1. Introduction
Air pollution and food contamination pose a serious risk to human health. The toxicity of some metals is well known, especially arsenic, mercury, lead and cadmium [
1] which have multiple technological applications and therefore are very present in everyday objects and waste . They can come into contact with humans in various forms, by inhalation, in the form of an aerosol, in contact with the skin and, finally, by ingestion through food. In particular, lead is present both in the air and in the water and tends to accumulate in the surface soil, compromising the development of crops [
2].It produces haematological and neurological diseases, particularly in children, even at low concentrations, and can induce several different diseases: anemia , cancer [
1], and renal failure[
3] , to name the most common.
Some other metals, however, such as iron or calcium and even strontium, are believed to be useful for various physiological activities and for general human well-being.
In particular, strontium, an alkaline metal considered non-toxic, finds various applications in medicine: it is quite similar to calcium and manganese as regards biological interactions, even if often, in humans, the response to strontium is weaker than that of calcim. This metal is then used like calcium for healing teeth, bones and tissue [
4].
However, recently there have been some warnings about possible effects on the heart in patients treated with strontium [
5]. Furthermore, potential risks to human health have been highlighted by a study conducted on strontium-contaminated water and this poses a serious vulnerability because this metal can be present in high quantities in volcanic rocks [
6] and therefore in drinking water.
Several types of bio-sensors have been developed to detect lead, some of which are based on aptamers [
7], i.e. small strands of DNA, RNA synthesized to bind a specific target [
8] with high affinity. Different techniques have been used to monitor the presence of pollutant ions in different substrates. Fluorescence was used to detect Pb
2+ in tea samples [
9], while an electrochemical measurement confirmed the selectivity of aptamers for lead and mercury [
3]. In [
10], a FET-based biosensor was developed in order to discriminate among these pollutants. The device exhibits high selectivity towards cations other than Pb
2+ thus allowing use in an array of complex samples. Their article explores the characteristics of the 8-17DNAzyme aptamer which shows a difference of about 9 orders of magnitude in the detection of Pb
2+ compared to other cations such as Na
+ ,K
+ , Mg
2+ , Ca
2+
Aptamers are small biomolecules (less than 50kDa) with a much more flexible structure than normal proteins. Revealing their 3D structure is still a challenge, both using physical-chemical methods such as NMR and X-ray crystallography [
11], and in silico methods [
12,
13,
14].
In recent years, much attention has been paid to the aptamer called TBA (5'-GGTTGGTGTGGTTGG-3') [
15] a guanine-rich oligomer that has a high affinity for a protein of great clinical interest , α- thrombin. Specifically, this aptamer is able to inhibit protein activity, reducing the formation of clots.
Regarding its 3D structure, most of the information comes from NMR analyses: they detected a G-quadruplex arrangement, which is a peculiar formation in G-rich oligomers: two G-quartets are linked by two TT loops at one end and a TGT loop on the other hand [
15].The G-quadruplex configuration has always been resolved in the presence of monovalent or divalent cations [
9,
15,
16,
17,
18,
19,
20,
21] which would seem to play a fundamental role in its stabilization.
The structure of TBA has been resolved by X-ray crystallography when bound to thrombin [
20] with which it forms a stable complex and it has been shown that the presence of two different cations, K
+ and Na
+, causes small but capable changes in the structure to induce a different ability to inhibit the target protein. More recently, X-ray analysis has been performed to resolve TBA in its free state in the presence of Pb
2+ [
9].
In each case studied, the specific G-quadruplex shape assumed by TBA depends on the size and charge of the cations, for example K
+ is larger than Na
+ and consequently the aptamer structure is more dilated [
20].
The binding of 15mer and 12mer oligomers with Mg
2+ has been explored in [
16]: both aptamers assume a stable structure in the presence of the cation and, in particular, while the 15mer acquires the characteristic G-quadruplex structure, the 12mer takes the form of a basket.
The first topological data on TBA stabilized in the presence of Sr
2+ are NMR data [
19]: in that paper, a mean distance greater than that of the TBA-K
+ complex was detected, also suggesting a different binding site.
In conclusion, the literature suggests that, in general, several cations are useful for stabilizing the free aptamer in the G-quadruplex structure, while producing small differences. These differences produce significant variations in the functioning of the TBA [
20] and therefore allow the TBA to be used, after a suitable calibration, as a very refined sensor of various toxic and non-toxic cations.
