Submitted:
11 August 2023
Posted:
15 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
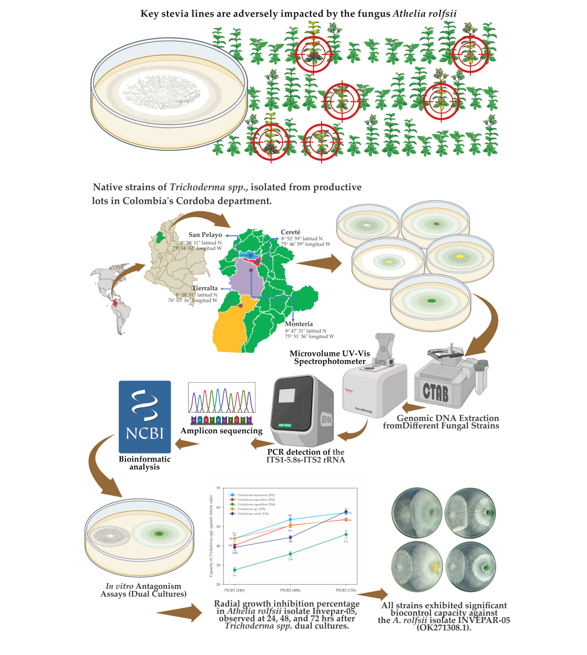
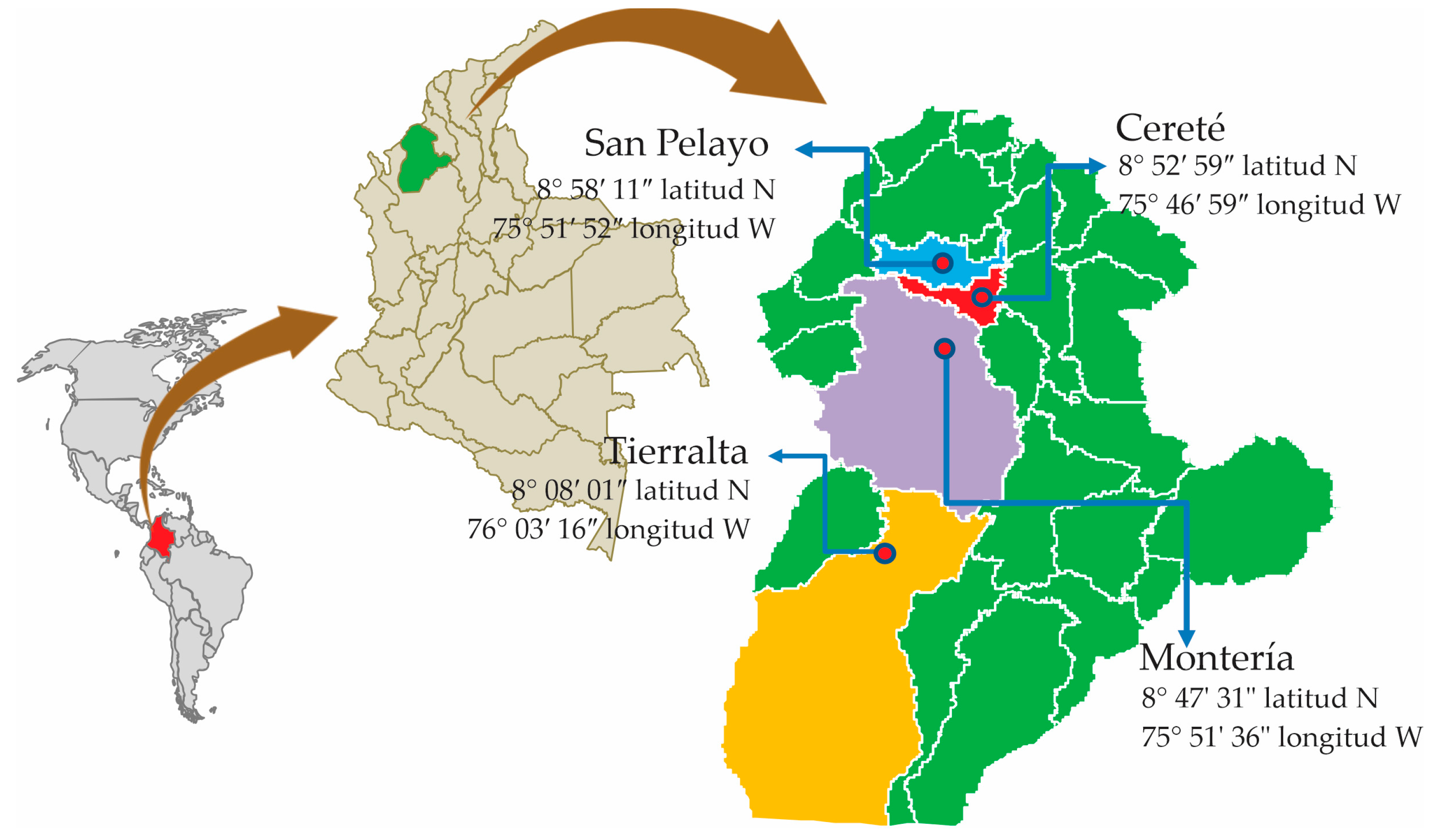
2.1. Location and Field Sampling
2.2. Laboratory Processing
2.3. Molecular identification of Trichoderma spp and A. rolfsii
