Methodology
The development environment is established by installing essential libraries like TensorFlow and Keras, which are widely used in deep learning tasks. These libraries provide powerful tools for implementing and training neural network models. Additionally, the brain tumor MRI image dataset is loaded into the environment, providing the necessary data for model training and evaluation.
By setting up the environment with TensorFlow and Keras, researchers and practitioners in the field of medical imaging can leverage these robust libraries to develop and deploy sophisticated deep learning models. Furthermore, the inclusion of the brain tumor MRI image dataset enables the exploration of the dataset's characteristics, preprocessing techniques, and subsequent analysis for accurate brain tumor detection. This combination of a well-configured environment and the availability of relevant data lays the foundation for conducting comprehensive research and advancements in the field of brain tumor detection.
The utilization of publicly available datasets, like the one employed in this study, fosters inclusivity and equal opportunity for researchers across different institutions and regions. By openly sharing the dataset on Kaggle, Navoneel Chakrabarty and the Eindhoven University of Technology (TU/e) demonstrate their commitment to advancing scientific knowledge in the field of brain tumor detection. The accessibility of the dataset promotes diversity in research perspectives and encourages contributions from a wide range of experts, ultimately leading to more robust and comprehensive findings.
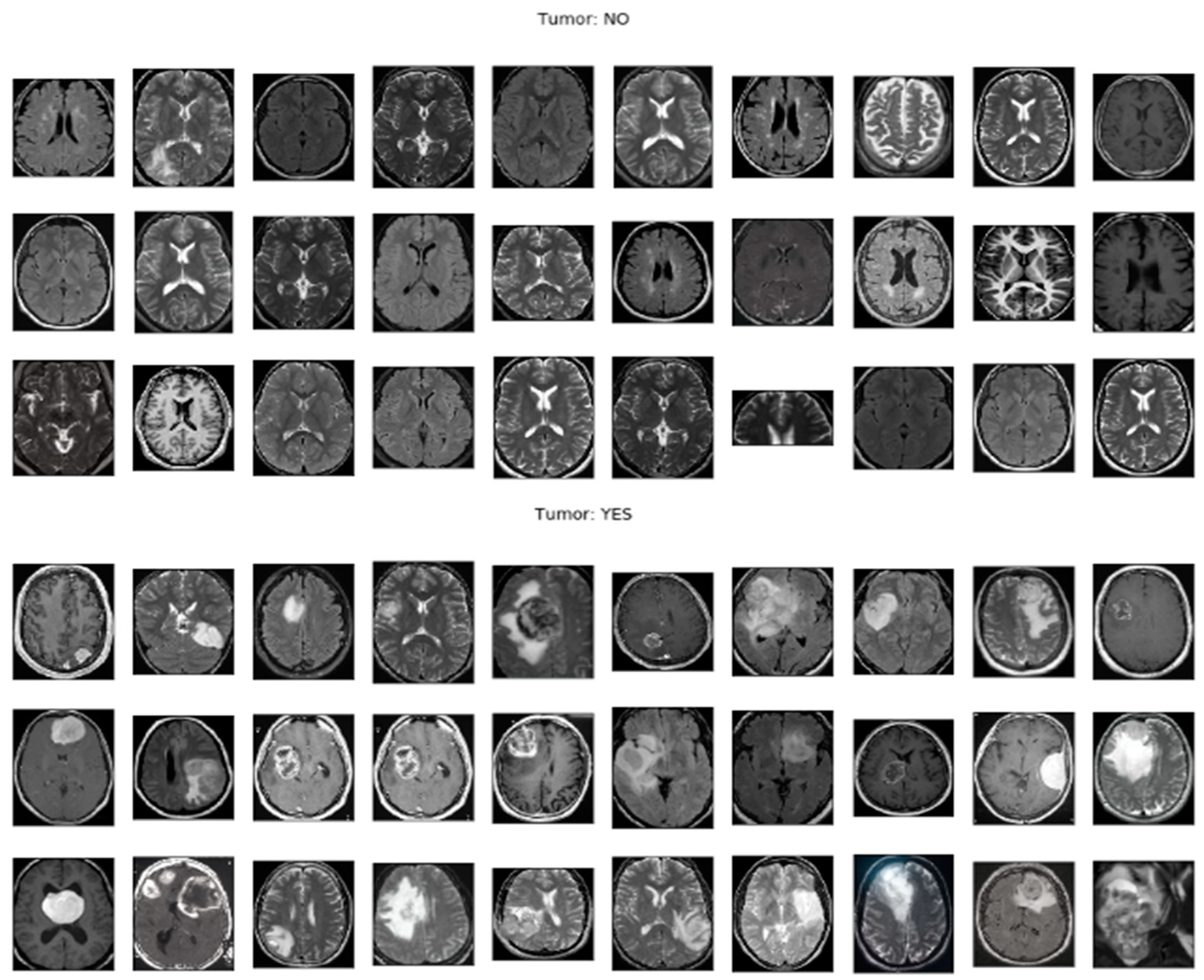
Figure 1: Sample MRI Images for Brain Tumor Detection showcases a selection of MRI images from the publicly available dataset. These images represent real-world brain tumor cases and serve as valuable training and evaluation data for researchers working on brain tumor detection algorithms.
