Submitted:
15 August 2023
Posted:
17 August 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Chemicals
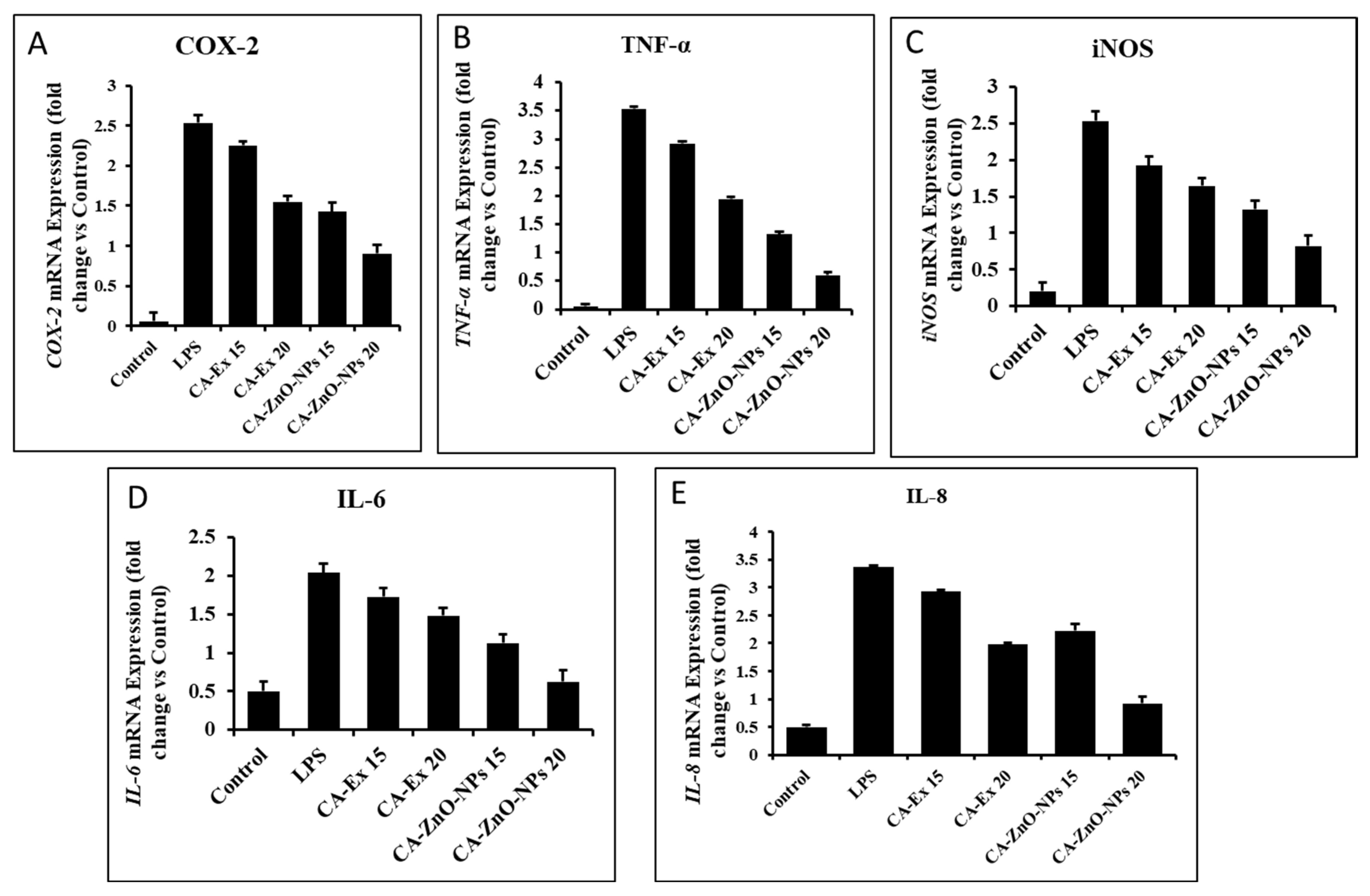
2.2. Preparation of Cissus Antractica Water Extract
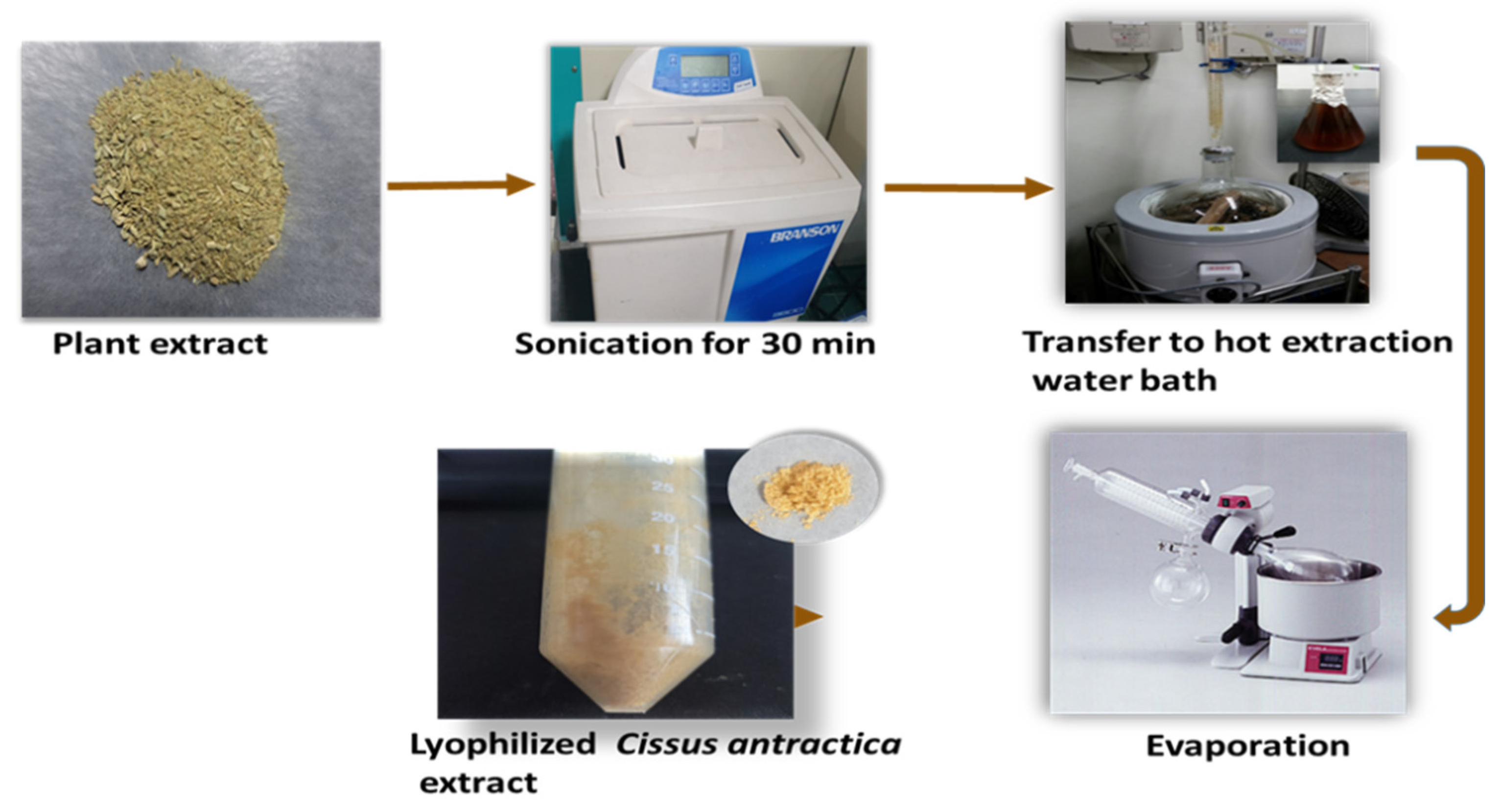
2.3. ZnO NPs Using Co-Precipitation Method
2.4. Cell Culture
2.5. Cytotoxicity Assay
2.6. Reactive Oxygen Species (ROS) Assay
2.7. Wound-Healing Assay
2.8. Hoechst Staining
2.9. PI Staining
2.10. Quantitative Reverse Transcription (qRT-PCR)
3. Results and Discussion
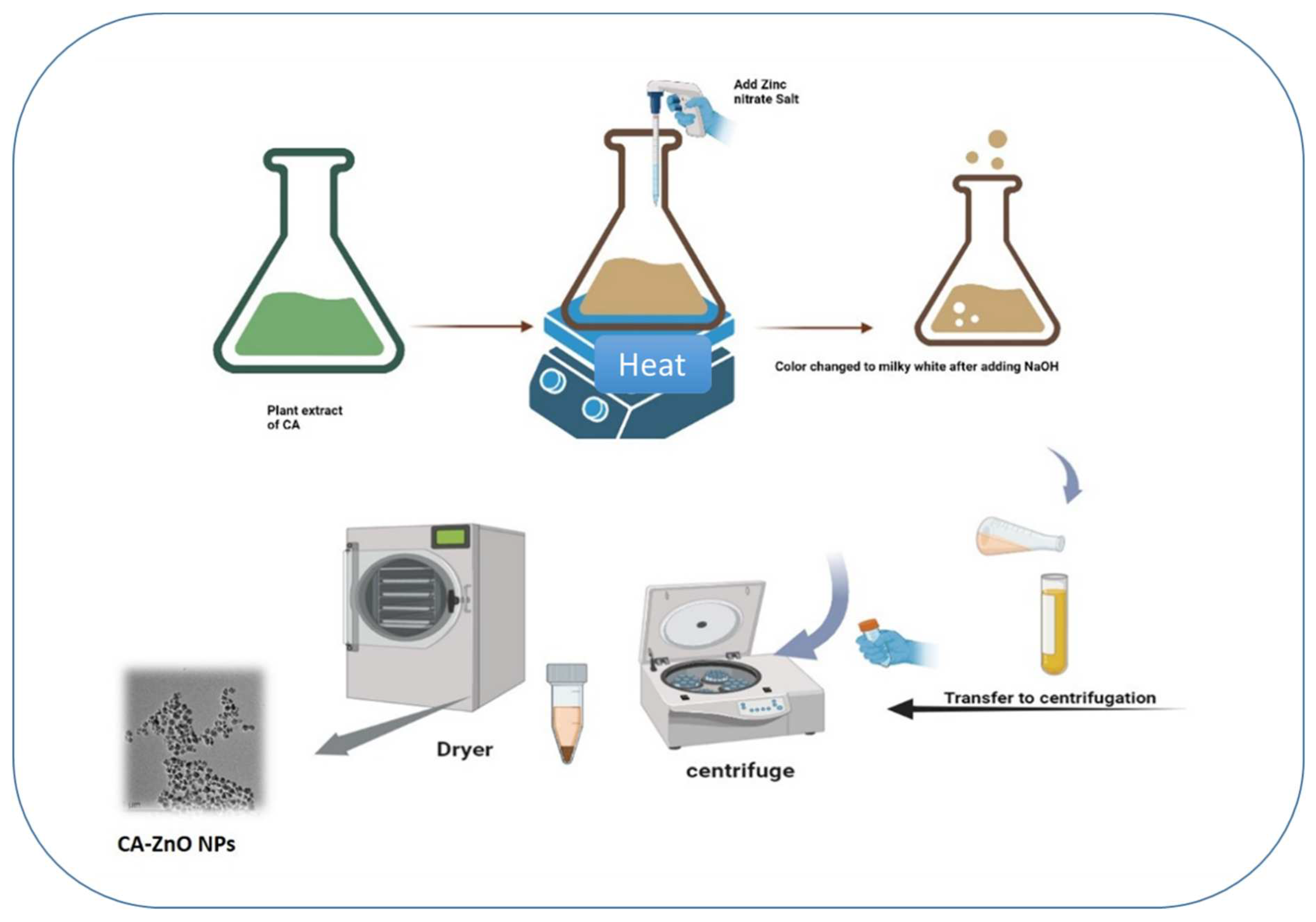
3.1. Synthesis of CA-ZnNps
3.2. Characterization of CA-ZnONps
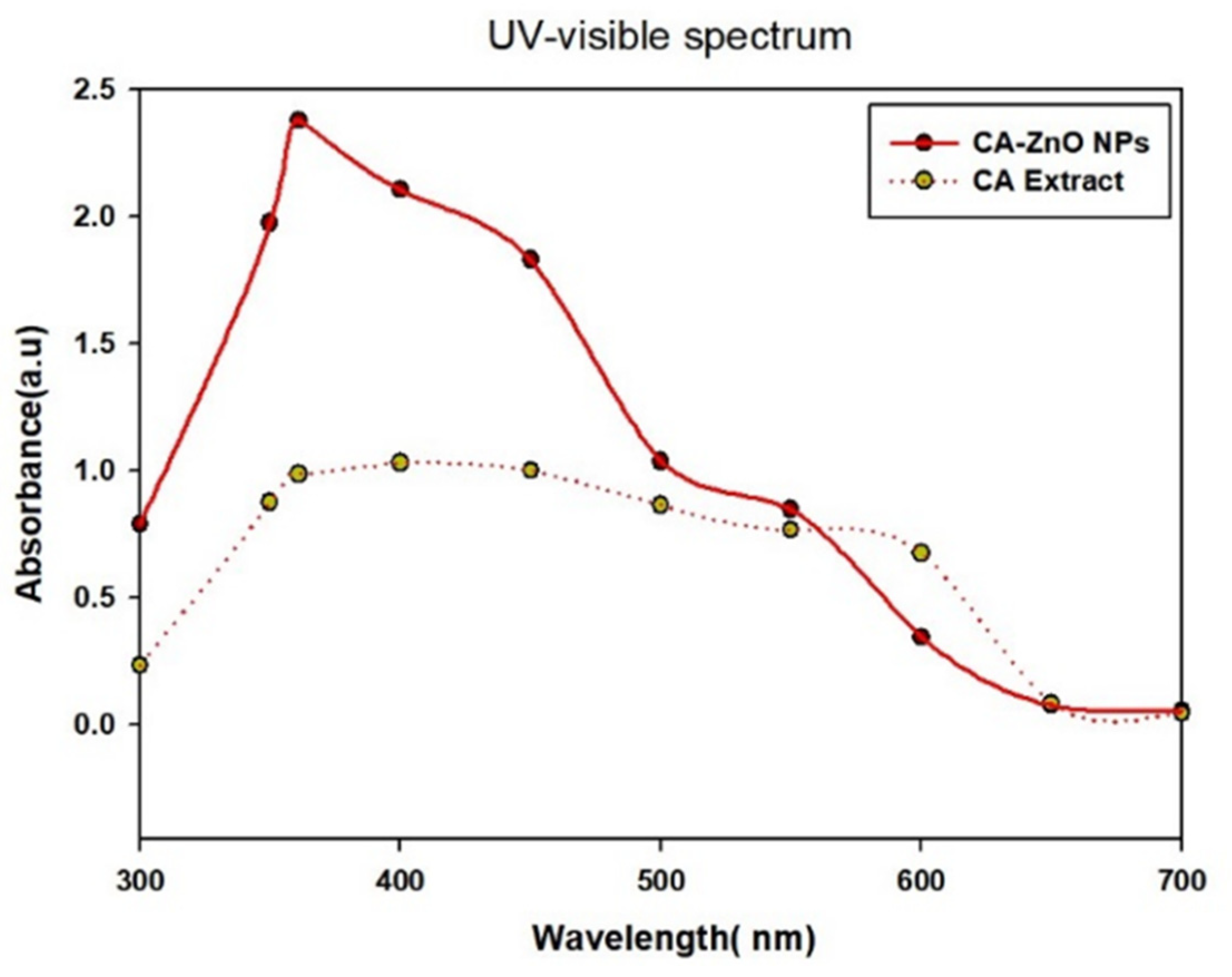
3.3. UV-Vis Spectral Analysis
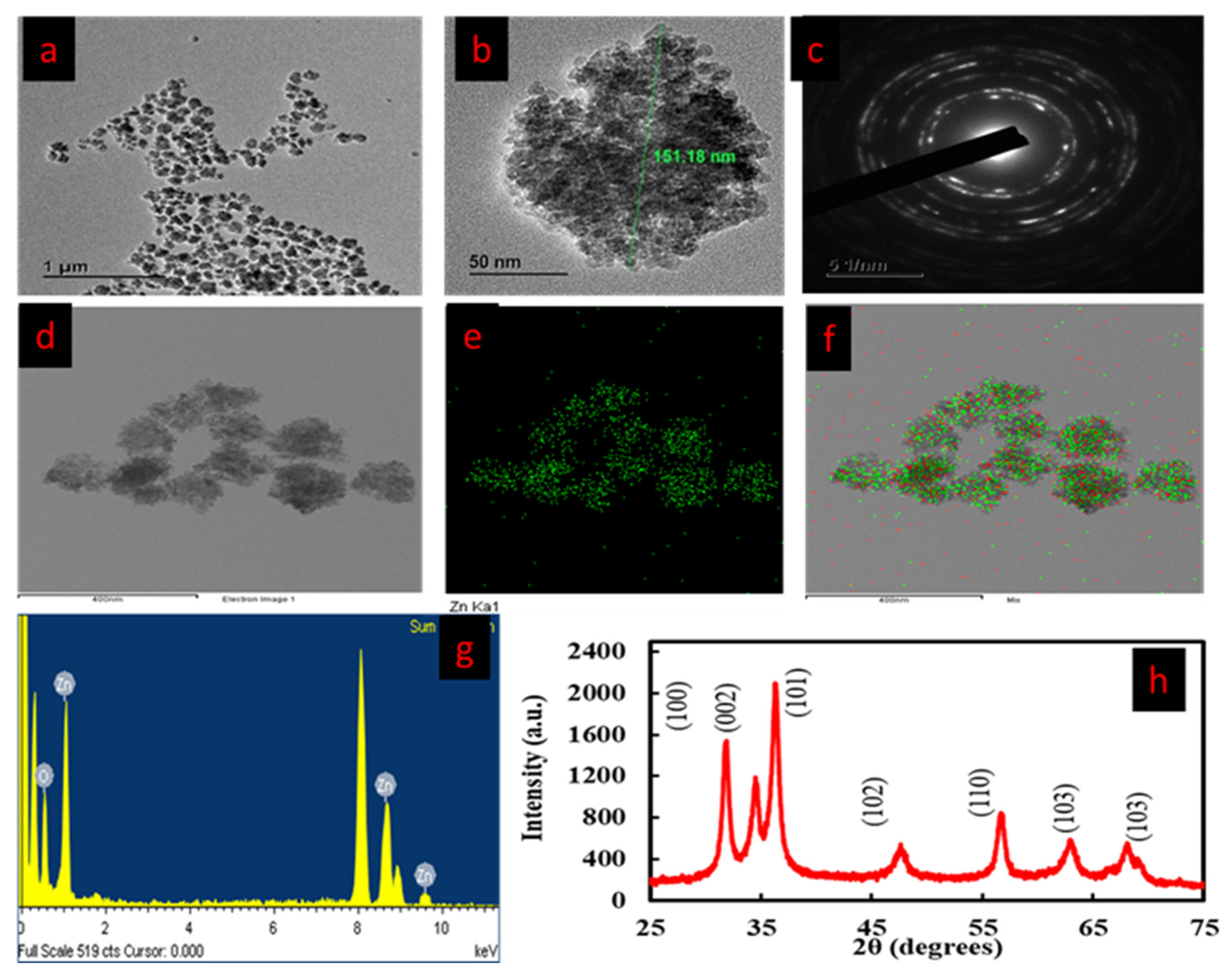
3.4. FE-TEM and EDX Analysis
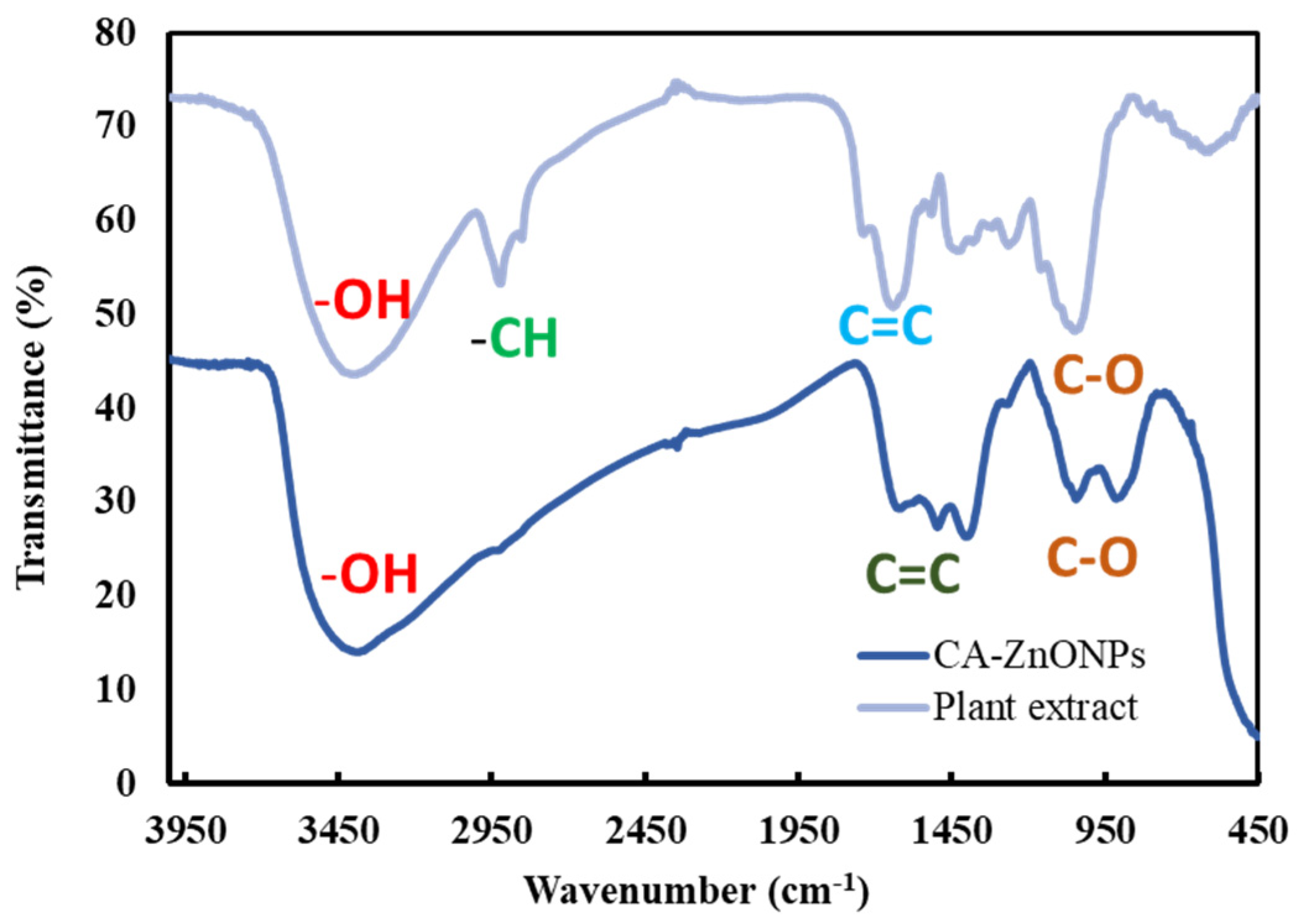
3.5. Fourier Transform-Infrared (FT-IR) Spectroscopy Analysis
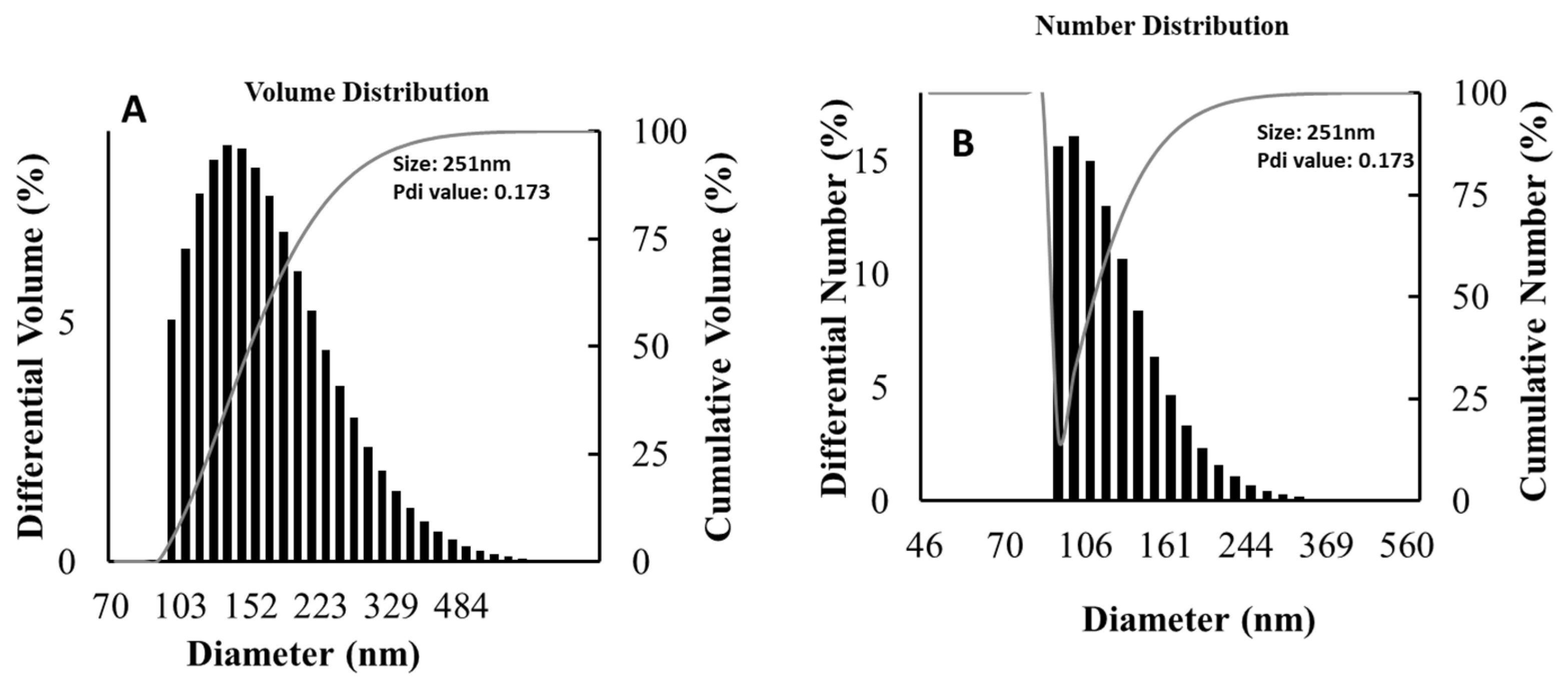