2.3.1. Genomic DNA Extraction from Different Fungal Strains
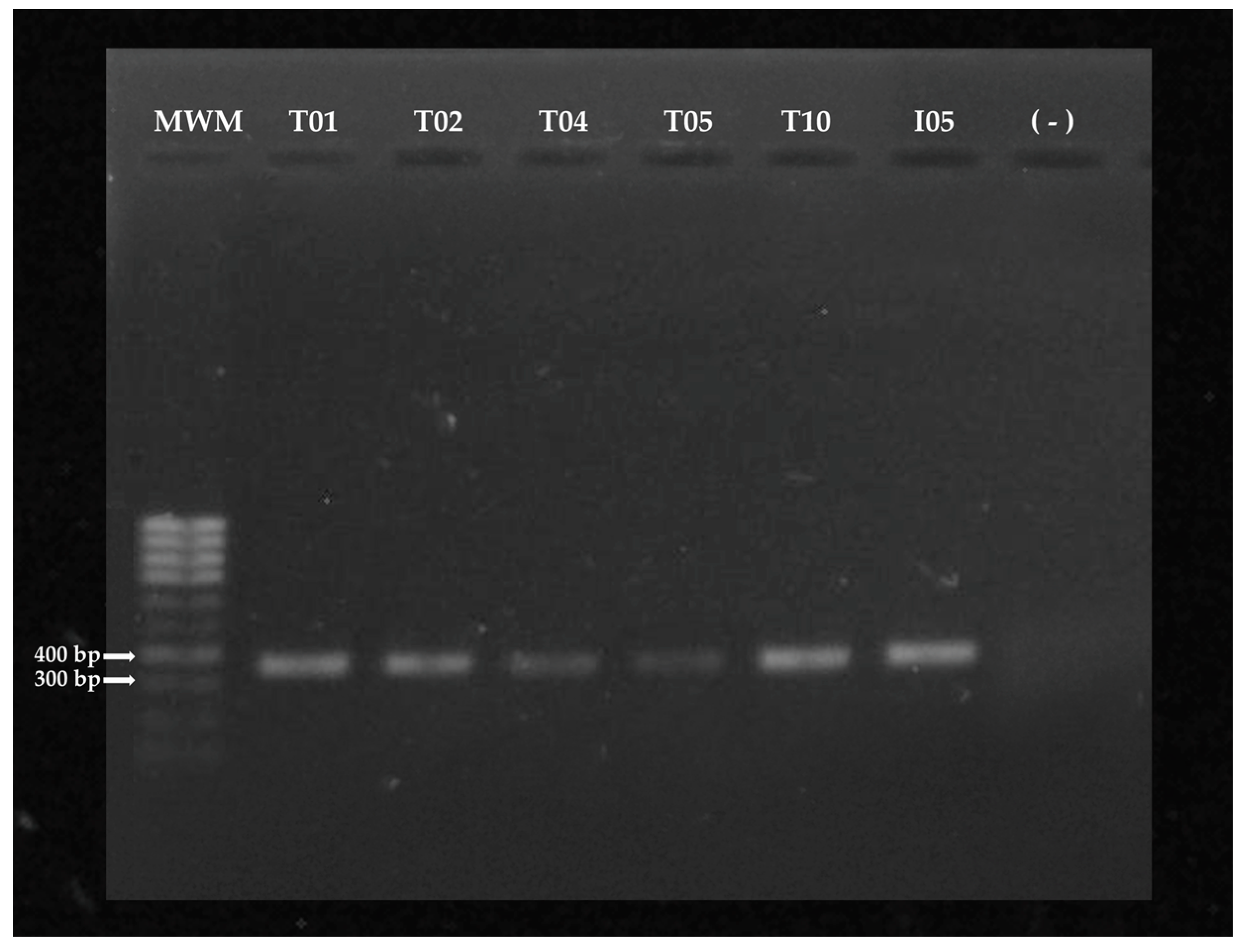
2.3.2. Amplification of the fungal ITS fragment
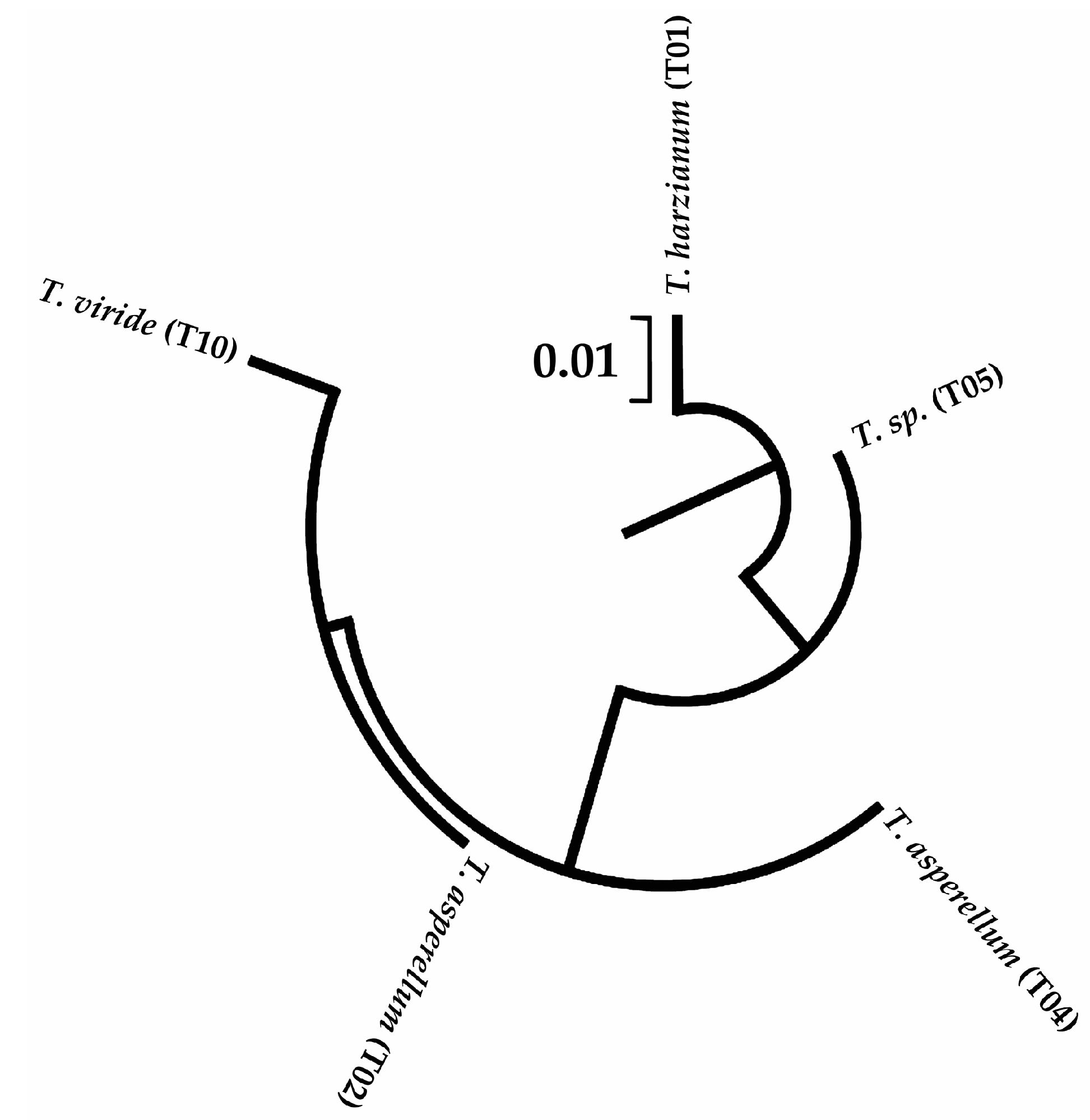
2.3.3. Sequencing of PCR Products and Sequence Analysis
2.4. In vitro Antagonism Assays (Dual Cultures)