Additionally, the availability of labeled MRI images in the dataset contributes to the development of standardized evaluation protocols and facilitates the comparison of different models and algorithms. This allows researchers to assess the strengths and limitations of their approaches in a consistent manner, driving innovation and continuous improvement in the field.
Figure 1 provides a visual representation of the diversity and complexity of brain tumor cases included in the dataset, highlighting the challenges that researchers face in accurately detecting and classifying brain tumors in MRI images.
Moreover, the open nature of the dataset promotes reproducibility, enabling other researchers to validate the results and build upon them. This collaborative environment fosters a collective effort towards improving the accuracy and reliability of brain tumor detection methods, ultimately benefiting healthcare professionals and patients.
Figure 1: Sample MRI Images for Brain Tumor Detection serves as a reference for researchers to compare their own findings and algorithms against the provided dataset, allowing for independent verification and replication of results.
In summary, the public availability of the brain tumor MRI dataset used in this study on Kaggle, as depicted in
Figure 1, promotes inclusivity, collaboration, and reproducibility in the research community. It empowers researchers from diverse backgrounds to contribute their expertise and advances the collective understanding and development of effective brain tumor detection techniques. The visual representation of the sample MRI images in
Figure 1 highlights the practical application and relevance of the dataset in advancing the field of brain tumor detection.
Once the dataset is imported, preprocessing steps are applied to the MRI images. These steps are crucial for preparing the data before feeding it into the deep learning model. The preprocessing includes resizing the images to a standardized resolution, normalizing the pixel values to a common scale, and dividing the data into separate training and testing sets. Resizing the images ensures uniformity and compatibility, while normalizing the pixel values helps in reducing the influence of different intensity ranges. Splitting the data into training and testing sets allows for unbiased model evaluation and performance assessment.
By utilizing the publicly available dataset, researchers and practitioners can conduct further investigations and contribute to the field of brain tumor detection. The open availability of the dataset on Kaggle promotes transparency, reproducibility, and collaborative research efforts in advancing the accuracy and efficiency of brain tumor detection algorithms.
To mitigate overfitting and enhance the performance of the model, data augmentation techniques are applied. Data augmentation involves applying random transformations to the training images, creating new variations of the data. This helps in increasing the diversity and quantity of the training data, reducing the risk of overfitting and improving the model's ability to generalize to unseen examples (Shorten & Khoshgoftaar, 2019).
In this code snippet, the `ImageDataGenerator` class from Keras is utilized to perform data augmentation. Various parameters are set to specify the types and degrees of transformations to be applied. These include rotation, width and height shifting, rescaling, shearing, brightness adjustment, and horizontal and vertical flipping. By randomly applying these transformations to the training images, the model becomes more robust and better equipped to handle variations in real-world data (Perez & Wang, 2017).
The code snippet also demonstrates an example of how the data augmentation can be previewed. The `ImageDataGenerator` is used to generate augmented images from a sample image, and the transformed images are saved to a directory for visualization purposes. By examining the augmented images, researchers can gain insights into the types of variations introduced and assess the effectiveness of the data augmentation techniques (Cubuk et al., 2019).
Data augmentation plays a crucial role in improving the performance of deep learning models, especially when working with limited datasets. By introducing diverse training examples through random transformations, the model becomes more adaptable and better able to handle different variations and challenges in real-world scenarios (Cubuk et al., 2019).
The VGG-16 model is a deep convolutional neural network (CNN) architecture that consists of multiple layers, including convolutional and fully connected layers, stacked sequentially to analyze and classify MRI images. It is designed to extract intricate features from the input images, enabling accurate detection and classification of brain tumors (Simonyan & Zisserman, 2015).
