elsayed, M.S.A.; Plescia, C.B.; Ravji, A.; Redon, C.E.; Kiselev, E.; Marchand, C.; Zeleznik, O.; Agama, K.; Pommier, Y.; Cushman, M. Synthesis and Biological Evaluation of the First Triple Inhibitors of Human Topoisomerase 1, Tyrosyl-DNA Phosphodiesterase 1 (Tdp1), and Tyrosyl-DNA Phosphodiesterase 2 (Tdp2). J Med Chem. 2017, 60, 3275–3288. [Google Scholar] [CrossRef]
- Beck, D.E.; Lv, W.; Abdelmalak, M.; Plescia, C.B.; Agama, K.; Marchand, C.; Pommier, Y.; Cushman, M. Synthesis and biological evaluation of new fluorinated and chlorinated indenoisoquinoline topoisomerase I poisons. Bioorg Med Chem. 2016, 24, 1469–1479. [Google Scholar] [CrossRef]
- Ivankin, D.I.; Dyrkheeva, N.S.; Zakharenko, A.L.; Ilina, E.S.; Zarkov, T.O.; Reynisson, J.; Luzina, O.A.; Volcho, K.P.; Salakhutdinov, N.F.; Lavrik, O.I. Monoterpene substituted thiazolidin-4-ones as novel TDP1 inhibitors: Synthesis, biological evaluation and docking. Bioorg. Med. Chem. Lett. 2022, 73, 128909. [Google Scholar] [CrossRef]
- Khomenko, T.M.; Zakharenko, A.L.; Kornienko, T.E.; Chepanova, A.A.; Dyrkheeva, N.S; Artemova, A.O.; Korchagina, D.V.; Achara, C.; Curtis, A.; Reynisson, J.; Volcho, K.P.; Salakhutdinov, N.F.; Lavrik, O.I. New 5-Hydroxycoumarin-Based Tyrosyl-DNA Phosphodiesterase I Inhibitors Sensitize Tumor Cell Line to Topotecan. Int J Mol Sci. 2023, 24, 9155. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).