1. Introduction
Google Flu Trends (GFT) was a service operated by Google to predict outbreaks of flu and was discontinued in 2015 due to inaccurate predictions. GFT trends overestimated flu prevalence by over 50% in 2011-2012, which some researchers blamed on the increased media coverage and google searches for “swine flu” and “bird flu” [
1]. A recent study indicated that a simple heuristic model predicted flu incidence better than the GFT black box algorithm [
2]. However, Google Trends may still have potential to be an affordable, timely, robust, and sensitive surveillance system [
3] given refinement of search terms, monitoring and updating of the algorithm, and use of additional data streams [
1,
4]. Google Trends data have been evaluated for their correlation with multiple zoonotic diseases, including Zika [
5], salmonellosis [
6], encephalitis [
7], and Lyme disease [
8]. These correlative studies show promise, although the use of Google Trends data for zoonotic disease prediction is underutilized. Lyme disease has been deemed a public health crisis and is reported at epidemic levels in certain geographic areas and is spreading to new geographic areas. Here, we demonstrate how Google Trends data can be used for prediction of Lyme disease cases. We build on previous work from Kim et al., 2020 [
9], who investigated the spatial-temporal associations of monthly Lyme disease incidence and Google Trend search data in the United States from 2011-2015 and found that there were similar patterns between the search patterns and incidence at the state-level and at the metro-level in Texas. However, the authors noted that validation of the method is needed due to the non-specific symptoms of Lyme that correspond to other conditions. In addition, the analysis was correlative rather than predictive. Therefore, we aimed to validate their findings by analyzing search terms for diseases with similar symptoms, including fibromyalgia, multiple sclerosis, and arthritis. In addition, we aimed to build predictive models for Lyme disease incidence by state to improve the utility of the models. The results of this paper serve as a case study for using Google Trends search data for prediction of zoonotic disease incidences. Due to the high predictive value of the models in this study, we recommend further testing Google Trends for its utility in predicting other zoonotic diseases.
2. Materials and Methods
2.1. Data retrieval
The Lincoln Memorial University Institutional Review Board approved the study protocol (1075 V.0). Monthly state-level Lyme disease case count data from 2010-2021 were requested from multiple state public health departments or obtained from online repositories. Only states with 10 or more cases in 2019 were considered [
10]. The final states included in the analysis were based on convenience, lack of missing or concerns regarding protection of individually identifiable health information, and data availability.
Google Trends search data was downloaded using the ‘
gtrendsR’ package in R version 4.0.2. [
11,
12]. Google Trends reports data as “interest over time,” which ranges from 0 to 100 and represents the terms current interest level compared to its highest interest level (at 100). Search terms were selected by evaluating previous research [
9] and through discussions of the primary literature and colloquial knowledge by the study team. The final list of search terms included terms for Lyme disease (“Lyme”, “Lyme disease”, and “Lymes”), tick (“seed tick”), symptoms of Lyme disease (“tick bite”, “bone pain”, “stiff neck”, “circular rash”, “brain fog”, tick fever”, “tick rash”, “bulls eye”, “droopy eye”, “muscle ache”, and “lethargy”), and diseases with similar symptoms as Lyme disease (“bells palsy”, “arthritis”, “fibromyalgia”, “multiple sclerosis”, “chronic fatigue”, “Summer Flu”, and “Rocky Mountain Spotted Fever”). The search terms for diseases with similar symptoms were used to test specificity of the search terms for Lyme disease and its symptoms for predicting Lyme disease case count.
2.2. Statistical analysis
Mixed negative binomial regression models were built using the ‘
menbreg’ command in Stata version 17.0 [
13] to predict the number of Lyme disease cases after determining the data were over-dispersed. Data were split into training (2010-2016) and test datasets (2017-2021). Separate models were built by search term, so in total, 22 models were tested. Monthly lags of search volumes were used as predictors (i.e., one month prior, two months prior, etc.) until statistical insignificance of the newest term was achieved. Random intercepts for state, year, and month were included to adjust for clustering of the data. Predictive ability was assessed in the test dataset via root mean squared error (RMSE) and through plots of the observed versus predicted counts. RMSE was calculated using the following equation for each observation (
i) within state (
j) within year (
k) within month (
l) [
14]:
where
O is the observed Lyme disease case count and
E is the expected, or predicted, case count. RMSE can be interpreted on the same scale as the outcome (Lyme disease case count) and is the average deviation of expected versus observed counts. Therefore, the lower the RMSE, the better the model is at predicting Lyme disease case count.