In this article we carry out a theoretical investigation regarding the presence of different cations in determining the final structure of the TBA. This allows us to predict the electrical response of this aptamer when used to construct a sensor for the detection of toxic or potentially toxic cations.
2. Results
We analyze some topological and electrical characteristics of the G-quadruplex structure of TBA in the presence of different cations. To this end, the theoretical methods described in
Section 3 have been adopted. The aptamer is described in terms of an equivalent network whose topology and response to electrical stimuli give a picture of what happens in real macromolecules.
2.1. Structural analysis
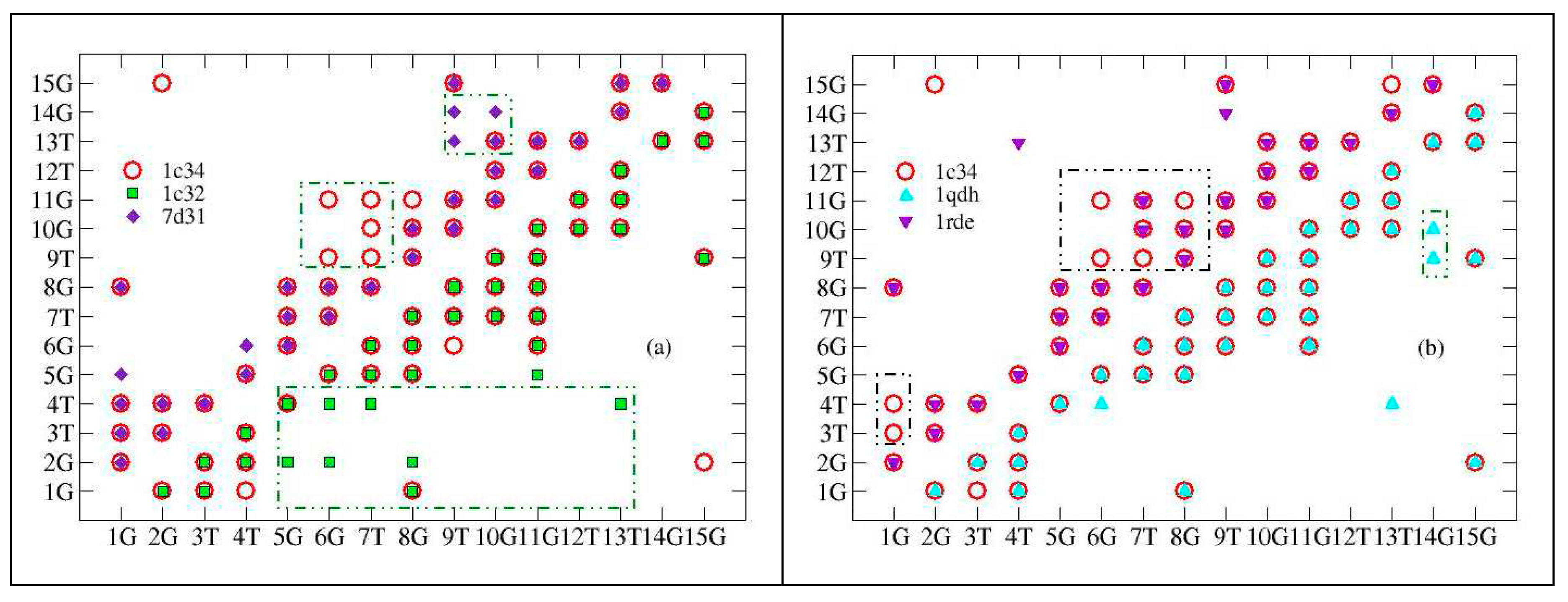
As detailed in subsection 3.1, the macromolecule is described in terms of an impedance network and the contact maps allow to represent its connections: each pair of connected nodes (
i, j) of the network is a symbol in the map and the maps are symmetric . The complete list of analyzed structures is reported in
Table 1 (subsection 3.2). The reference structure, 1c34, solved in the presence of K
+ , is graphically represented both above and below the diagonal, while the other structures are drawn above and below the diagonal (see
Figure 1).