2.4.1. Evaluation of the Antagonistic Capacity of Trichoderma spp. vs. A. rolfsii isolate INVEPAR-05
2.5. Statistical Analysis
3. Results
3.1. Molecular identification of Trichoderma spp and A. rolfsii
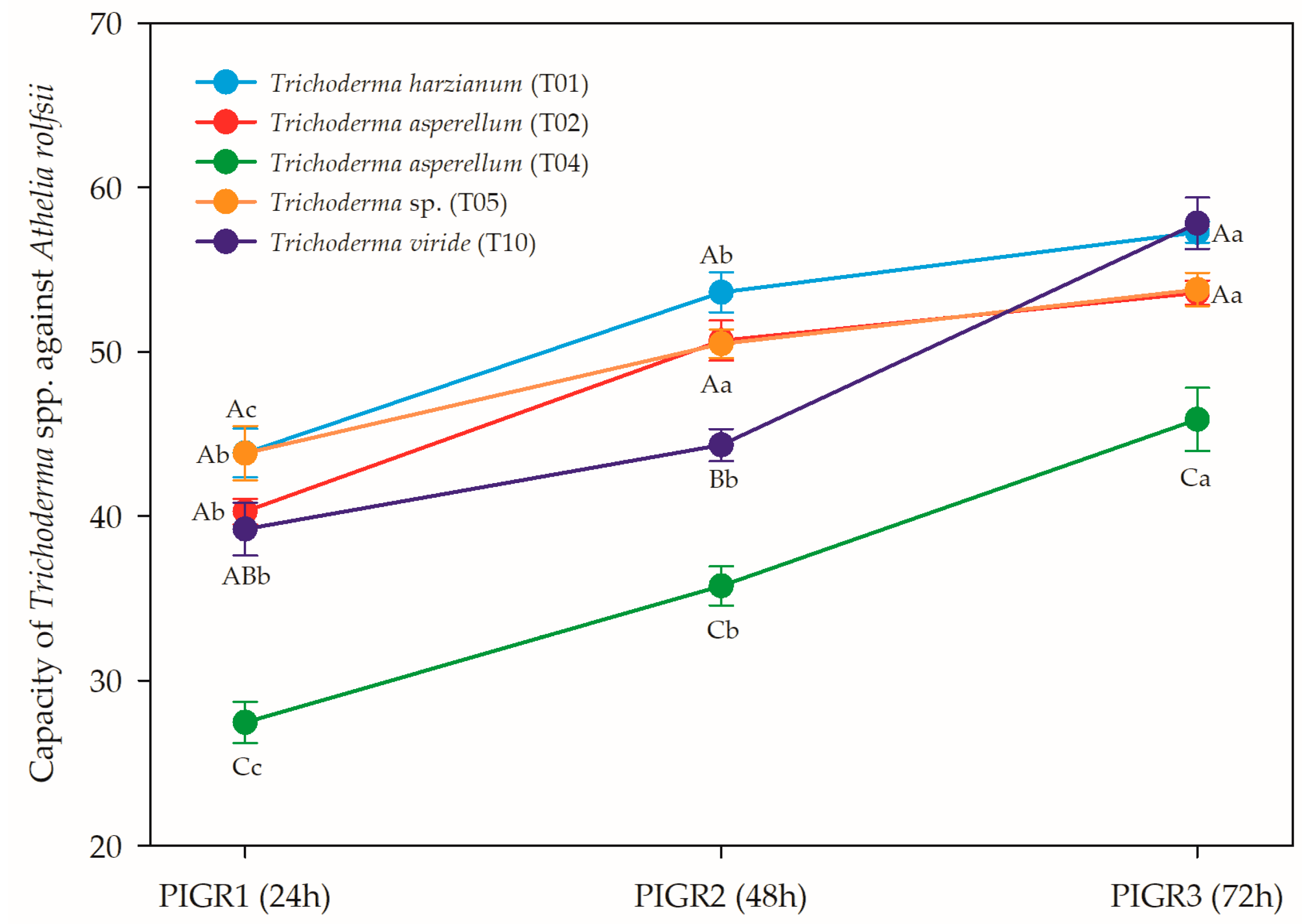
3.2. In vitro Antagonistic Capacity of Trichoderma spp. Strains against Athelia rolfsii
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Yadav, A.K.; Singh, S.; Dhyani, D.; P.S., A. A review on the improvement of stevia [Stevia rebaudiana (Bertoni)]. Can J Plant Sci 2011, 91, 1-27. [CrossRef]
- Soejarto, D.D.; Compadre, C.M.; Kinghorn, D. Ethnobotanical notes in Stevia. Bot Mus Leafl 1983, 29, 1-25.