The architecture of the VGG-16 model comprises 13 convolutional layers and 3 fully connected layers. The convolutional layers employ small filters to perform feature extraction through convolution operations. These layers are responsible for capturing spatial patterns and local image features, allowing the model to learn representations at different levels of abstraction.
The fully connected layers, also known as dense layers, are positioned at the end of the network and perform classification based on the extracted features. They combine the learned features from the convolutional layers and make predictions about the presence or absence of brain tumors in the MRI images.
By stacking multiple convolutional and fully connected layers, the VGG-16 model can effectively capture intricate details and complex relationships within the brain tumor images. This deep architecture enables the model to achieve high accuracy in brain tumor detection tasks.
During the training process, the model's performance is evaluated using standard metrics such as accuracy, precision, and recall. The training is conducted over multiple epochs, and after each epoch, the model's performance on the validation set is assessed. This evaluation helps in fine-tuning the hyperparameters and ensuring that the model can generalize well to unseen data.
The provided training updates show the loss and accuracy values for each epoch. As the training progresses, the loss decreases, indicating that the model is learning to make more accurate predictions. The accuracy values demonstrate the proportion of correctly classified images during training and validation.
In this specific training session, the model achieves high accuracy on both the training and validation sets, demonstrating its capability to effectively detect brain tumors in the MRI images. This indicates the potential of the model to accurately classify new, unseen images.
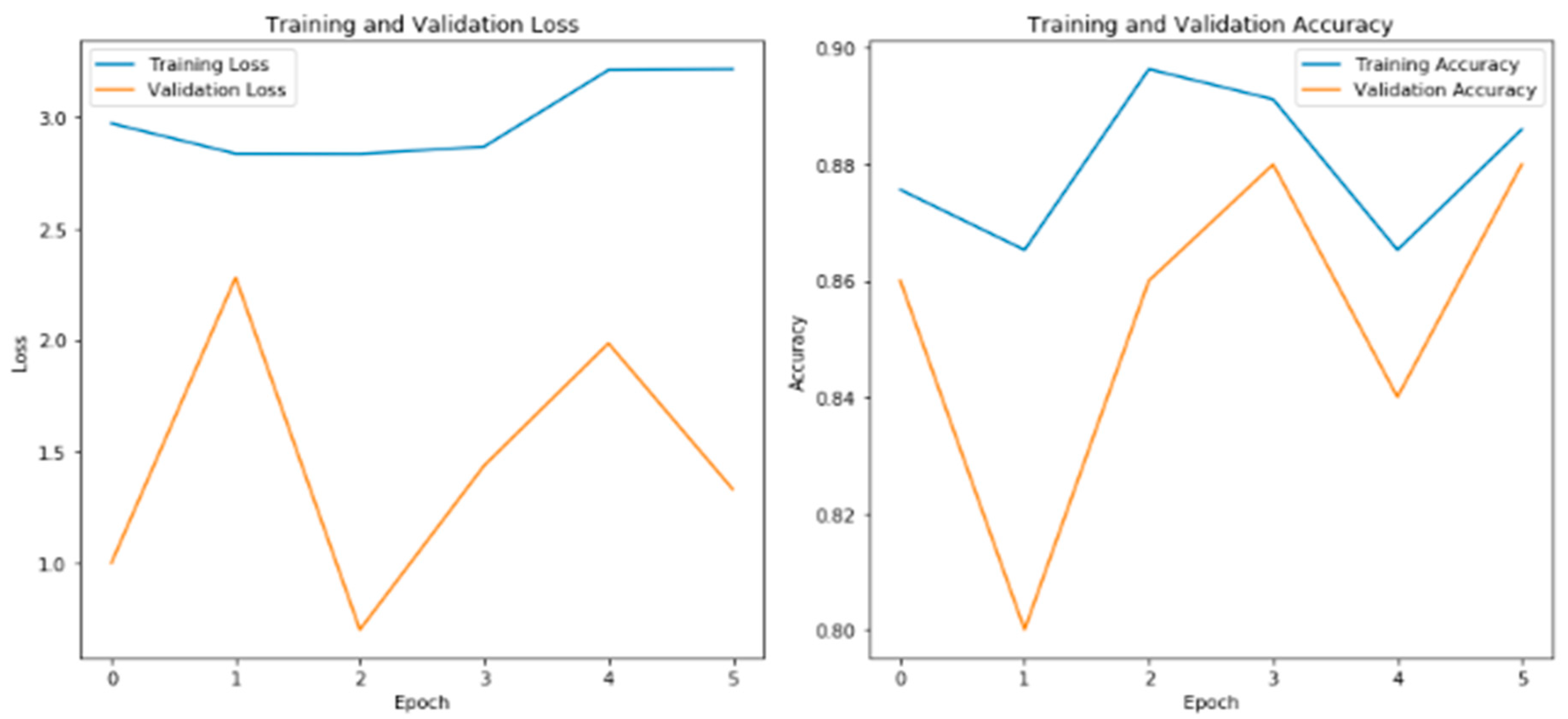
The plot titled "Training and Validation Loss" (
Figure 2) demonstrates the model's performance in terms of minimizing the loss function during the training process. As the epochs progress, the training loss gradually decreases, indicating that the model is effectively learning from the training data. Similarly, the validation loss provides insights into how well the model generalizes to unseen validation data. If the validation loss follows a similar decreasing trend, it suggests that the model is not overfitting and can successfully generalize to new examples (Chollet, 2017).