Pb2+ and Sr2+ (pdb entries :7d31 and 1rde, respectively) modify the structure more than K+ . The differences are localized in the central part of the aptamer (6G-8G): here some contacts are missing, indicating a dilatation of the structure. Furthermore, Pb 2+ adds some contacts between 9T, 10G and the aptamer tip, suggesting a folding in this region. The structure obtained using potassium with 2:1 stoichiometry (file pdb : 1c32 ) produces significant differences with respect to the 1:1 stoichiometry, in fact it affects the whole aptamer, producing many more bonds between 2G and 4T and the other nucleobases. Finally, the structure obtained in the presence of Mg2+ (pdb entry: 1qdh) does not present substantial differences with respect to the reference structure.
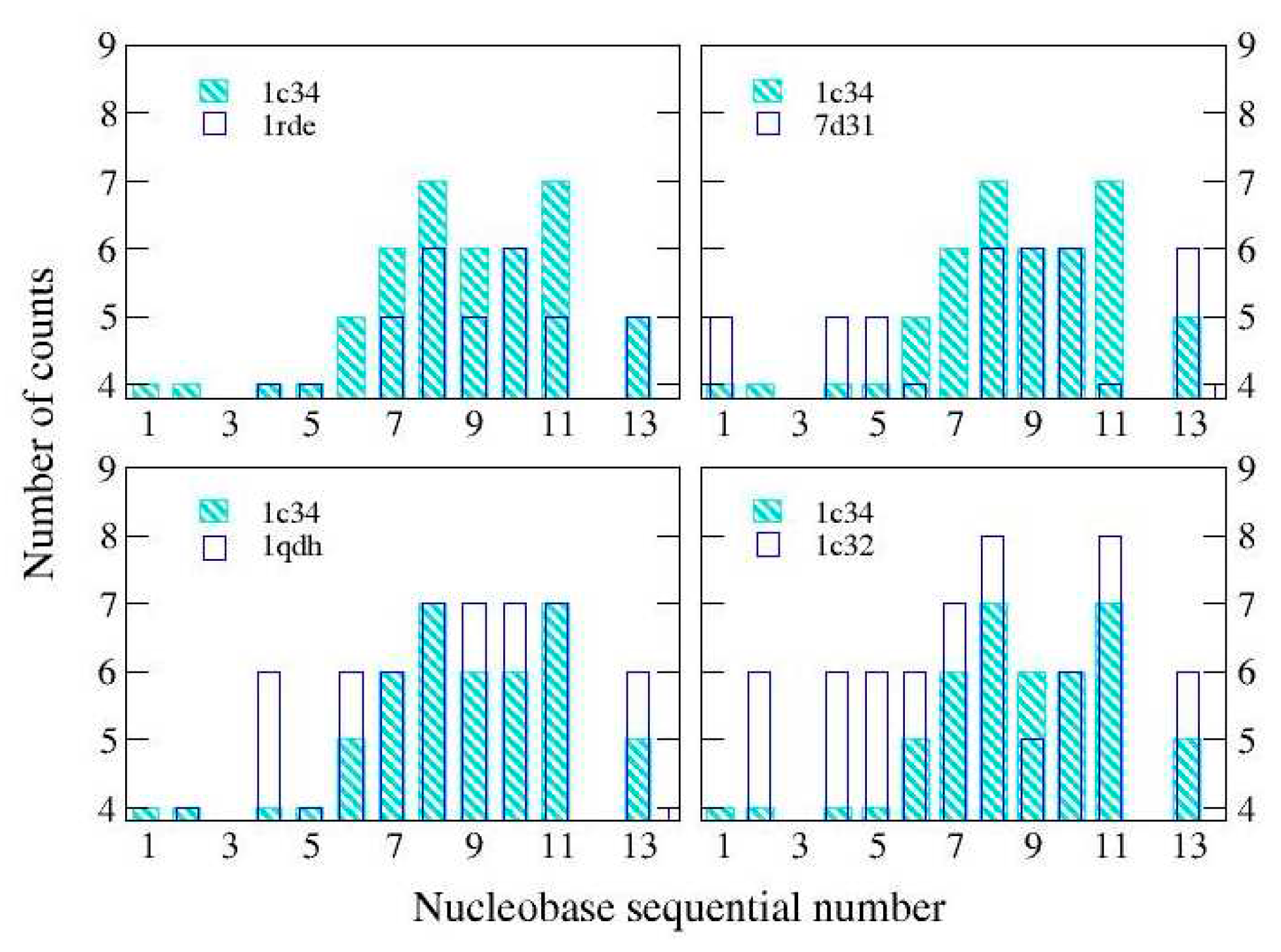
Looking globally at the linkage distribution, (
Figure 2) we can see that for both the reference structure and 1qdh (Mg
2+), most of the links are found around the central nucleobases of the aptamer, (6G-11G) . Both 1rde (Sr
2+) and 7d31 (Pb
2+) show a reduction of the bonds in this region, without significant variations compared to the other nucleobases, therefore the global effect is an expansion. Finally, the structure with potassium (2:1), 1c32, shows a greater number of bonds and a more uniform distribution: this should be related to the presence of the second cation located in a different binding site. In this way, the structure is globally more connected.