3. Results
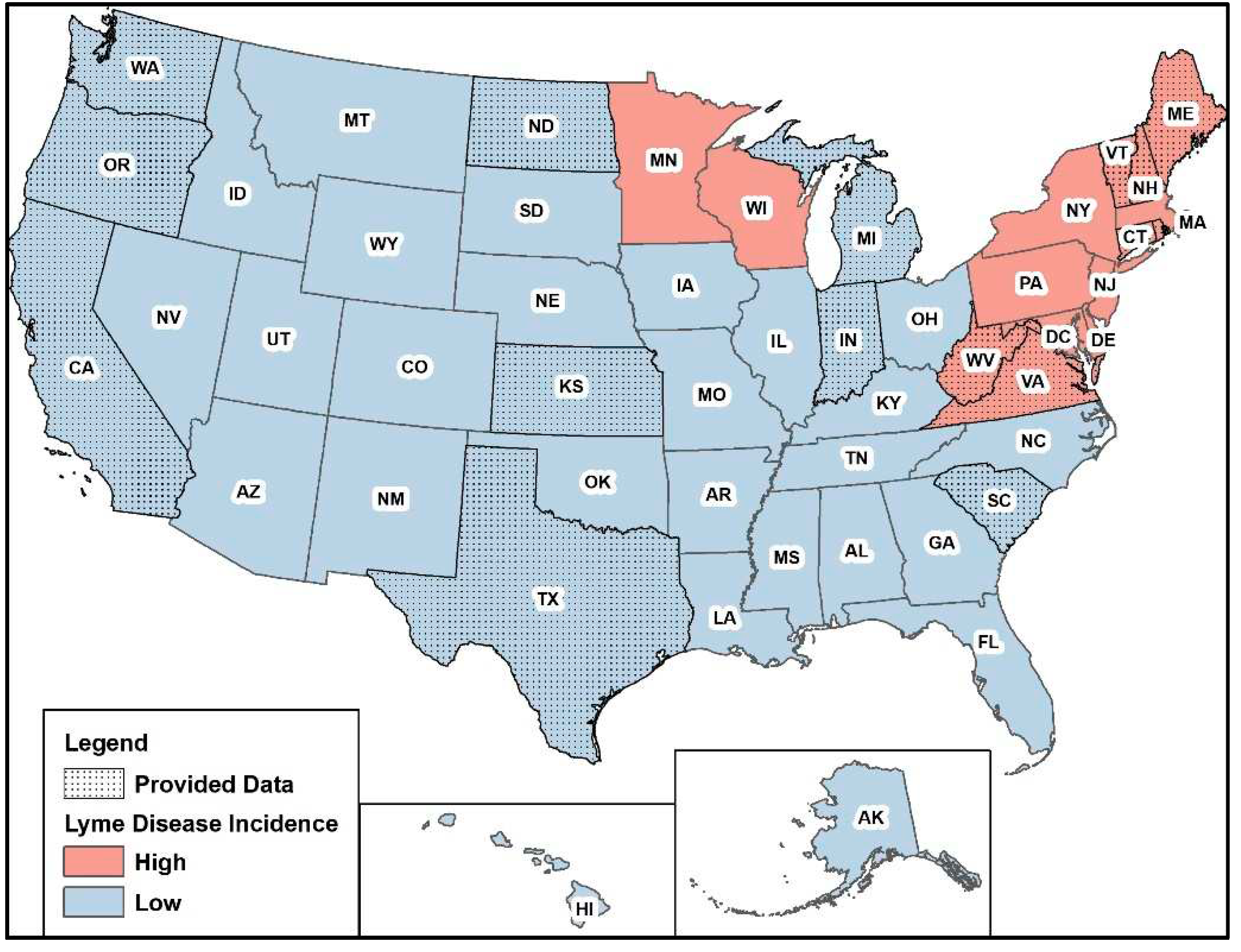
The final sample included data from 16 states (
Figure 1). Seven of the 16 states are considered high incidence states according to the CDC (
https://www.cdc.gov/lyme/datasurveillance/lyme-disease-maps.html). All available data provided from 2010-2021 was used for the analysis and states had variable levels of missing data (
Table 1). Data notes and caveats supplied from health departments are listed in Supplementary File 1. Washington had the lowest amount of missing data and Virginia had the highest amount of missing data. Descriptive statistics of the average monthly Lyme disease case counts stratified by state are summarized in
Table 1.