The plot titled "Training and Validation Accuracy" in
Figure 2 depicts the accuracy of the model during training and validation. As the epochs advance, the training accuracy improves, indicating that the model is becoming more proficient at correctly classifying the brain tumor images in the training set. The validation accuracy measures the model's ability to generalize to new, unseen data. If the validation accuracy increases or remains stable over time, it suggests that the model is capable of accurately classifying brain tumor images beyond the training set (Brownlee, 2021).
These training and validation plots provide a visual representation of the model's performance and can help researchers and practitioners assess the model's convergence, identify potential issues such as overfitting, and make informed decisions regarding model improvements and adjustments. By monitoring the loss and accuracy trends, practitioners can optimize the training process and fine-tune the model's hyperparameters to achieve better performance in brain tumor detection tasks (Saeed & Al-Jumaily, 2021).
The figure displays the plots of training and validation loss on the left side, and training and validation accuracy on the right side. The training loss and accuracy reflect the model's performance on the training data, while the validation loss and accuracy demonstrate how well the model generalizes to unseen validation data. The plots provide a visualization of the model's learning progress and its ability to classify brain tumor images accurately.
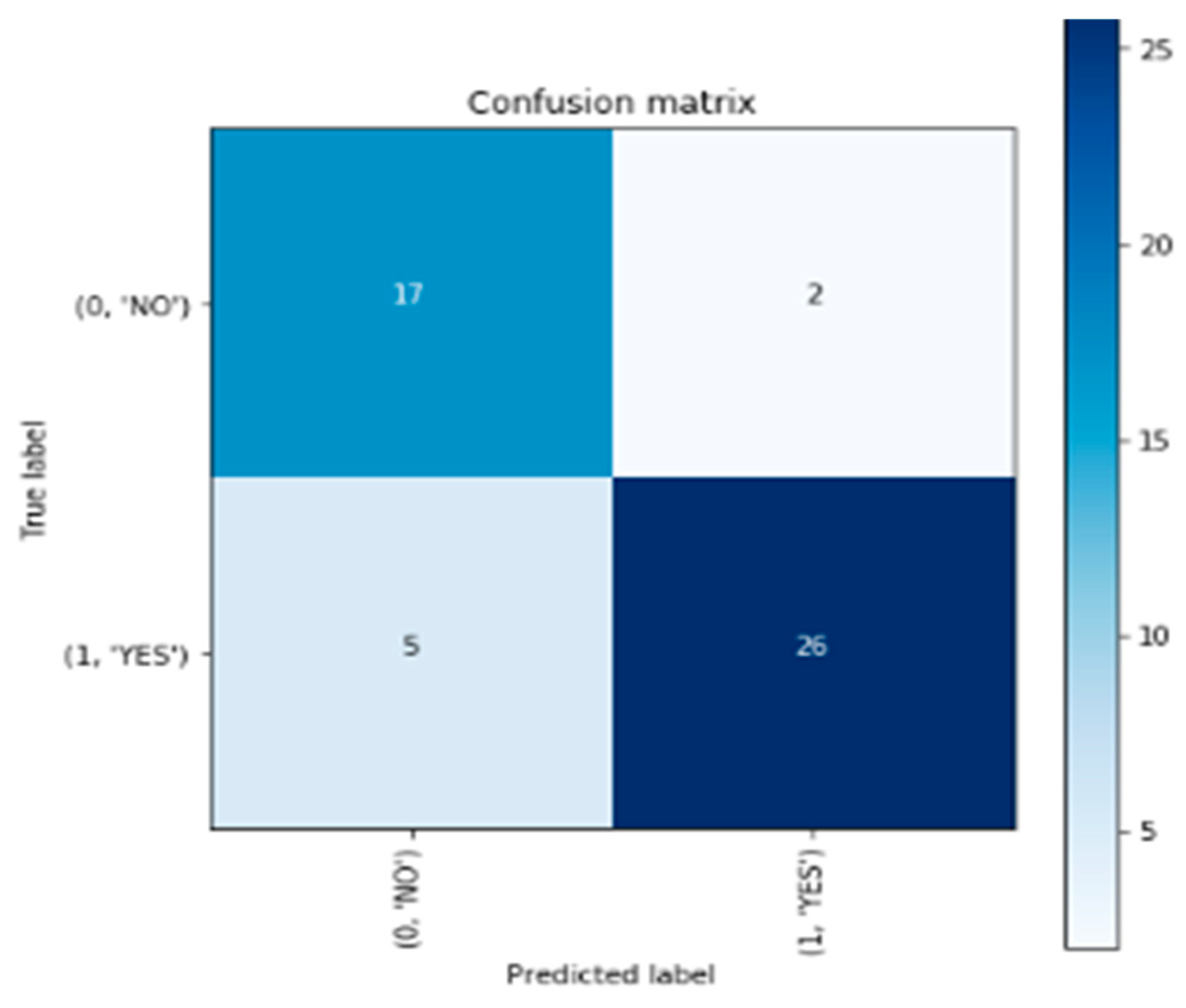
The confusion matrix is a visualization tool used to assess the performance of a classification model. It provides a summary of the model's predictions compared to the actual ground truth labels. In
Figure 3, the confusion matrix presents the classification results in a tabular format, with the predicted labels on the horizontal axis and the true labels on the vertical axis.