3.6. Particle Size Distribution Analysis
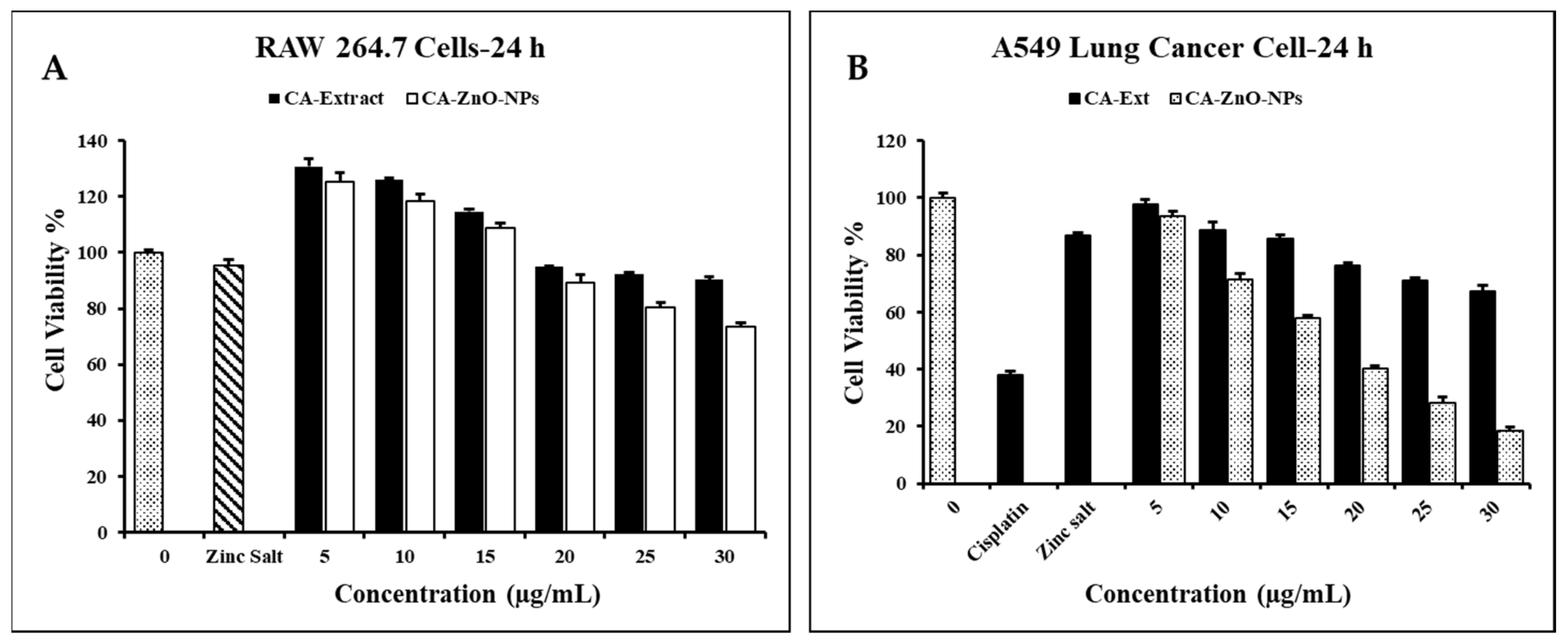
3.7. Evaluation of Cell Cytotoxicity
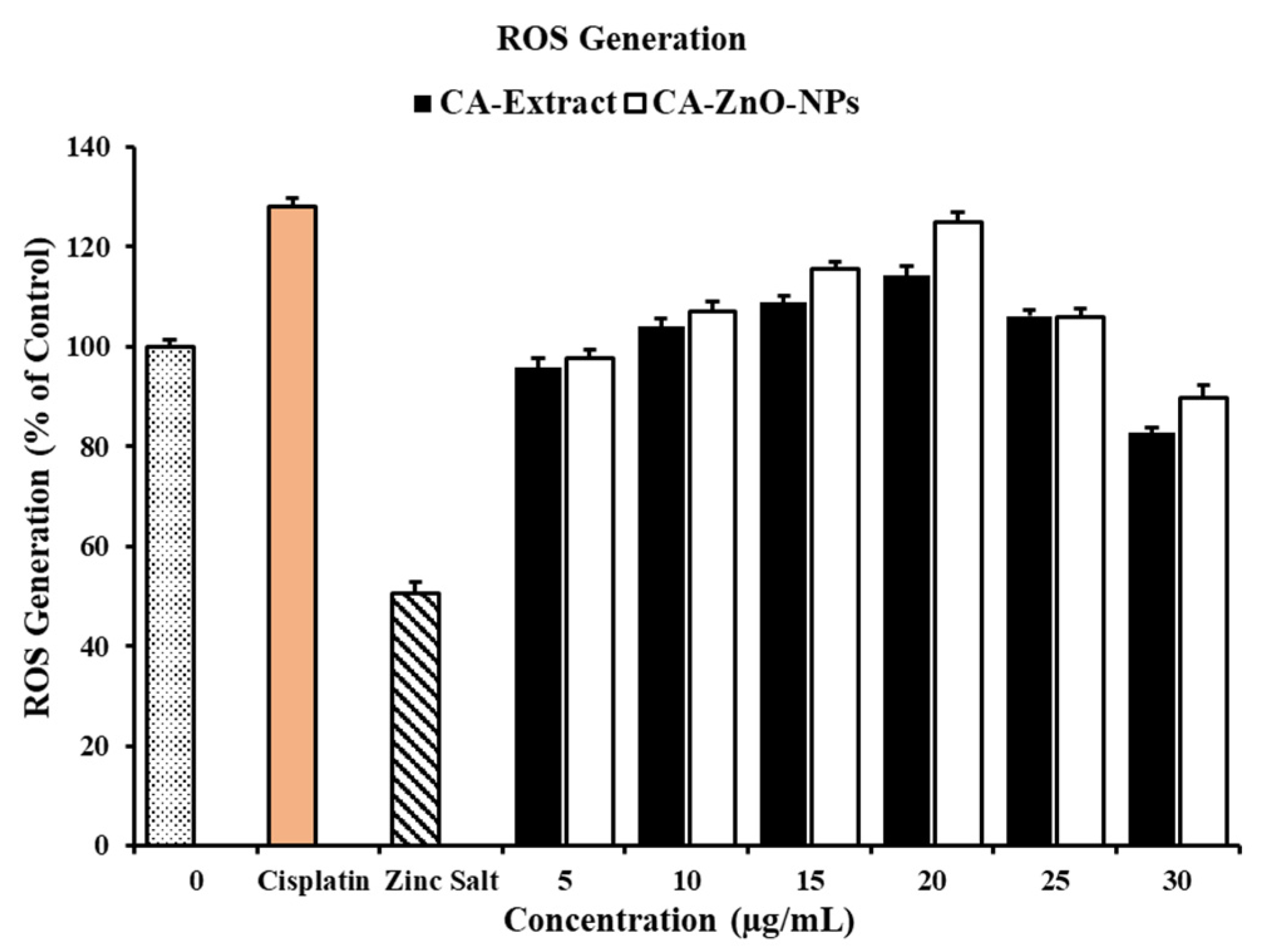
3.8. In Vitro ROS Induced by on CA-ZnO-NPs Cancer Cells
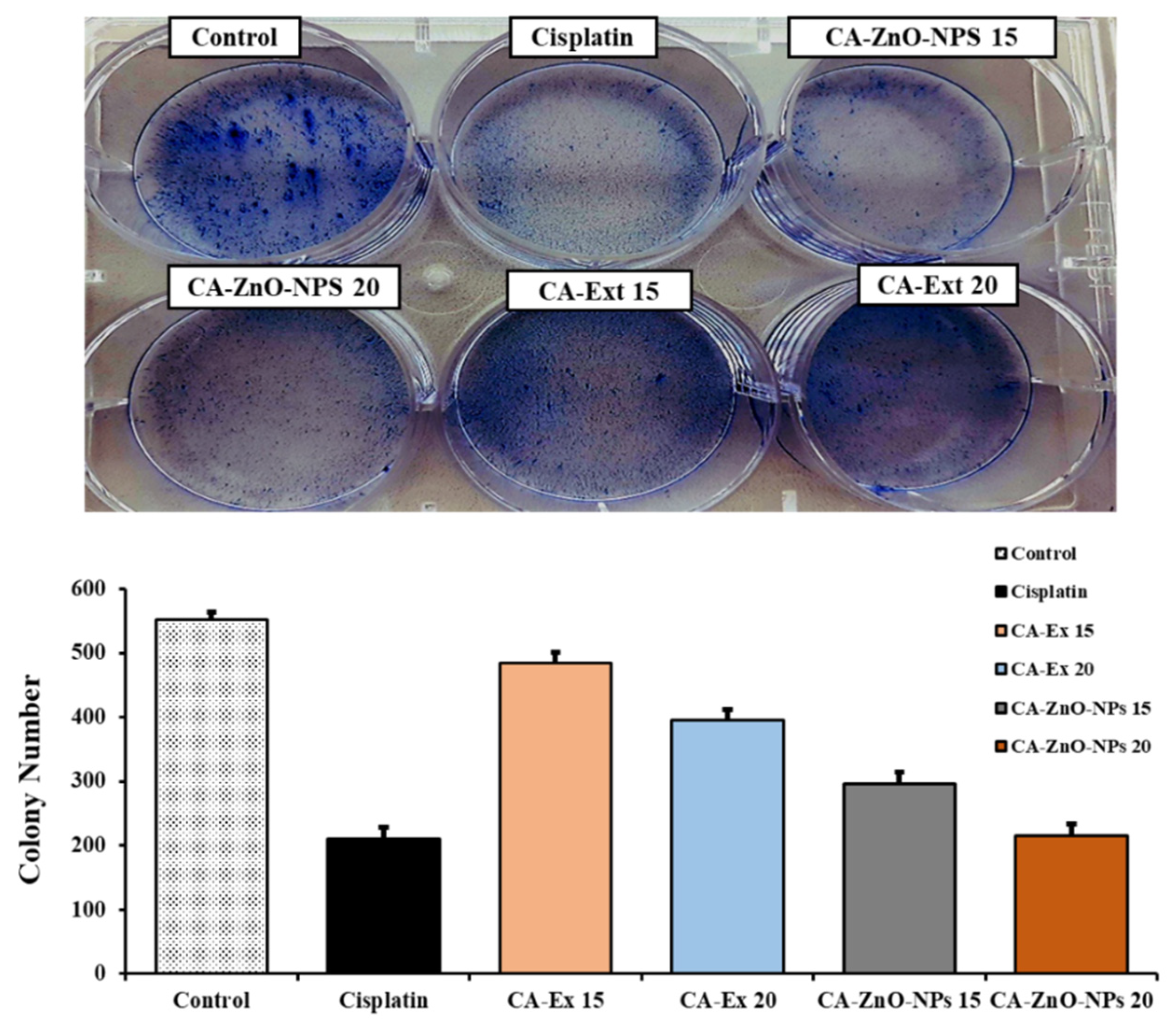
3.9. Inhibition of Colony Formation on Cancer Cells
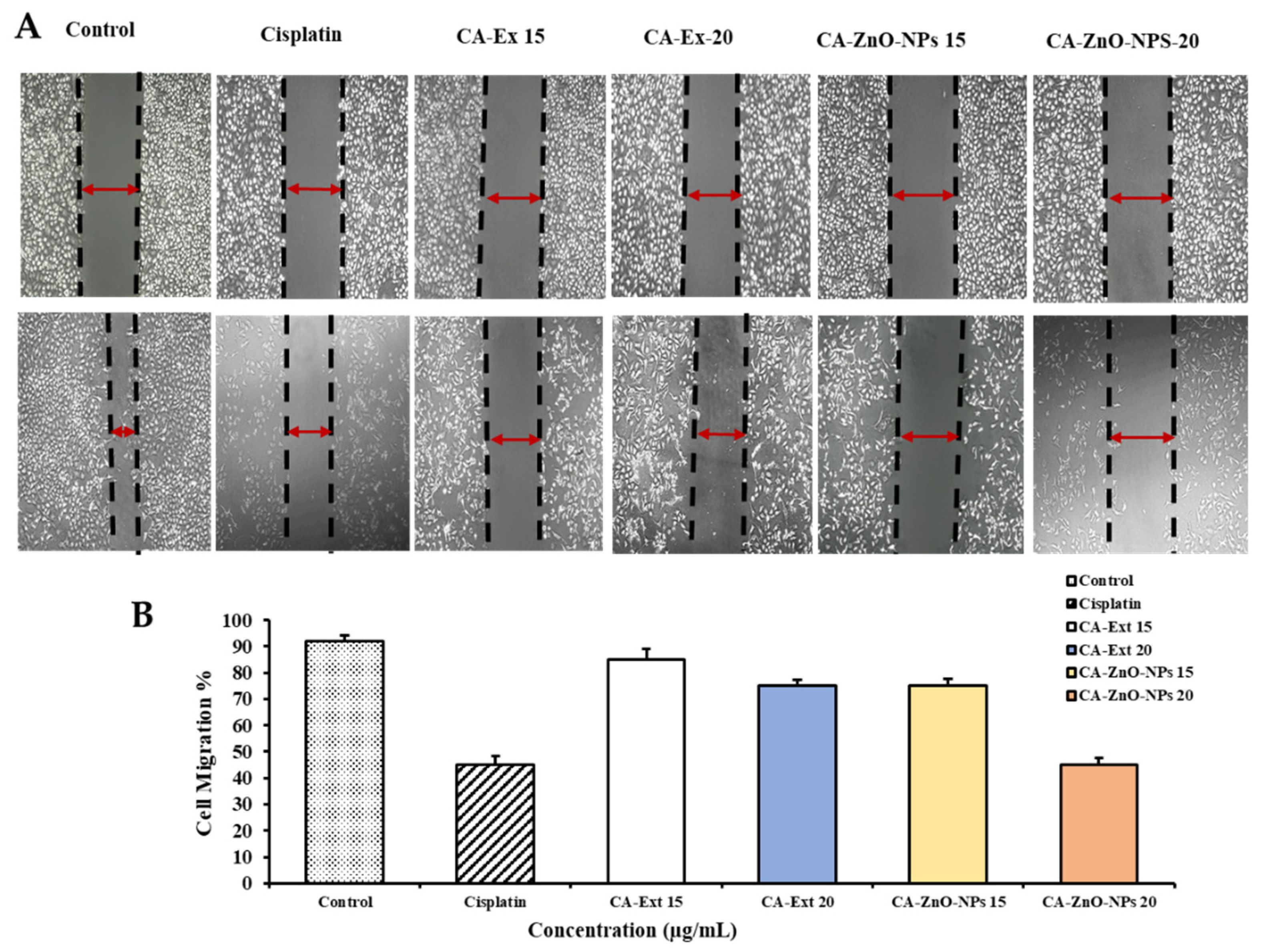
3.10. CA-ZnO-NPs Inhibit Migration of Cancer Cells
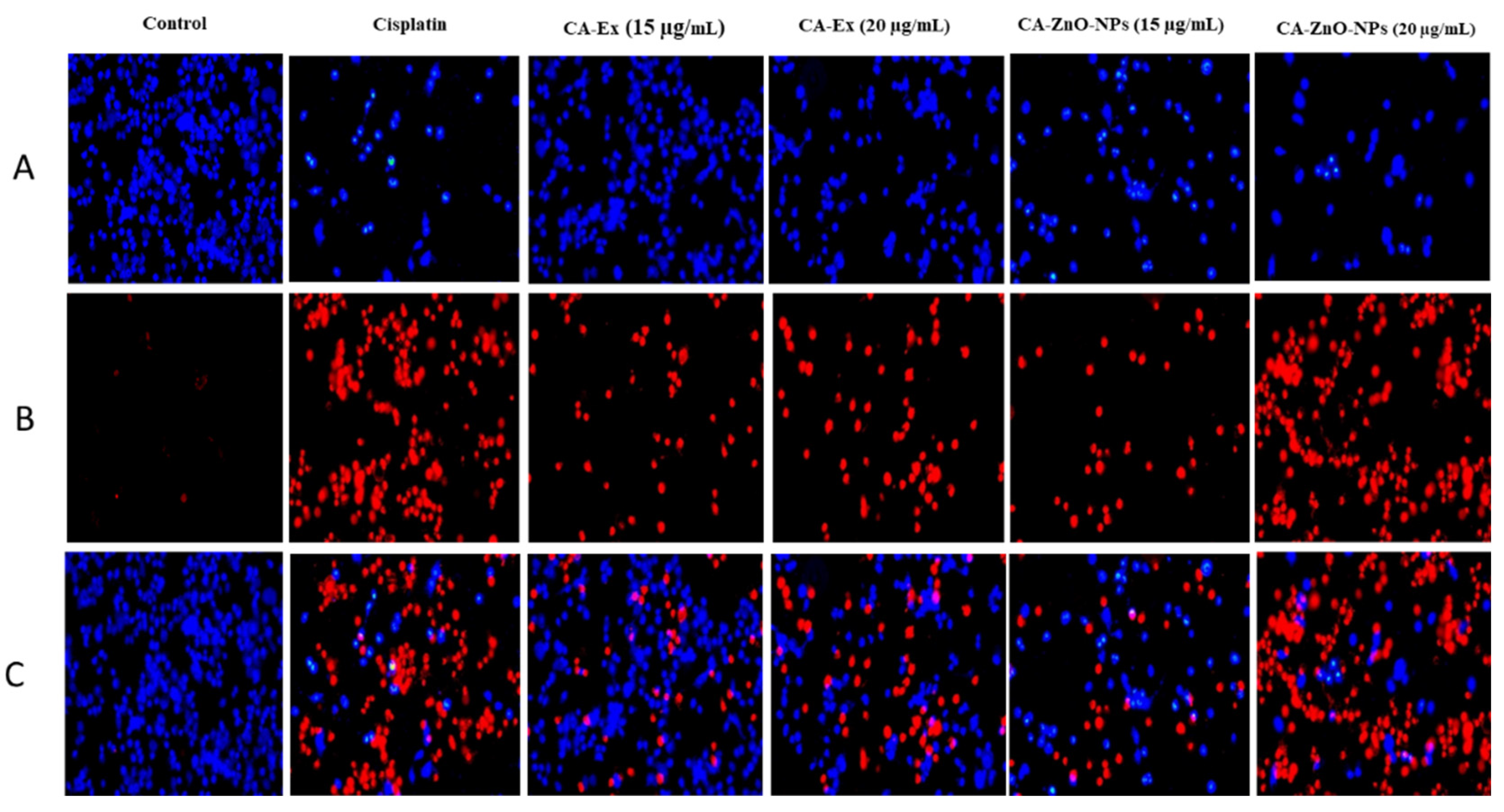
3.11. Detection of HGRCm-ZnO NP-Induced Apoptosis by Hoechst-33342/PI Dye Staining
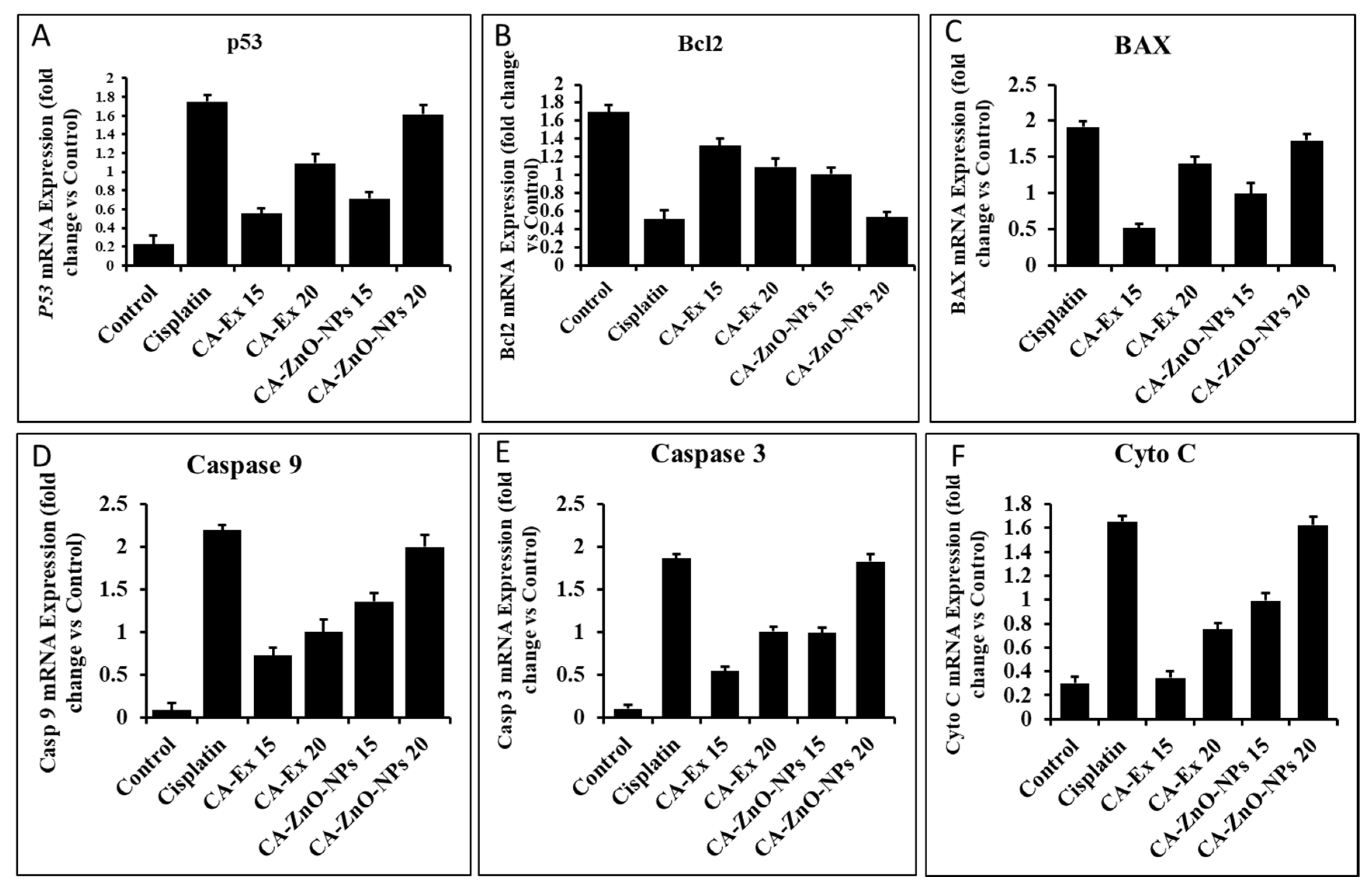
3.12. By Controlling Apoptotic Gene Expression, CA-ZnO-NPs Induced Apoptosis
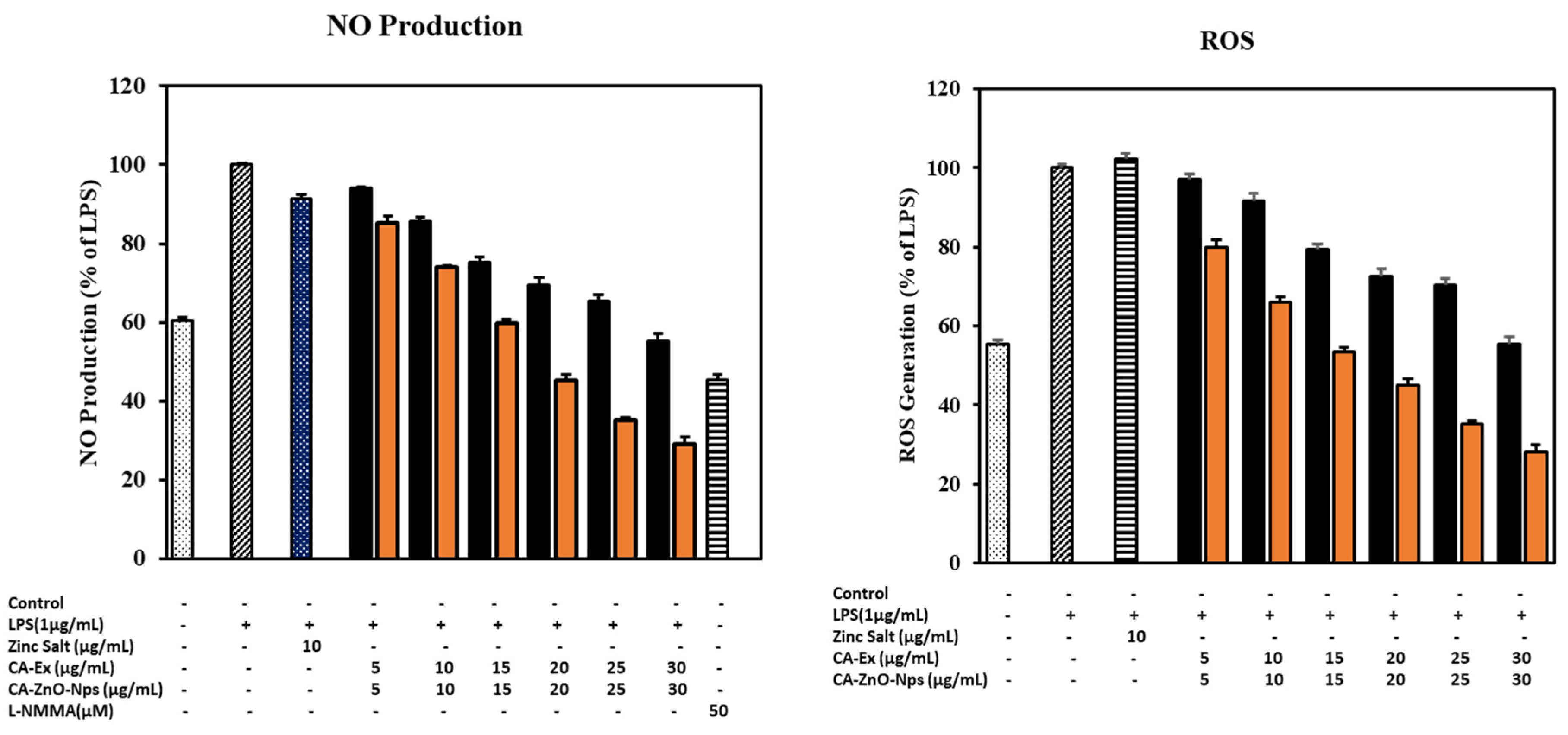
3.13. CA-ZnO-NPs Extract Increased NO Production and Inhibited ROS Generation Induced by LPS
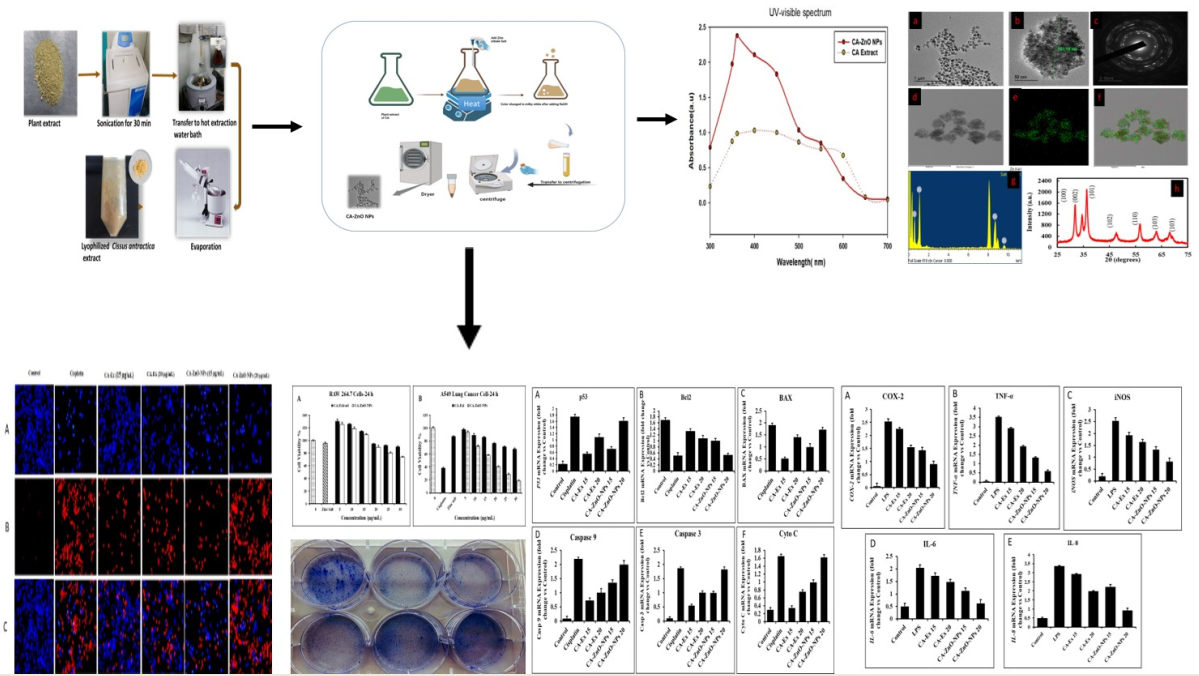
3.14. Outcome of CA-ZnO-NPs on Inflammatory Cytokines