3.1. Predictive models
Multiple terms were significantly associated with Lyme disease case count (
Table 2), including all terms for Lyme diseases and multiple terms for symptoms. However, terms for diseases with similar symptoms were also significant, including “arthritis”, “Rocky Mountain Spotted Fever,” and “Summer Flu”, which indicates low specificity of these selected terms. The strongest predictive term for Lyme disease case count was “Summer Flu”, which had the lowest overall RMSE value (Table 3). The RMSE for “Summer Flu” was 1.7, which can be interpreted as: on average, the model with search terms for “Summer Flu” predicted within 1.7 cases of the actual case count. Even for the highest incidence state, Connecticut, the model predicted within 7 cases of the actual case count on average.
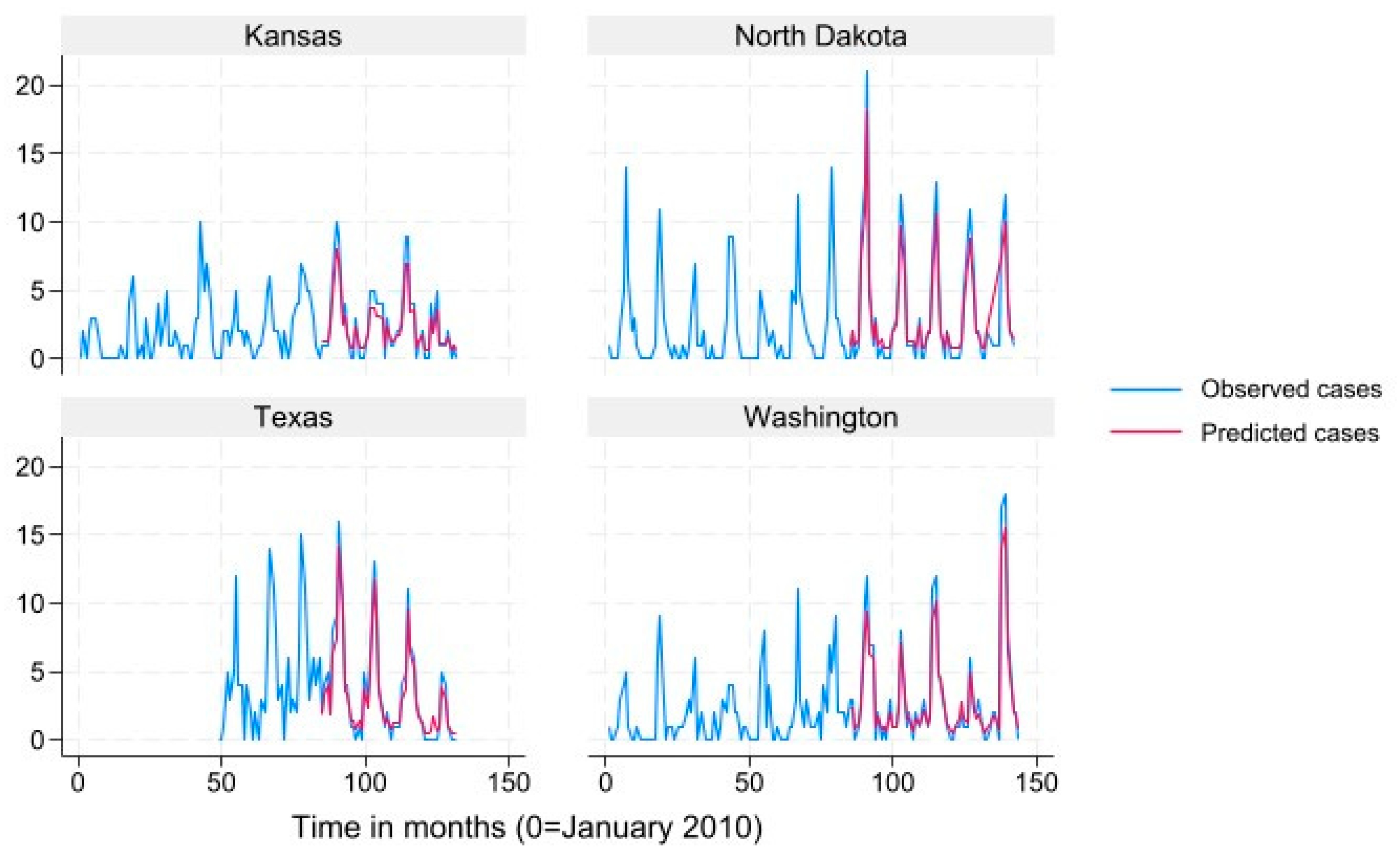
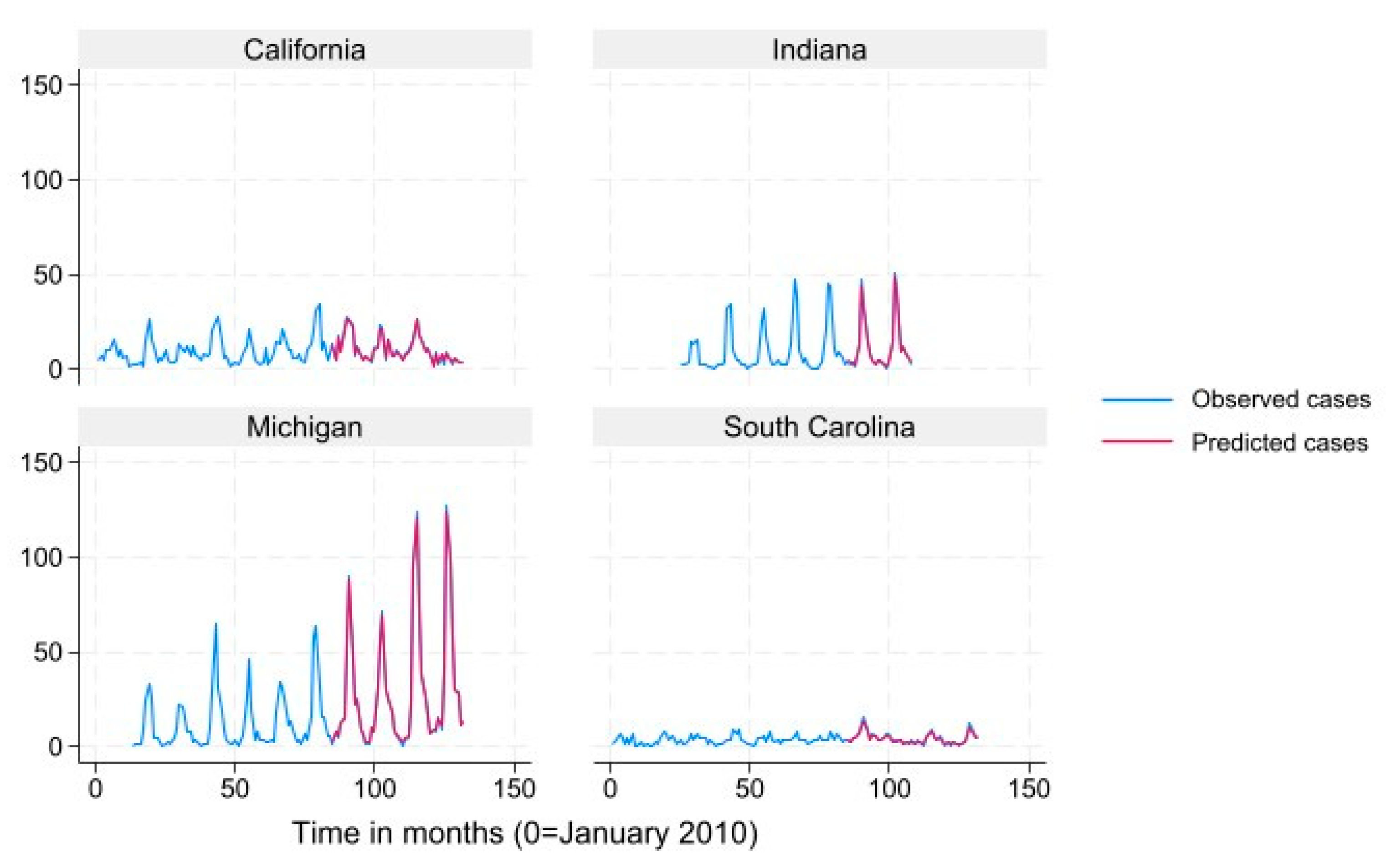
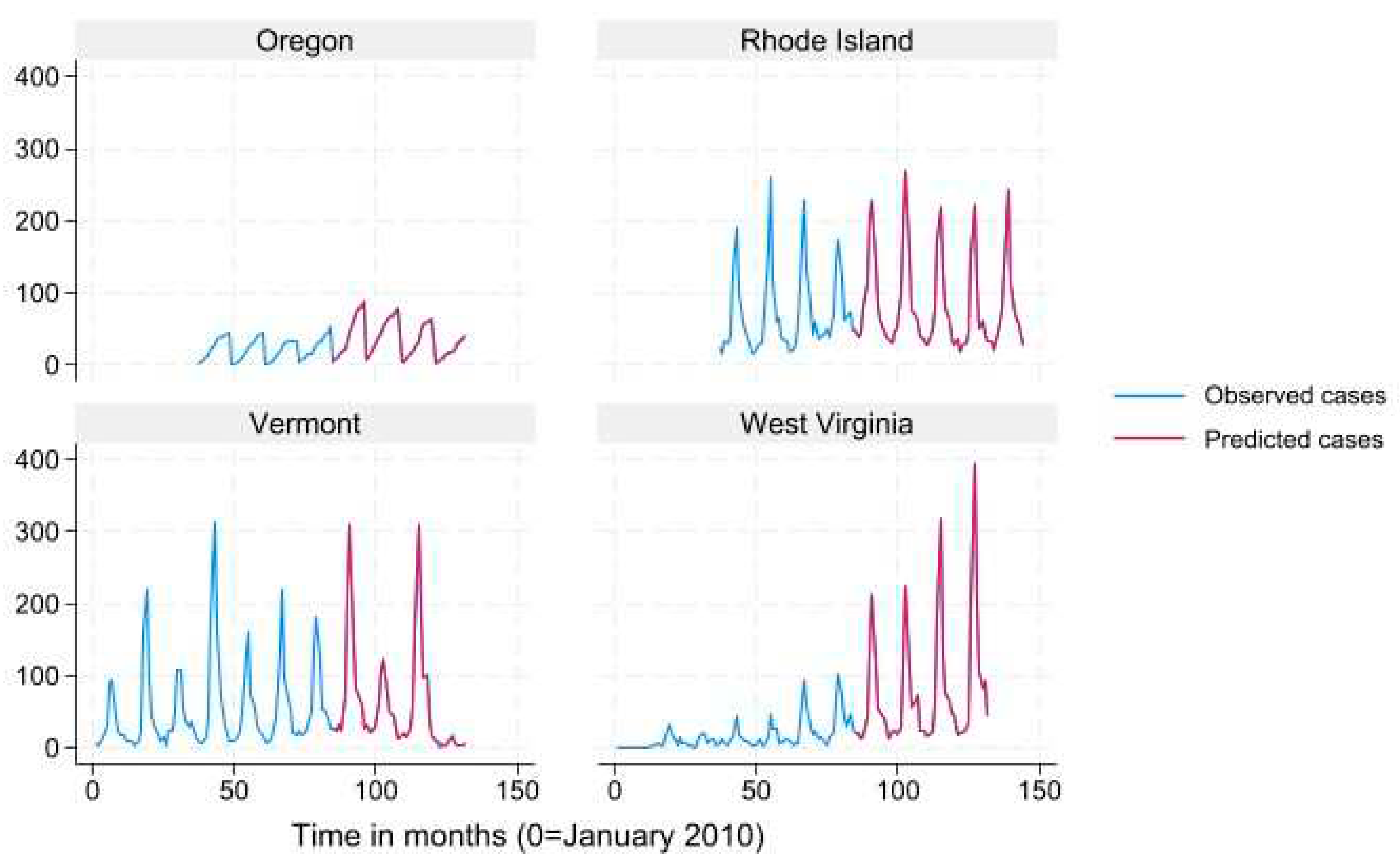
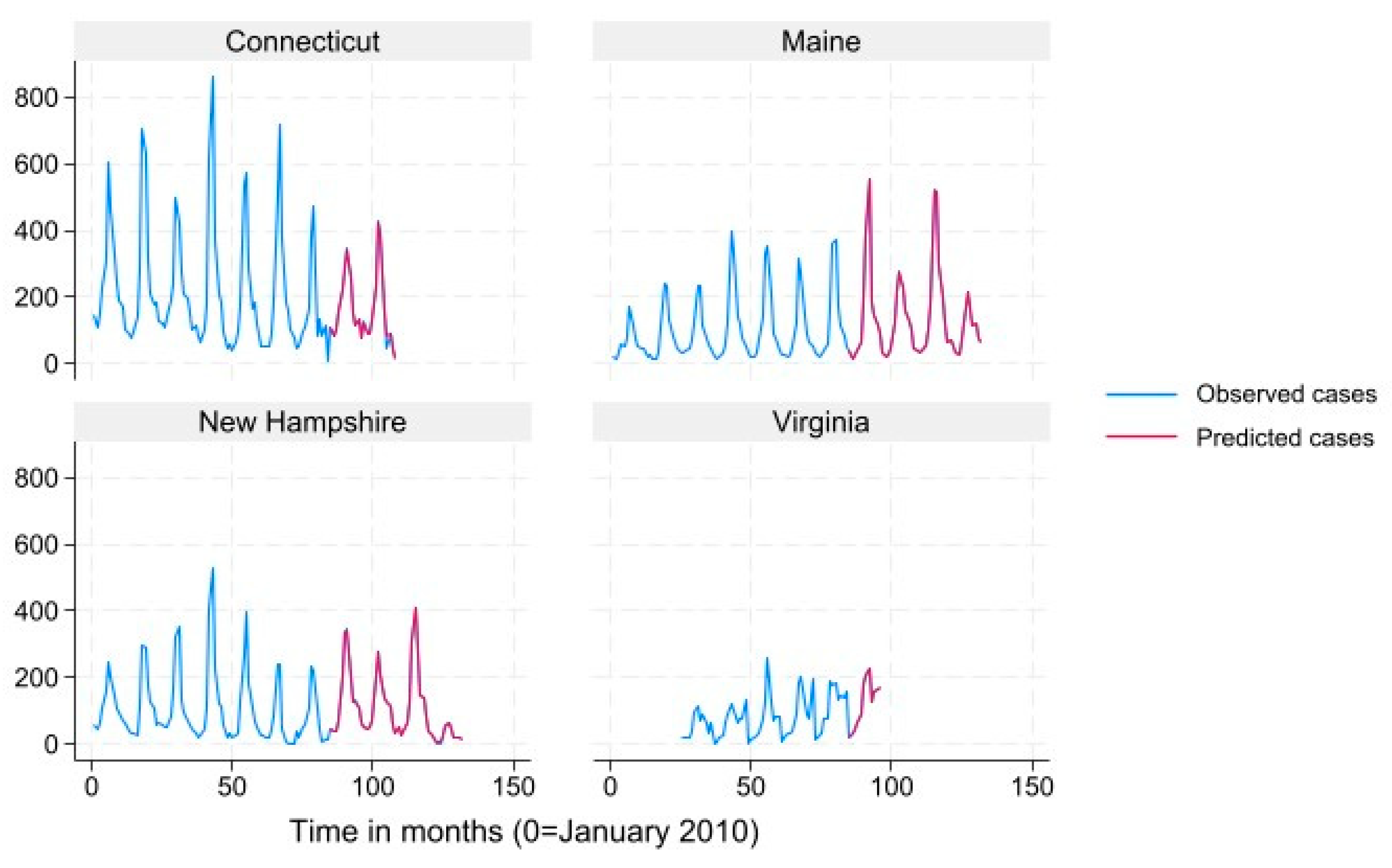
We used mean monthly Lyme disease case count as calculated from the data to define states into “very high incidence” (>78.6), “high incidence” (19.3-78.6) “low incidence” (3.9-19.2) and “very low incidence” (<3.9) categories for data presentation in
Figure 2,
Figure 3,
Figure 4 and
Figure 5. Results for the term “Summer Flu” are presented. The predicted case counts closely follow the observed case counts for all states and incidence levels, which indicates high predictive ability.
4. Discussion
Google Trends data are freely available and downloadable, which provides accessibility for researchers, epidemiologists, and health departments. Google Trends were used by the CDC for prediction of yearly influenza cases, but eventually they discontinued use due to low predictive ability [
1]. In this study, we assessed the predictive ability of Google search terms for monthly Lyme disease case count at the state level. We found that the models produced accurate predictions, as demonstrated by the closeness of the predicted and observed case counts. We conclude that Google Trends data have potential to be a tool for zoonotic disease incidence prediction.