- Rodriguez-Paez, L.A.; Jimenez-Ramirez, A.M.; Pompelli, M.F.; ineda-Rodriguez, Y.Y.; Jarma-Orozco, A.; Jaraba-Navas, J.D.; Aramendiz-Tatis, H.; Combatt-Caballero, E.; Oloriz-Ortega, M.I.; Rodríguez, N.V. Physiological and enzymatic evaluation of selected genotypes of Stevia rebaudiana Bertoni. Agronomy 2023, 13, 403. [CrossRef]
- Mordue, J.E.M. Descriptions of pathogenic fungi and bacteria. No. 410; CMI: Kew, U.K, 1974.
- Tamayo-Molano, P.J. Tecnologia para el cultivo de estevia. Rionegro, Antioquia: Corporación colombiana de investigación agropecuaria. Agrosavia 2006, Availabe in https://repository.agrosavia.co/handle/20.500.12324/17960. Accessed in Aug 01, 2023.
- Koehler, A.M.; Shew, H.D. Enhanced overwintering survival of Stevia by QoI gungicides used for management of Sclerotium rolfsii. Plant Dis 2017, 101, 1417-1421. [CrossRef]
- Chang, K.F.; Howard, R.J.; Gaudiel, R.G.; Hwang, S.F. First report of Stevia as a host of Sclerotinia sclerotiorum. Plant Dis 1997, 81, 311-311. [CrossRef]
- Hilal, A.A.; Baiuomy, M.A. First record of fungal diseases of stevia (Stevia rebaudiana Bertoni) in Egypt. Egypt J Agric Res 2000, 78, 1435-1448. [CrossRef]
- Kamalakannan, A.; Valluvaparidasan, V.; Chitra, K.; Rajeswari, E.; Salaheddin, K.; Ladhalakshmi, D.; Chandrasekaran, A. First report of root rot of stevia caused by Sclerotium rolfsii in India. Plant Pathol 2007, 56, 350-350. [CrossRef]
- Koehler, A.; Shew, H. First report of stem and root rot of Stevia caused by Sclerotium rolfsii in North Carolina. Plant Dis 2014, 98, 1005. [CrossRef]
- Erper, I.; Ozer, G.; Yildirim, E.; Ozgen, T.; Turkkan, M. First report of southern blight caused by Athelia rolfsii on candyleaf in Turkey. J Plant Pathol 2020, 102, 245-246. [CrossRef]
- Le Bihan, Z.; Gaudin, J.; Robledo-Garcia, F.; Cosson, P.; Hastoy, C.; Rolin, D.; Schurdi-Levraud, V. First report of Sclerotium stem rot caused by Athelia rolfsii on Stevia rebaudiana in southwestern France. Plant Dis 2020, 104, 584. [CrossRef]
- Vélez-Olmedo, J.B.; Vélez-Zambrano, S.; Bonfim, B.A.; Cuenca, E.C.; García, S.; Cedeño, A.G.; Pinho, D.B. First report of Athelia rolfsii (Curzi) causing stem and root rot on stevia (Stevia rebaudiana Bertoni) in Ecuador. J Plant Pathol 2021, 103, 743. [CrossRef]
- Carrieri, R.; Cozzolino, E.; Tarantino, P.; Cerrato, D.; Lahoz, E. First report of southern blight on candyleaf (Stevia rebaudiana) caused by Sclerotium rolfsii in Italy. Plant Dis 2016, 100, 220. [CrossRef]
- Shwetha, G.S.; Hegde, Y.R. Management of Sclerotium wilt of Stevia rebaudiana through biorationals. Int J Plant Prot 2012, 5, 248-251.
- Sanabria-Velazquez, A.D.; Enciso-Maldonado, G.A.; Maidana-Ojeda, M.; Diaz-Najera, J.F.; Ayvar-Serna, S.; Thiessen, L.D.; Shew, H.D. Integrated pathogen management in Stevia using anaerobic soil disinfestation combined with different fungicide programs in USA, Mexico, and Paraguay. Agronomy 2023, 13, 1358. [CrossRef]
- Schludecker, M. Tiempo Montería. Metroblue 2023, Available on https://www.meteoblue.com/es/tiempo/semana/monter%C3%ADa_colombia_3674453. Acessed on Aug 01 2023.
- Martyniuk, S.; Martyniuk, M. Occurrence of Azotobacter spp. in some Polish soils. Pol J Environ Stud 2003, 12, 371-374.
- Tejera, N.; Lluch, C.; Martìnez-Toledo, M.V.; Gonzàlez-López, J. Isolation and characterization of Azotobacter and Azospirillum strains from the sugarcane rhizosphere. Plant Soil 2005, 270, 223-232. [CrossRef]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium species: an illustrated manual for identification; Pennsylvania State University Press: Harrisburg, Pennsylvania, USA, 1983.