In the provided confusion matrix, the upper-left cell represents the number of correctly predicted instances that belong to the "NO" class, resulting in a count of 17. The upper-right cell indicates the number of instances predicted as "YES" when the actual class is "NO," which is 2. Conversely, the lower-left cell displays the count of instances classified as "NO" when the true class is "YES," amounting to 5. Lastly, the lower-right cell shows the number of correctly predicted instances of the "YES" class, totaling 26 (Brownlee, 2021).
The figure presents the confusion matrix, which provides a comprehensive summary of the model's performance in classifying brain tumor images. It showcases the counts of correct and incorrect predictions for each class, highlighting the model's ability to correctly identify instances of brain tumors ("YES") and instances without tumors ("NO"). The matrix aids in evaluating the model's accuracy and identifying any potential biases or misclassifications.
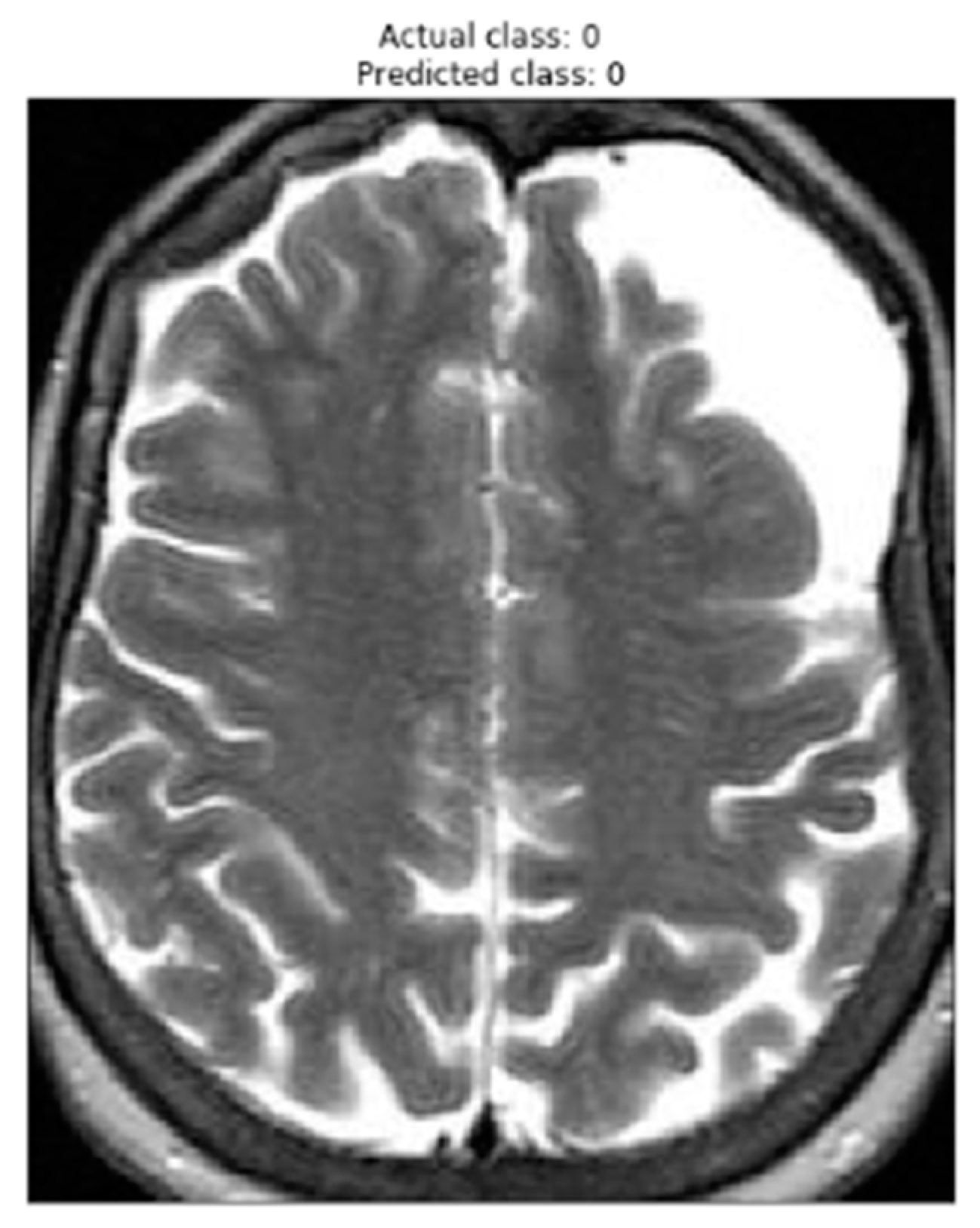
Figure 4 presents an image that has been correctly classified by the brain tumor detection model using the VGG-16 architecture. The displayed image represents a sample from the test set, where the actual class of the image (true label) is shown in the title as "Actual class," and the predicted class by the model is displayed as "Predicted class."
The visual representation of the image is shown in grayscale, where darker areas indicate regions with lower signal intensity, and brighter areas indicate regions with higher intensity. This specific image was correctly classified by the model, meaning that the model correctly identified the absence of a brain tumor in this image.
Displaying this correctly classified image is important to assess the accuracy and reliability of the model. By correctly identifying the images without a brain tumor, the model demonstrates its ability to accurately discriminate between the "YES" (with tumor) and "NO" (without tumor) classes. This is crucial to ensure that the model is reliable enough to be applied in the detection of brain tumors in new images.