Interestingly, the most predictive term for Lyme disease case count was “Summer Flu.” “Summer Flu” performed better than terms for Lyme disease and common Lyme disease symptoms, which indicates low specificity of the search terms. Searches for “Summer Flu” may be more of an indicator of season and temperature than actual Lyme disease symptoms. However, this term still could still be useful in predicting Lyme disease risk, due to the large environmental influence on Lyme disease risk, such as the effect of temperature and humidity on nymph and adult tick metamorphosis and activity [
15,
16]. “Summer Flu” searches may also indicate undiagnosed or misdiagnosed Lyme disease [
17].
One of the challenges of this type of research is obtaining Lyme disease case data. In the United States, each state health department tracks and reports Lyme disease data and there is not a centralized data system. The health departments then report yearly data to the CDC. The system for requesting data in each state varies. Some states have data readily available for use on their official websites, whereas others require full Institutional Review Board review. In addition, case definitions are not consistent across state or even across time, although this did not seem to impact the performance of the models. Some states with low case counts censored small cell sizes, so we were unable to include those states in the analysis. Another challenge is the geographical units of the Google Trends search data. Google Trends data does not report at the county level, likely due to search volume and data privacy issues. The smallest geographical unit reported is at the metro-level, which are geographical areas that correspond to metropolitan areas. Unfortunately, this does not correspond directly to county-level data, which is how most health departments report case data. Another challenge is selecting search terms. In the future, we recommend considering regional differences in terminology when selecting Google trends search terms. In addition, we recommend considering search volume. In less-populated states, some of our selected Google Trends search terms did not reach an adequate search volume to use in the models.
A nationwide, centralized data reporting system for monthly Lyme disease cases would improve the feasibility of utilizing Google Trends for Lyme disease prediction. Currently, the CDC maintains a Lyme disease data dashboard, although the units reported are at the yearly level, which makes finer prediction not possible. Lyme disease cases are now reported at epidemic levels in some areas and there should be urgency in improving access to data [
18].
In our study, we used one- and two-month lags of search terms, which leaves little time for immediate intervention. Future studies can determine how early we can predict increases in Lyme disease case counts. For example, we can build models that predict a season or a year in advance. In addition, future studies should investigate the inclusion of environmental, tick, and companion animal data for model refinement and to consider the full One Health triad. Future studies can also validate the findings of this case study in other zoonotic diseases and determine if the Lyme models continue to be accurate over time. There is a risk that with more media attention on Lyme disease, the models will be less predictive.
5. Conclusions
In the study, we demonstrate the use of Google Trends search data for prediction of monthly Lyme disease case counts at the state-level. The models produced accurate predictions for both low and high incidence states. We outline challenges for Google Trends disease prediction, such as data availability and mismatch of Google Trends geographical units with county case counts. However, there are many opportunities for utilizing Google Trends data, as it is a free, publicly available resource and has not yet been tested for predictive ability for many zoonotic diseases. Integration of environmental, tick, and companion animal data is the next step to make it a true One Health model.
Author Contributions
Conceptualization, L.W.; methodology, L.W., K.G., V.F., B.S.; formal analysis, L.W.; data curation, L.W.; writing—original draft preparation, L.W.; writing—review and editing, L.W., K.G., V.F., B.S.; visualization, L.W., K.G.; funding acquisition, L.W., K.G., V.F., B.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Richard A. Gillespie College of Veterinary Medicine intramural research funds.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Lincoln Memorial University (protocol code 1075 V.0. approved 2/15/2022).
Informed Consent Statement
Not applicable. Secondary analysis of de-identified case data was used for the analysis.
Data Availability Statement
Restrictions apply to the availability of Lyme disease case data. Supplementary File 1 includes links to data repositories for Connecticut, Indiana, Oregon, and Virginia. Data from the remaining states can be requested from individual health departments. Google trends search volume data available on request from the corresponding author.