- Pineda-Rodriguez, Y.Y.; Pompelli, M.F.; Jarma-Orozco, A.; Rodríguez, N.V.; Rodriguez-Paez, L.A. A new and profitable protocol to DNA extraction in Limnospira maxima. Methods and Protocols 2023, 6, 62. [CrossRef]
- Wilfinger, W.W.; Mackey, K.; Chomczynski, P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. BioTechniques 1997, 22, 474-481. [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; J.W., T. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, Innis, M.A., Gelfand, D.H., Sninsky, J.J., T.J., W., Eds.; Academic Press, Inc.: New York, 1990.
- Bell, D.K. In vitro antagonism of Trichoderma species against six fungal plant pathogens. Phytopathology 1982, 72, 379. [CrossRef]
- Skidmore, A.M.; Dickinson, C.H. Colony interactions and hyphal interference between Septoria nodorum and Phylloplane Fungi. Trans Br Mycol Soc 1976, 66, 57-64. [CrossRef]
- Rodriguez-Paez, L.A.; Pineda-Rodriguez, Y.Y.; Jaraba-Navas, J.D.D.; Jarma-Orozco, A.; Combatt-Caballero, E.M.; Jimenez-Ramirez, A.M.; Oloriz-Ortega, M.I.; Veitia-Rodriguez, N. Athelia rolfsii isolate UCLV-IBP 5.8S ribosomal RNA gene, partial sequence; internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, Partial Sequence. Available on https://www.ncbi.nlm.nih.gov/nuccore/OM345235. Accessed on August 03 2023. NCBI 2022.
- Schuster, A.; Schmoll, M. Biology and biotechnology of Trichoderma. Appl Microbiol Biotechnol 2010, 87, 787-799. [CrossRef]
- Hewedy, O.A.; Abdel Lateif, K.S.; Seleiman, M.F.; Shami, A.; Albarakaty, F.M.M.; El-Meihy, R. Phylogenetic diversity of Trichoderma strains and their antagonistic potential against soil-borne pathogens under stress conditions. Biology 2020, 9, 189. [CrossRef]
- Benttoumi, N.; Colagiero, M.; Sellami, S.; Boureghda, H.; Keddad, A.; Ciancio, A. Diversity of nematode microbial antagonists from Algeria shows occurrence of nematotoxic Trichoderma spp. Plants 2020, 9, 941. [CrossRef]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The current status of Its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int J Mol Sci 2022, 23 2329. [CrossRef]
- Shrestha, U.; Dee, M.E.; Piya, S.; Ownley, B.H.; Butler, D.M. Soil inoculation with Trichoderma asperellum, T. harzianum or Streptomyces griseoviridis prior to anaerobic soil disinfestation (ASD) does not increase ASD efficacy against Sclerotium rolfsii germination. Appl Soil Ecol 2020, 147, 103383,. [CrossRef]
- Raja, M.; Sharma, R.K.; Jambhulkar, P.P.; Pandian, R.T.P.; Sharma, P. Comparative evaluation of native Trichoderma species from groundnut rhizosphere against stem rot caused by Sclerotium rolfsii Sacc. . Indian Phytopathol 2023, 76, 459-471. [CrossRef]
- Ayyandurai, M.; Akila, R.; Manonmani, K.; Mini, M.L.; Vellaikumar, S.; Brindhadevi, S.; Theradimani, M. Combined application of Trichoderma longibrachiatum T(SP)-20 and Trichoderma asperellum T(AR)-10 in the management of stem rot of groundnut. Legume Res 2022, 46, 215-221. [CrossRef]
- Guzmán-Guzmán, P.; Kumar, A.; de los Santos-Villalobos, S.; Parra-Cota, F.I.; Orozco-Mosqueda, M.d.C.; Fadiji, A.E.; Hyder, S.; Babalola, O.O.; Santoyo, G. Trichoderma species: our best fungal allies in the biocontrol of plant diseases - a review. Plants 2023, 12, 432. [CrossRef]
- Khan, R.A.A.; Najeeb, S.; Mao, Z.; Ling, J.; Yang, Y.; Li, Y.; Xie, B. Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic bacteria and root-knot nematode. Microorganisms 2020, 8, 401. [CrossRef]
- Bulgari, D.; Fiorini, L.; Gianoncelli, A.; Bertuzzi, M.; E., G. Enlightening gliotoxin biological system in agriculturally relevant Trichoderma spp. Front Microbiol 2020, 11, 200. [CrossRef]
- Vinale, F.; Sivasithamparam, K. Beneficial effects of Trichoderma secondary metabolites on crops. Phytother Res 2020, 34, 2835-2842. [CrossRef]
- Kai, K.; Mine, K.; Akiyama, K.; Ohki, S.; Hayashi, H. Anti-plant viral activity of peptaibols, trichorzins HA II, HA V, and HA VI, isolated from Trichoderma harzianum HK-61. J Pest Sci 2018, 43, 283-286. [CrossRef]
- Deng, J.J.; Huang, W.Q.; Li, Z.W.; Lu, D.L.; Zhang, Y.; Luo, X.C. Biocontrol activity of recombinant aspartic protease from Trichoderma harzianum against pathogenic fungi. Enzyme Microb Technol 2018, 112, 35-42. [CrossRef]
- Peng, K.-C.; Lin, C.-H.; Liao, C.-F.; Yu, H.-C.; Lo, C.-T.; Yang, H.-H.; Lin, K.-C. Expression of L-amino acid oxidase of Trichoderma harzianum in tobacco confers resistance to Sclerotinia sclerotiorum and Botrytis cinerea. Plant Sci 2021, 303, 110772. [CrossRef]
- Prajapati, B.K.; Patel, J.K.; Patil, R.K. Bioeficacy of Trichoderma spp. against Sclerotium rolfsii Sacc., an incitant of collar rot of chick pea in vitro. Bioscan 2015, 10, 1745-1748.