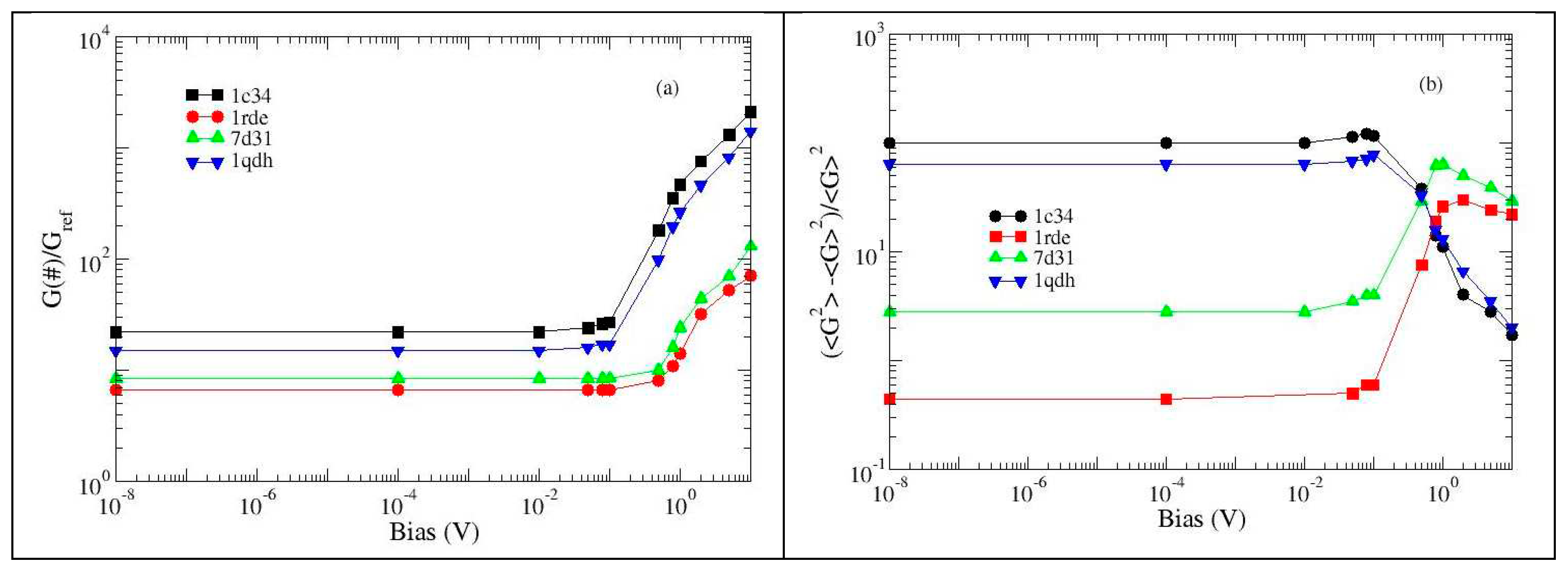
2.2. Resistance analysis (low bias regime)
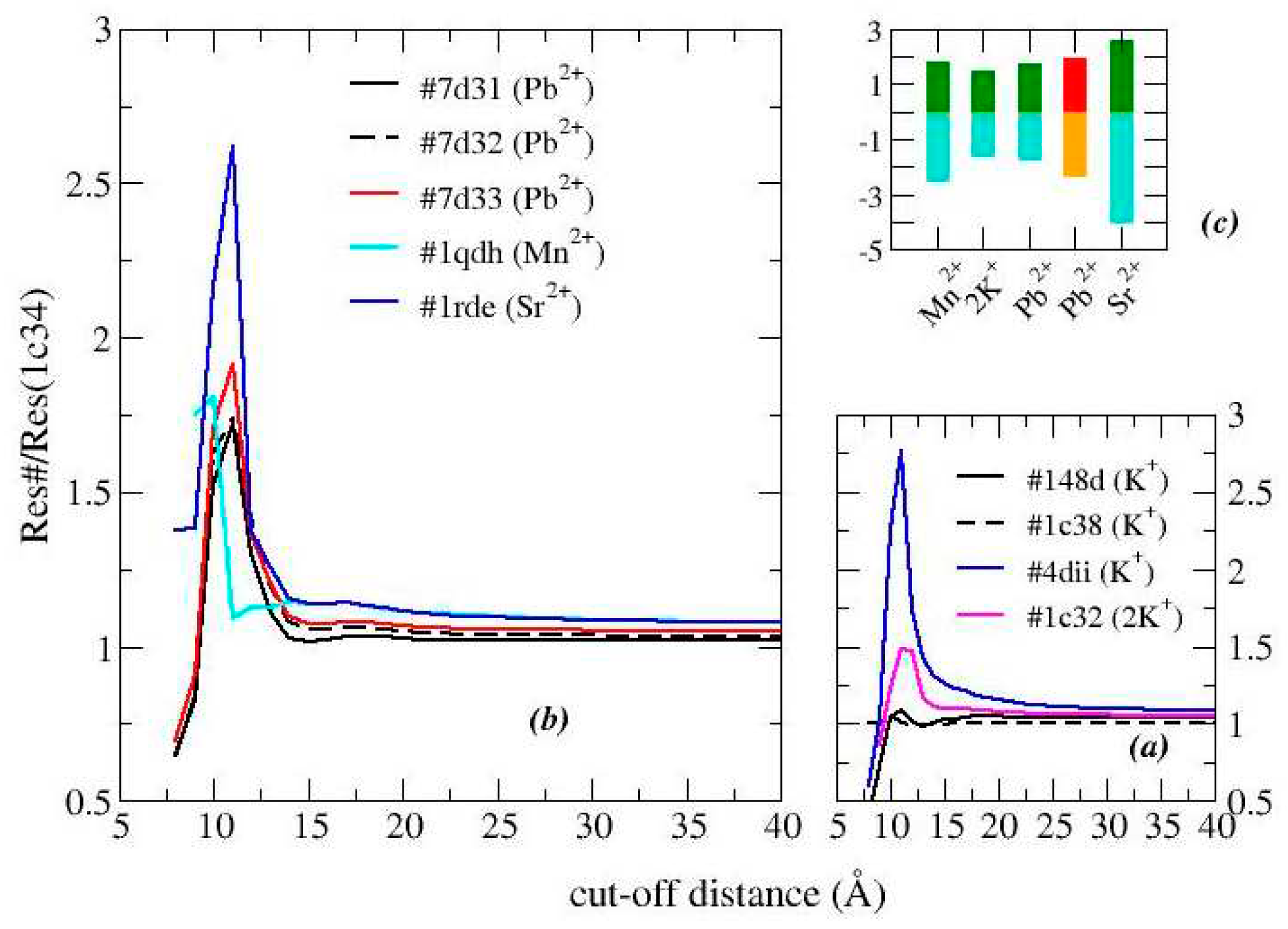
As detailed in
Section 3, the effective resistance of various G-quadruplex structures has been calculated, in the low voltage regime (less than 100 mV), over a wide range of cut-off values (
D), i.e. the parameter which determines the degree of connection to the network. The analysis of the free aptamers and also of two TBA structures complexed with thrombin (although lacking the protein) is shown in
Figure 3. The resistance of the network decreases as the values of
D increase [
22,
23]: in fact, a more connected network is equivalent to having more channels for charge transfer, thus widening the current flow. This is true for all types of networks, so it doesn't give much information.
However, by comparing the resistances of two different structures it is possible to collect some interesting data: in particular, it is possible to detect whether the differences concern the entire structure or if they are localized [
22,
23]. Analyzing the effects of the different cations, it is observed that the reference structure produces the least resistance, which is why
Figure 3 shows the relationships between the resistances of the various structures and the reference one.