Acknowledgments
We would like to thank our funding source, the Richard A. Gillespie College of Veterinary Medicine at Lincoln Memorial University. We would like to thank the following health departments for providing the Lyme disease case data for this project: California Department of Public Health; Kansas Department of Health and Environment; Maine Department of Health and Human Services; Michigan Department of Health and Human Services; New Hampshire Division of Public Health Services; North Dakota Department of Health; Rhode Island Department of Health, Division of Preparedness, Response, Infectious Disease and Emergency Medical Services, Center for Acute Infectious Disease Epidemiology; South Carolina Department of Health and Environmental Control; Texas Department of State Health Services; Vermont Department of Health; Washington State Department of Health; West Virginia Department of Health and Human Resources. We would also like to thank the following health departments with online data portals and reports that we used to obtain case data: Connecticut State Department of Public Health; Indiana Department of Health; Oregon Health Authority; Virginia Department of Health.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Lazer, D.; Kennedy, R.; King, G.; Vespignani, A. The parable of Google Flu: traps in big data analysis. Science 2014, 343, 1203–1205. [Google Scholar] [CrossRef] [PubMed]
- Katsikopoulas, K.V.; Şimşek, Ö.; Buckmann, M.; Gigerenzer, G. Transparent modeling of influenza incidence: Big data or a single data point from psychological theory? Intern J of Forecasting 2022, 38, 613–619. [Google Scholar] [CrossRef]
- Carneiro, H.A.; Mylonakis, E. Google Trends: A web-based tool for real-time surveillance of disease outbreaks. Clin Infect Dis 2009, 49, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bambrick, H.; Mengersen, K.; Tong, S.; Hu, W. Using Google Trends and ambient temperature to predict seasonal influenza outbreaks. Environ Intern 2018, 117, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Morsy, S.; Dang, T.N.; Kamel, M.G.; Zayan, A.H.; Makram, O.M.; Elhady, M.; Hirayama, K.; Huy, N.T. Prediction of Zika-confirmed cases in Brazil and Colombia using Google Trends. Epi Infect 2018, 146, 1625–1627. [Google Scholar] [CrossRef] [PubMed]
- Wang, M-Y. ; Tang, N-j. The correlation between Google Trends and salmonellosis. BMI Pub Heal 2021, 21, 1575. [Google Scholar] [CrossRef] [PubMed]
- Sulyok, M.; Richter, H.; Sulyok, Z.; Kapitány-Fövény, M.; Walker, M.D. Predicting tick-borne encephalitis using Google Trends. Ticks and Tick-borne Diseases 2020, e101306. [Google Scholar] [CrossRef] [PubMed]
- Kapitány-Fövény, M.; Ferenci, T.; Sulyok, Z.; Kegele, J.; Richter, H.; Vályi-Nagy, I.; Sulyok, M. Can Google Trends data improve forecasting of Lyme disease incidence? Zoonoses Pub Heal 2018, 66, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Maxwell, S.; Le, Q. Spatial and temporal comparison of perceived risks and confirmed cases of Lyme Disease: An exploratory study of google trends. Front Pub Heal 2020, 8, 395. [Google Scholar] [CrossRef] [PubMed]
- Surveillance data. Available online: https://www.cdc.gov/lyme/datasurveillance/surveillance-data.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Flyme%2Fstats%2Fgraphs.html (accessed on 3 August 2023).
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2021.
- Massicotte, P.; Eddelbuettel, D. gtrendsR: Perform and Display Google Trends Queries. R package version 1.5.1, 2022.
- StataCorp. Stata Statistical Software: Release 17. 2021. College Station, TX: StataCorp LLC.
- Kuhn, M.; Johnson, K. Applied Predictive Modeling. Springer Nature, New York, NY, 2013; pp. 95–100.
- Burtis, J.C.; Sullivan, P.; Levi, T.; Oggenfuss, K.; Fahey, T.J. , Ostfeld, R.S. The impact of temperature and precipitation on blacklegged tick activity and Lyme disease incidence in endemic and emerging regions. Paras Vect 2016, 9, e606. [Google Scholar] [CrossRef] [PubMed]
- Heaney, C.D.; Moon, K.A.; Ostfeld, R.S.; Pollak, J.; Poulsen, M.N.; Hirsch, A.G.; DeWalle, J.; Aucott, J.N.; Schwartz, B.S. Relations of peri-residential temperature and humidity in tick-life-cycle-relevant time periods with human Lyme disease risk in Pennsylvania, USA. Sci Tot Envirn 2021, e148697. [Google Scholar] [CrossRef] [PubMed]
- Aucott, J.N.; Seifter, A. Misdiagnosis of early Lyme disease as the summer flu. Ortho Rev 2011, 3, e14. [Google Scholar] [CrossRef]
- Stricker, R.B.; Johnson, L. Lyme disease: Call for a “Manhattan Project” to combat the epidemic. PLOS Path 2014, 10, e1003796. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).