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Medzhitov, R.J.C. Inflammation 2010: new adventures of an old flame. 2010, 140, 771–776.
- Ferrero-Miliani, L.; Nielsen, O.; Andersen, P.; Girardin, S.J.C.; Immunology, E. Chronic inflammation: importance of NOD2 and NALP3 in interleukin-1β generation. 2007, 147, 227–235.
- Nathan, C.; Ding, A.J.C. Nonresolving inflammation. 2010, 140, 871–882.
- Takeuchi, O.; Akira, S.J.C. Pattern recognition receptors and inflammation. 2010, 140, 805–820.
- Kumar, D.; Bhat, Z.A.; Singh, P.; Bhujbal, S.S.; Mudgade, S.C.; Deoda, R.S.J.L.A.J.o.P. Antiallergic and anti-inflammatory properties of methanolic extract of stem bark of Ailanthus excelsa Roxb. 2010, 29. 29.
- Kumar, V.; Bhat, Z.A.; Kumar, D.; Khan, N.; Chashoo, I.J.A.P.J.o.T.B. Evaluation of anti-inflammatory potential of leaf extracts of Skimmia anquetilia. 2012, 2, 627–630.
- Reddy, D.B.; Reddanna, P.J.B.; Communications, B.R. Chebulagic acid (CA) attenuates LPS-induced inflammation by suppressing NF-κB and MAPK activation in RAW 264. 7 macrophages. 2009, 381, 112–117. [Google Scholar]
- Lavanya, R.; Maheshwari, S.U.; Harish, G.; Raj, J.B.; Kamali, S.; Hemamalani, D.; Varma, J.B.; Reddy, C.U.J.R.j.o.p., biological; sciences, c. Investigation of in-vitro anti-inflammatory, anti-platelet and anti-arthritic activities in the leaves of Anisomeles malabarica Linn. 2010, 1, 745–752.
- Ginwala, R.; Bhavsar, R.; Chigbu, D.G.I.; Jain, P.; Khan, Z.K.J.A. Potential role of flavonoids in treating chronic inflammatory diseases with a special focus on the anti-inflammatory activity of apigenin. 2019, 8, 35. 8.
- Siegel, R.L.; Miller, K.D.; Jemal, A.J.C.a.c.j.f.c. Cancer statistics, 2018. 2018, 68, 7–30.