We observe that for the structures analyzed, as
D increases, the resistances converge towards similar values, which means that the differences between the structures are limited to small regions. This confirms what was previously observed (
Figure 1 and
Figure 2), namely that the binding sites of the various each cation are very close and the deformations of the structure are concentrated there.
The exception is 1c32 which has 2 cations. Indeed potassium appears, when compared to lead and strontium, to produce a folding of the structure rather than an expansion and this is most evident with this structure. On the other hand, its resistance is greater than that of the reference structure, which signals that the shrinkage does not concern the region through which the current passes the most.
As a first test, we compare the structural data of TBA stabilized in the presence of the same cation but classified differently in the PDB: 148d, 1c34,1c38 for K+ (1:1); 1c32 and 1c35 for K+ (2:1) 7d31,7d32, 7d33 (see
Figure 3a,b). We observe that their answers are almost superimposable. A slight increase in resistance is observed for the G8C mutant (cysteine replaces guanine 8).
The binding with thrombin further deforms the structure obtained in the presence of K+ (PDB entry: 4dii), here deprived of the protein. The relative resistance curve is similar to that given by TBA free although with a much higher maximum value. This result confirms that protein docking further deforms the aptamer, although the region of deformation remains confined to that of the free aptamer.The value of D that maximizes the differences is 11 Å.
Ultimately, ions other than potassium produce larger differences than the reference structure. In particular, in the presence of Pb2+, and even more of Sr2+ , the relative resistance is higher than that of K+ . Manganese appears to produce a similar result even when maximized over D=10Å.
Further information is given by considering the entire region in which the difference occurs, so we can compare the different areas under the curve. The data are reported in
Figure 3c and confirm that Sr
2+ produces the greatest deformation not only locally.
2.3. Impedance analysis (low bias regime)
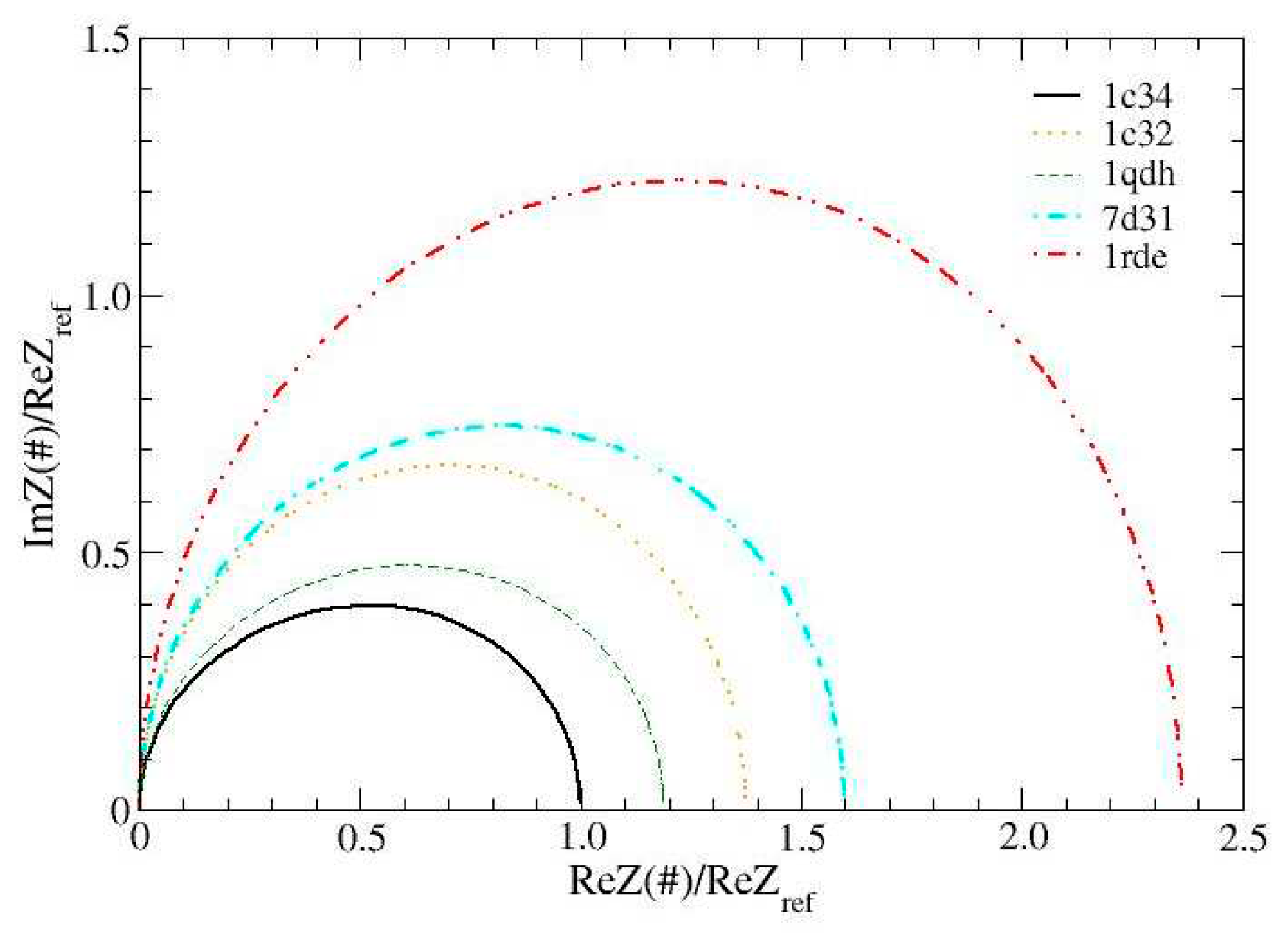
The global impedance of the aptamer is calculated as described in
Section 3. This quantity, measured over a wide range of frequencies, is the typical output of an investigation conducted with electrochemical impedance spectroscopy (EIS) [
22,
23,
25]. It is reported in the so-called Nyquist graphs, which, in our case, represent calculated and not measured data.The Nyquist graphs, normalized to the maximum impedance of the reference structure are shown in
Figure 4. These data strongly suggest that the measurements carried out with EIS are able to resolve, after an appropriate calibration, the presence of different cations. In fact, each of the cations considered produces an almost semicircular pattern and the structures chelated with Pb