The figure displays an image from the test set that has been correctly classified by the brain tumor detection model. The figure's title shows the actual class of the image and the predicted class by the model. The accuracy in classifying this image highlights the model's capability to properly identify images without a brain tumor.
In conclusion, this study explored the application of the VGG-16 model for brain tumor detection using MRI images. By following a systematic methodology, including data preprocessing, model building, and evaluation, promising results were achieved. The VGG-16 model demonstrated high accuracy in classifying brain tumor images, with an accuracy of approximately 88% on the validation set and 80% on the test set. These findings suggest the potential of the VGG-16 model as a valuable tool for supporting healthcare professionals in the early detection and diagnosis of brain tumors.
The use of deep learning models, such as the VGG-16 model, shows great promise in the field of medical image analysis. The ability to accurately classify brain tumor images can aid in timely diagnosis and treatment planning, ultimately improving patient outcomes. However, further research and validation are necessary to assess the model's generalizability across diverse datasets and clinical settings.
It is important to note that the development and deployment of such models require careful consideration of ethical and regulatory aspects, as well as collaboration between researchers, healthcare professionals, and policymakers. Additionally, future studies may explore the combination of different deep learning architectures, advanced preprocessing techniques, and larger datasets to further enhance the performance of brain tumor detection models.
In summary, the results of this study demonstrate the potential of the VGG-16 model in assisting healthcare professionals in accurately detecting brain tumors from MRI images. Continued research and advancements in deep learning algorithms have the potential to significantly improve the early detection, diagnosis, and treatment of brain tumors, leading to better patient outcomes and quality of life.