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A.J.C.a.c.j.f.c. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. 2018, 68, 394–424.
- Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Kubik, M.J.J. Screening for lung cancer: US Preventive Services Task Force recommendation statement. 2021, 325, 962–970.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F.J.C.a.c.j.f.c. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. 2021, 71, 209–249.
- Tan, D.; Tan, S.Y.; Lim, S.T.; Kim, S.J.; Kim, W.-S.; Advani, R.; Kwong, Y.-L.J.T.L.O. Management of B-cell non-Hodgkin lymphoma in Asia: resource-stratified guidelines. 2013, 14, e548–e561.
- Alsharairi, N.A.J.N. The effects of dietary supplements on asthma and lung cancer risk in smokers and non-smokers: A review of the literature. 2019, 11, 725.
- Kalaivani, T.; Rajasekaran, C.; Suthindhiran, K.; Mathew, L.J.E.-B.C.; Medicine, A. Free radical scavenging, cytotoxic and hemolytic activities from leaves of Acacia nilotica (L. ) Wild. ex. Delile subsp. indica (Benth.) Brenan. 2011, 2011. [Google Scholar]
- Ko, E.C.; Raben, D.; Formenti, S.C.J.C.C.R. The integration of radiotherapy with immunotherapy for the treatment of non–small cell lung cancer. 2018, 24, 5792–5806.
- Jamkhande, P.G.; Ghule, N.W.; Bamer, A.H.; Kalaskar, M.G.J.J.o.d.d.s. ; technology. Metal nanoparticles synthesis: An overview on methods of preparation, advantages and disadvantages, and applications. 2019, 53, 101174. [Google Scholar]
- Thakkar, K.N.; Mhatre, S.S.; Parikh, R.Y.J.N.n. , biology; medicine. Biological synthesis of metallic nanoparticles. 2010, 6, 257–262. [Google Scholar]
- Zhang, D.; Ma, X.-l.; Gu, Y.; Huang, H.; Zhang, G.-w.J.F.i.C. Green synthesis of metallic nanoparticles and their potential applications to treat cancer. 2020, 8, 799. 8.