- Sab, J.; Nagaraja, A.; Nagamma, G. Efficacy of bio-pesticides against Sclerotium rolfsii Sacc. causing collar rot of chickpea (Cicer arietinum L.). Availabe on https://api.semanticscholar.org/CorpusID:86373077. Accessed August 03, 2023 2014.
- Xiao, Z.; Zhao, Q.; Li, W.; Gao, L.; Liu, G. Strain improvement of Trichoderma harzianum for enhanced biocontrol capacity: Strategies and prospects. Front Microbiol 2023, 14, 1146210. [CrossRef]
- Pasquoto-Stigliani, T.; Guilger-Casagrande, M.; Campos, E.V.R.; Germano-Costa, T.; Bilesky-José, N.; Migliorini, B.B.; Feitosa, L.O.; Sousa, B.T.; de Oliveira, H.C.; Fraceto, L.F.; et al. Titanium biogenic nanoparticles to help the growth of Trichoderma harzianum to be used in biological control. J Nanobiotechnology 2023, 21, 166. [CrossRef]
- Hossain, M.A.; Swarna, F.T.; Arabi, R.A.; Hamim, I. Trichoderma asperellum suppresses viral diseases and promotes the growth and yield of country bean. Front Agron 2023, 5, 1150359. [CrossRef]
- Zhang, Y.; Xiao, J.; Yang, K.; Wang, Y.; Tian, Y.; Liang, Z. Transcriptomic and metabonomic insights into the biocontrol mechanism of Trichoderma asperellum M45a against watermelon Fusarium wilt. PLoS ONE 2022, 17, e0272702. [CrossRef]
- Schirmböck, M.; Lorito, M.; Wang, Y.L.; Hayes, C.K.; Arisan-Atac, I.; Scala, F.; Harman, G.E.; Kubicek, C.P. Parallel formation and synergism of hydrolytic enzymes and peptaibol antibiotics, molecular mechanisms involved in the antagonistic action of Trichoderma harzianum against phytopathogenic fungi. Appl Environ Microbiol 1994, 60, 4364-4370. [CrossRef]
- Matas-Baca, M.A.; García, C.U.; Pérez-Álvarez, S.; Flores-Córdova, M.A.; Escobedo-Bonilla, C.M.; Magallanes-Tapia, M.A.; Chávez, E.S. Morphological and molecular characterization of a new autochthonous Trichoderma sp. isolate and its biocontrol efficacy against Alternaria sp. Saudi J Biol Sci 2022, 29, 2620-2625. [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma–plant–pathogen interactions. Soil Biol Biochem 2008, 40, 1-10. [CrossRef]
- Cao, Z.-J.; Qin, W.-T.; Zhao, J.; Liu, Y.; Wang, S.-X.; Zheng, S.-Y. Three new Trichoderma species in Harzianum clade associated with the contaminated substrates of edible fungi. J Fungi 2022, 8, 1154. [CrossRef]
- Tamizi, A.A.; Mat-Amin, N.; Weaver, J.A.; Olumakaiye, R.T.; Akbar, M.A.; Jin, S.; Bunawan, H.; Alberti, F. Genome sequencing and analysis of Trichoderma (Hypocreaceae) isolates exhibiting antagonistic activity against the papaya dieback pathogen, Erwinia mallotivora. J Fungi 2022, 8, 246. [CrossRef]
- Šašić Zorić, L.; Janjušević, L.; Djisalov, M.; Knežić, T.; Vunduk, J.; Milenković, I.; Gadjanski, I. Molecular approaches for detection of Trichoderma green mold disease in edible mushroom production. Biology 2023, 12, 299. [CrossRef]
- Sarria, G.; Garcia, A.; Mestizo, Y.; Medina, C.; Varón, F.; Mesa, E.; Hernandez, S. Antagonistic interactions between Trichoderma Spp. and Phytophthora palmivora (Butler) from oil palm in Colombia. Eur J Plant Pathol 2021, 161, 751-768. [CrossRef]