- Hashemi, S.F.; Tasharrofi, N.; Saber, M.M.J.J.o.M.s. Green synthesis of silver nanoparticles using Teucrium polium leaf extract and assessment of their antitumor effects against MNK45 human gastric cancer cell line. 2020, 1208, 127889.
- Xie, B.; Yang, J.; Yang, Q.; Yuan, W.J.J.o.M.C.B.E. Enantioselective reduction of fluorenones in surfactant-aqueous solution by fruits and vegetables. 2009, 61, 284–288.
- Hameed, S.; Iqbal, J.; Ali, M.; Khalil, A.T.; Abbasi, B.A.; Numan, M.; Shinwari, Z.K.J.M.R.E. Green synthesis of zinc nanoparticles through plant extracts: establishing a novel era in cancer theranostics. 2019, 6, 102005.
- Chandrasekaran, S.; Anusuya, S.; Anbazhagan, V.J.J.o.M.S. Anticancer, anti-diabetic, antimicrobial activity of zinc oxide nanoparticles: A comparative analysis. 2022, 1263, 133139.
- Velsankar, K.; Venkatesan, A.; Muthumari, P.; Suganya, S.; Mohandoss, S.; Sudhahar, S.J.J.o.M.S. Green inspired synthesis of ZnO nanoparticles and its characterizations with biofilm, antioxidant, anti-inflammatory, and anti-diabetic activities. 2022, 1255, 132420.
- Eswari, K.M.; Asaithambi, S.; Karuppaiah, M.; Sakthivel, P.; Balaji, V.; Ponelakkia, D.; Yuvakkumar, R.; Kumar, P.; Vijayaprabhu, N.; Ravi, G.J.C.I. Green synthesis of ZnO nanoparticles using Abutilon Indicum and Tectona Grandis leaf extracts for evaluation of anti-diabetic, anti-inflammatory and in-vitro cytotoxicity activities. 2022, 48, 33624–33634.
- Dey, A.; Ray, P.G.; Dhara, S.; Neogi, S.J.A.S.S. Optically engineered ZnO Nanoparticles: Excitable at visible wavelength and lowered cytotoxicity towards bioimaging applications. 2022, 592, 153303.
- Sathappan, S.; Kirubakaran, N.; Gunasekaran, D.; Gupta, P.K.; Verma, R.S.; Sundaram, J.J.P.o.t.N.A.o.S. , India Section B: Biological Sciences. Green synthesis of zinc oxide nanoparticles (ZnO NPs) using cissus quadrangularis: Characterization, antimicrobial and anticancer studies. 2021, 91, 289–296. [Google Scholar]
- Prashanth, G.; Prashanth, P.; Nagabhushana, B.; Ananda, S.; Krishnaiah, G.; Nagendra, H.; Sathyananda, H.; Rajendra Singh, C.; Yogisha, S.; Anand, S.J.A.c. , nanomedicine,, et al. Comparison of anticancer activity of biocompatible ZnO nanoparticles prepared by solution combustion synthesis using aqueous leaf extracts of Abutilon indicum, Melia azedarach and Indigofera tinctoria as biofuels. 2018, 46, 968–979. [Google Scholar]
- Gurib-Fakim, A.J.M.a.o.M. Medicinal plants: traditions of yesterday and drugs of tomorrow. 2006, 27, 1–93.
- Balandrin, M.F.; Kinghorn, A.D.; Farnsworth, N.R. Plant-derived natural products in drug discovery and development: an overview. 1993.
- Chen, H.; Yang, H.; Fan, D.; Deng, J.J.e. The anticancer activity and mechanisms of ginsenosides: an updated review. 2020, 1, 226–241.
- Jung, D.-H.; Nahar, J.; Mathiyalagan, R.; Rupa, E.J.; Ramadhania, Z.M.; Han, Y.; Yang, D.-C.; Kang, S.C.J.A.-c.A.i.M.C. Focused Review on Molecular Signalling Mechanisms of Ginsenosides on Anti-lung cancer and Anti-inflammatory Activities. 2022.
- Liu, X.-Q.; Ickert-Bond, S.M.; Chen, L.-Q.; Wen, J.J.M.p. ; evolution. Molecular phylogeny of Cissus L. of Vitaceae (the grape family) and evolution of its pantropical intercontinental disjunctions. 2013, 66, 43–53. [Google Scholar]
- Gerrath, J.M.; Posluszny, U.J.C.j.o.b. Morphological and anatomical development in the Vitaceae. VI. Cissus antarctica. 1994, 72, 635–643. [Google Scholar]
- Balasubramanian, P.; Rajasekaran, A.; Prasad, S.J.A.S.o.L. Folk medicine of the Irulas of Coimbatore forests. 1997, 16, 222.
- Manokari, M.; Shekhawat, M.S.J.W.N.o.N.S. An updated review on Cissus vitiginea L. (Family: Vitaceae)-An important medicinal climber. 2019, 22. [Google Scholar]
- Chidambara Murthy, K.; Vanitha, A.; Mahadeva Swamy, M.; Ravishankar, G.J.J.o.M.F. Antioxidant and antimicrobial activity of Cissus quadrangularis L. 2003, 6, 99–105.
- Mate, G.; Naikwade, N.; Magdum, C.; Chowki, A.; Patil, S.J.I.J.o.G.P. Evaluation of anti-nociceptive activity of Cissus quadrangularis on albino mice. 2008, 2. 2.
- Vijay, P.; Vijayvergia, R.J.J.o.P.S. ; Technology. Analgesic, anti-inflammatory and antipyretic activity of Cissus quadrangularis. 2010, 2, 111–118. [Google Scholar]
- Nocedo-Mena, D.; Garza-González, E.; González-Ferrara, M.; del Rayo Camacho-Corona, M.J.C.T.i.M.C. Antibacterial activity of Cissus incisa extracts against multidrug-resistant bacteria. 2020, 20, 318–323.
- Vijayalakshmi, A.; Kumar, P.; Sakthi Priyadarsini, S.; Meenaxshi, C.J.J.o.C. In vitro antioxidant and anticancer activity of flavonoid fraction from the aerial parts of Cissus quadrangularis Linn. against human breast carcinoma cell lines. 2013, 2013. [Google Scholar]
- Jin, Y.; Huynh, D.T.N.; Myung, C.-S.; Heo, K.-S. Ginsenoside Rh1 Prevents Migration and Invasion through Mitochondrial ROS-Mediated Inhibition of STAT3/NF-κB Signaling in MDA-MB-231 Cells. International journal of molecular sciences 2021, 22, 10458. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Mazumdar, M.; Mukherjee, S.; Bhattacharjee, P.; Adhikary, A.; Manna, A.; Chakraborty, S.; Khan, P.; Sen, A.; Das, T. Restoration of p53/miR-34a regulatory axis decreases survival advantage and ensures Bax-dependent apoptosis of non-small cell lung carcinoma cells. FEBS letters 2014, 588, 549–559. [Google Scholar] [CrossRef]
- Lu, H.-F.; Chie, Y.-J.; Yang, M.-S.; Lee, C.-S.; Fu, J.-J.; Yang, J.-S.; Tan, T.-W.; Wu, S.-H.; Ma, Y.-S.; Ip, S.-W. Apigenin induces caspase-dependent apoptosis in human lung cancer A549 cells through Bax-and Bcl-2-triggered mitochondrial pathway. International journal of oncology 2010, 36, 1477–1484. [Google Scholar]
- Wang, J.-P.; Hsieh, C.-H.; Liu, C.-Y.; Lin, K.-H.; Wu, P.-T.; Chen, K.-M.; Fang, K. Reactive oxygen species-driven mitochondrial injury induces apoptosis by teroxirone in human non-small cell lung cancer cells. Oncology Letters 2017, 14, 3503–3509. [Google Scholar] [CrossRef] [PubMed]
- Fowsiya, J.; Madhumitha, G.; Al-Dhabi, N.A.; Arasu, M.V.J.J.o.P.; Biology, P.B. Photocatalytic degradation of Congo red using Carissa edulis extract capped zinc oxide nanoparticles. 2016, 162, 395–401.
- Goutam, S.P.; Yadav, A.K.; Das, A.J.J.J.o.N. ; Technology. Coriander extract mediated green synthesis of zinc oxide nanoparticles and their structural, optical and antibacterial properties. 2017, 249-252.
- Yedurkar, S.; Maurya, C.; Mahanwar, P.J.O.J.o.S.T. ; Applications. Biosynthesis of zinc oxide nanoparticles using ixora coccinea leaf extract—a green approach. 2016, 5, 1–14. [Google Scholar]
- Rupa, E.J.; Kaliraj, L.; Abid, S.; Yang, D.-C.; Jung, S.-K.J.N. Synthesis of a zinc oxide nanoflower photocatalyst from sea buckthorn fruit for degradation of industrial dyes in wastewater treatment. 2019, 9, 1692.
- Kim, W.J.; Soshnikova, V.; Markus, J.; Oh, K.H.; Anandapadmanaban, G.; Mathiyalagan, R.; Perez, Z.E.J.; Kim, Y.J.; Yang, D.C.J.O. Room temperature synthesis of germanium dioxide nanorods and their in vitro photocatalytic application. 2019, 178, 664–668.
- Shen, C.; James, S.A.; de Jonge, M.D.; Turney, T.W.; Wright, P.F.; Feltis, B.N.J.T.S. Relating cytotoxicity, zinc ions, and reactive oxygen in ZnO nanoparticle–exposed human immune cells. 2013, 136, 120–130.
- Mishra, P.K.; Mishra, H.; Ekielski, A.; Talegaonkar, S.; Vaidya, B.J.D.d.t. Zinc oxide nanoparticles: a promising nanomaterial for biomedical applications. 2017, 22, 1825–1834.
- Truong-Tran, A.Q.; Carter, J.; Ruffin, R.; Zalewski, P.D.J.I.; Biology, c. New insights into the role of zinc in the respiratory epithelium. 2001, 79, 170–177.
- Sana, S.S.; Kumbhakar, D.V.; Pasha, A.; Pawar, S.C.; Grace, A.N.; Singh, R.P.; Nguyen, V.-H.; Le, Q.V.; Peng, W.J.M. Crotalaria verrucosa leaf extract mediated synthesis of zinc oxide nanoparticles: Assessment of antimicrobial and anticancer activity. 2020, 25, 4896.
- Sarmiento-Salinas, F.L.; Perez-Gonzalez, A.; Acosta-Casique, A.; Ix-Ballote, A.; Diaz, A.; Treviño, S.; Rosas-Murrieta, N.H.; Millán-Perez-Peña, L.; Maycotte, P.J.L.S. Reactive oxygen species: Role in carcinogenesis, cancer cell signaling and tumor progression. 2021, 284, 119942.
- Tanino, R.; Amano, Y.; Tong, X.; Sun, R.; Tsubata, Y.; Harada, M.; Fujita, Y.; Isobe, T.J.M.c.t. Anticancer activity of ZnO nanoparticles against human small-cell lung cancer in an orthotopic mouse ModelZnO nanoparticles inhibit growth of small-cell lung cancer. 2020, 19, 502–512.
- Yi, C.; Yu, Z.; Ren, Q.; Liu, X.; Wang, Y.; Sun, X.; Yin, S.; Pan, J.; Huang, X.J.P.; therapy, p. Nanoscale ZnO-based photosensitizers for photodynamic therapy. 2020, 30, 101694.
- Dröse, S.; Brandt, U.J.M.O.P.N.-E.G. , Enzyme Regulation,; Pathophysiology. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. 2012, 145–169. [Google Scholar]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C.J.N.p. Clonogenic assay of cells in vitro. 2006, 1, 2315–2319.
- Hanahan, D.; Weinberg, R.A.J.c. Hallmarks of cancer: the next generation. 2011, 144, 646–674.
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A.J.C. Emerging biological principles of metastasis. 2017, 168, 670–691.
- Sikdar, S.; Lallemand, B.; Dubois, J.J.P. ; Pharmacy. Induction of phase II enzymes glutathione-s-transferase and NADPH: quinone oxydoreductase 1 with novel sulforaphane derivatives in human keratinocytes: evaluation of the intracellular GSH level. 2014, 5, 937. [Google Scholar]
- Yarrow, J.C.; Perlman, Z.E.; Westwood, N.J.; Mitchison, T.J.J.B.b. A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. 2004, 4, 1–9. 4.
- Rani, N.; Saini, K. Biogenic metal and metal oxides nanoparticles as anticancer agent: a review. Proceedings of IOP Conference Series: Materials Science and Engineering; p. 012043.
- Valko, M.; Rhodes, C.; Moncol, J.; Izakovic, M.; Mazur, M.J.C.-b.i. Free radicals, metals and antioxidants in oxidative stress-induced cancer. 2006, 160, 1–40.
- Ismail, N.I.; Othman, I.; Abas, F. ; H. Lajis, N.; Naidu, R.J.I.j.o.m.s. Mechanism of apoptosis induced by curcumin in colorectal cancer. 2019, 20, 2454. [Google Scholar]
- George, B.P.; Abrahamse, H.J.O.m.; longevity, c. Increased oxidative stress induced by rubus bioactive compounds induce apoptotic cell death in human breast cancer cells. 2019, 2019. 2019.
- Johnson, T.M.; Yu, Z.-X.; Ferrans, V.J.; Lowenstein, R.A.; Finkel, T.J.P.o.t.N.A.o.S. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. 1996, 93, 11848–11852.
- Liu, B.; Chen, Y.; Clair, D.K.S.J.F.R.B. ; Medicine. ROS and p53: a versatile partnership. 2008, 44, 1529–1535. [Google Scholar]
- Omoyeni, O.A.; Hussein, A.; Meyer, M.; Green, I.; Iwuoha, E.J.B.C.; Medicine, A. Pleiocarpa pycnantha leaves and its triterpenes induce apoptotic cell death in Caco-2 cells in vitro. 2015, 15, 1–7.
- Kang, M.H.; Reynolds, C.P.J.C.c.r. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. 2009, 15, 1126–1132.
- Zheng, J.H.; Viacava Follis, A.; Kriwacki, R.W.; Moldoveanu, T.J.T.F.j. Discoveries and controversies in BCL-2 protein-mediated apoptosis. 2016, 283, 2690–2700.
- Julien, O.; Wells, J.A.J.C.D. ; Differentiation. Caspases and their substrates. 2017, 24, 1380–1389. [Google Scholar]
- Li, W.; Zihan, X.; Yizhe, W.; Yanyang, L.; Zhixi, L.; Xi, Y.J.N. ; Cancer. Trilobatin induces apoptosis and attenuates stemness phenotype of acquired gefitinib resistant lung cancer cells via suppression of NF-κB pathway. 2022, 74, 735–746. [Google Scholar]
- Li, Y.; Wang, Y.; Yu, X.; Yu, T.; Zheng, X.; Chu, Q.J.N. ; Cancer. Radix tetrastigma inhibits the non-small cell lung cancer via Bax/Bcl-2/Caspase-9/Caspase-3 pathway. 2022, 74, 320–332. [Google Scholar]
- Jayappa, M.D.; Ramaiah, C.K.; Kumar, M.A.P.; Suresh, D.; Prabhu, A.; Devasya, R.P.; Sheikh, S.J.A.n. Green synthesis of zinc oxide nanoparticles from the leaf, stem and in vitro grown callus of Mussaenda frondosa L. : characterization and their applications. 2020, 10, 3057–3074. [Google Scholar]
- Agarwal, H.; Shanmugam, V.J.B.c. A review on anti-inflammatory activity of green synthesized zinc oxide nanoparticle: Mechanism-based approach. 2020, 94, 103423.
- Ilves, M.; Palomäki, J.; Vippola, M.; Lehto, M.; Savolainen, K.; Savinko, T.; Alenius, H.J.P.; toxicology, f. Topically applied ZnO nanoparticles suppress allergen induced skin inflammation but induce vigorous IgE production in the atopic dermatitis mouse model. 2014, 11, 1–12.
- Rajakumar, G.; Thiruvengadam, M.; Mydhili, G.; Gomathi, T.; Chung, I.-M.J.B.; engineering, b. Green approach for synthesis of zinc oxide nanoparticles from Andrographis paniculata leaf extract and evaluation of their antioxidant, anti-diabetic, and anti-inflammatory activities. 2018, 41, 21–30.
- Mayer, B.; John, M.; Böhme, E.J.F.l. Purification of a Ca2+/calmodulin-dependent nitric oxide synthase from porcine cerebellum: Cofactor-role of tetrahydrobiopterin. 1990, 277, 215–219.
- Clancy, R.M.; Amin, A.R.; Abramson, S.B.J.A. ; Rheumatism. The role of nitric oxide in inflammation and immunity. 1998, 41, 1141–1151. [Google Scholar]
- Salvemini, D.J.C.; CMLS, M.L.S. Regulation of cyclooxygenase enzymes by nitric oxide. 1997, 53, 576–582.
- Wang, Z.; Liu, X.; Bao, Y.; Wang, X.; Zhai, J.; Zhan, X.; Zhang, H.J.C.P. Characterization and anti-inflammation of a polysaccharide produced by Chaetomium globosum CGMCC 6882 on LPS-induced RAW 264. 7 cells. 2021, 251, 117129. [Google Scholar]
- Šoltés, L.; Kogan, G.J.E.o.P.; Chemicals Complexity Volume II: New Approaches, L. ; Control. Hyaluronan: A Harbinger of the Status and Functionality of the Joint. Apple Academic Press/Taylor & Francis Group: Canada/United States: 2014; pp 259-286.
- Cao, J.; Naeem, M.; Noh, J.-K.; Lee, E.H.; Yoo, J.-W.J.M.R. Dexamethasone phosphate-loaded folate-conjugated polymeric nanoparticles for selective delivery to activated macrophages and suppression of inflammatory responses. 2015, 23, 485–492.
- Yang, M.; Wang, Y.; Patel, G.; Xue, Q.; Njateng, G.S.S.; Cai, S.; Cheng, G.; Kai, G.J.J.o.E. In vitro and in vivo anti-inflammatory effects of different extracts from Epigynum auritum through down-regulation of NF-κB and MAPK signaling pathways. 2020, 261, 113105.
- Park, E.; Lee, S.-M.; eun Lee, J.; Kim, J.-H.J.J.o.F.F. Anti-inflammatory activity of mulberry leaf extract through inhibition of NF-κB. 2013, 5, 178–186.
| Gene | Primer Sequences (5′-3′) |
|---|---|
| p53 | F: TCT TGGGCC TGT GTT ATC TCC |
| R: CGC CCA TGC AGG AAC TGT TA | |
| Bcl2 | F: GAA GGG CAG CCG TTA GGAAA |
| R: GCG CCC AAT ACG ACC AAA TC | |
| BAX | F: GGT TGC CCT CTT CTA CTT T |
| R: AGC CAC CCT GGT CTT G | |
| CASPASE 3 | F: GAA GGA ACA CGC CAG GAA AC |
| R: GCA AAG TGA AAT GTA GCA CCA A | |
| CASPASE 9 | F: GCC CGA GTT TGA GAG GAA AA |
| R: CAC AGC CAG ACC AGG AC | |
| COX-2 | F: CCT GAG CAT CTA CGG TTT GC |
| R: ACT GCT CAT CAC CCC ATT CA | |
| TNF-α | F: GCCAGAATGCTGCAGGACTT |
| R: GGCCTAAGGTCCACTTGTGTCA | |
| iNOS | F: CCT GAG CAT CTA CGG TTT GC |
| R: ACT GCT CAT CAC CCC ATT CA | |
| IL-6 | F: AGGGTTGCCAGATGCAATAC |
| R: AAACCAAGGCACAGTGGAAC | |
| IL-8 | F: CCGGAGAGGAGACTTCACAG |
| R: GGAAATTGGGGTAGGAAGGA | |
| GAPDH | F: CAA GGT CAT CCA TGA CAA CTT TG |
| R: GTC CAC CAC CCT GTT GCT GTA G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