- Guerra-Mateo, D.; Gené, J.; Baulin, V.; Cano-Lira, J.F. Phylogeny and taxonomy of the genus Amphichorda (Bionectriaceae): an update on beauveria-like strains and description of a novel species from marine sediments. Diversity 2023, 15, 795. [CrossRef]
- Cai, F.; Druzhinina, I.S. In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Divers 2021, 107, 1-69. [CrossRef]
- Zhang, S.; Xiang, D.; Sun, C.; Han, K.; Li, T.; Zhou, J.; Xu, B. Morphological and molecular identification of peach brown rot disease in Tibet and exploration of the biocontrol efficiency of Trichoderma. J Fungi 2022, 8, 1174. [CrossRef]
- Noman, E.A.; Al-Gheethi, A.A.; Talip, B.A.; Mohamed, R.M.S.R.; Almoheer, R.; Al-Wrafy, F.A.; Al-Shorgani, N.; El Enshasy, H.A. New fungal strains from peat soil in Malaysia: morphological and molecular characteristics. Sustainability 2023, 15, 5902. [CrossRef]
- Zehra, A.; Dubey, M.K.; Meena, M.; Upadhyay, R. Effect of different environmental conditions on growth and sporulation of some Trichoderma species. J Environ Biol 2017, 38, 197-203. [CrossRef]
- Nieto-Jacobo, M.F.; Steyaert, J.M.; Salazar-Badillo, F.B.; Nguyen, D.V.; Rostás, M.; Braithwaite, M.; De Souza, J.T.; Jimenez-Bremont, J.F.; Ohkura, M.; Stewart, A.; et al. Environmental growth conditions of Trichoderma spp. affects indole acetic acid derivatives, volatile organic compounds, and plant growth promotion. Front Plant Sci 2017, 8, 102. [CrossRef]
- Rimkus, A.; Namina, A.; Dzierkale, M.T.; Grigs, O.; Senkovs, M.; Larsson, S. Impact of growth conditions on the viability of Trichoderma asperellum during storage. Microorganisms 2023, 11, 1084. [CrossRef]
| Solution | Final Concentration | 10 mL |
|---|---|---|
| dH2O (Merck KGaA, part number: 38796) | 5.6 mL | |
| 1 M Tris-HCl- 7.5 (Merck KGaA, part number: T3253) | 100 mM | 1.0 mL |
| 5 M NaCl (Merck KGaA, part number: 1064041000) | 1400 mM | 2.8 mL |
| 0.5 M EDTA-8.0 (Merck KGaA, part number: 324506) | 20 mM | 0.4 mL |
| CTAB (Merck KGaA, part number: H6269-500G) | 2% (w/v) | 0.2 g |
| PVP 10% (w/v) (Merck KGaA, part number: T3253-500G) | 1% (v/v) | 0.1 mL |
| Degree | antagonistic capacity |
|---|---|
| 0 | No invasion of the antagonist on the colony of A. rolfsii |
| 1 | Invasion of 25% of the colony of the fungus A. rolfsii |
| 2 | Invasion of 50% of the fungal colony A. rolfsii |
| 3 | Invasion of 100% of the colony of the fungus A. rolfsii |
| 4 | Invasion of 100% of the colony of the fungus A. rolfsii, sporulating on it. |
| Strain | URL GenBank |
|---|---|
| Trichoderma asperellum (T02) | https://www.ncbi.nlm.nih.gov/nuccore/ON237703 |
| Trichoderma harzianum (T01) | https://www.ncbi.nlm.nih.gov/nuccore/OK310695 |
| Trichoderma sp. (T05) | https://www.ncbi.nlm.nih.gov/nuccore/ON237737 |
| Trichoderma viride (T10) | https://www.ncbi.nlm.nih.gov/nuccore/ON237923 |
| Trichoderma asperellum (T04) | https://www.ncbi.nlm.nih.gov/nuccore/ON238106 |
| Athelia rolfsii isolate INVEPAR-05 | https://www.ncbi.nlm.nih.gov/nuccore/OK271308 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