2+ and Sr
2+ have the largest radius. Furthermore, as a distinctive trait, actual departures from the semicircular shape are mainly observed for the reference structure 1c34 and 1qdh and become even less marked for the other structures.
This behavior signals the presence of different characteristic times and thus appears to be due to the different organization of the structures which, in 1c34 and 1qdh are very connected in the central part (hub), while the others have much more uniformly distributed links (see
Figure 2).
3. Materials and Methods
3.1. Methods
The analyzes were conducted in the context of Proteotronics [
13,
22,
23,
24,
25,
30] a method of theoretical investigation which analyzed the topological properties of biological macromolecules, associating them with a network of electrical elements.
The structural and electrical characteristics of the network depend on the macromolecule topology and its chemical composition. The deformation of structure produces changes in the electrical response of the network. Briefly, the oligomer is mapped into a set of nodes, each representing a single nucleobase and into a set of links which connect a couple of nodes when they are closer together than an assigned cut-off distance,
D . Increasing the value of
D means increasing the number of nearest neighbors and, in turn, making the network more connected. Too small values of
D produce too simple networks (almost 1D) which do not allow to appreciate the structure of the macromolecule, and
vice versa too large values of
D produce a completely connected network, which is useless to appreciate the specific topology and the electrical characteristics of the aptamer . The best choice is in the range of 5-12 A [
22]. This topological network, far from being regular, has a small-world structure [
22]. The main information relating to the results described in
Section 2 is presented below.
The structural properties of the network are the mirroring of those of the macromolecule. In particular, for an assigned value of D, it is represented by a Boolean (symmetric) matrix of elements 1/0, being the number of nucleobases; the value 1 is attributed to a couple of connected nodes, otherwise it is zero. The graphical representation of this matrix is the contact map of subsection 2.1.
The electrical response of macromolecules when brought into contact with an appropriate bias, has been analyzed in previous investigations [
22,
24,
25] which produce results in good agreement with experiments. The technique consists in associating each link of the network that simulates the macromolecule a circuit element chosen to describe the main characteristics of living matter: charge transfer (high resistance) and charge separation (polarization).
Specifically, each link connecting a pair of nodes (i,j) is associated with:
- i)
an elementary resistance to represent the charge transfer:
where
is the resistivity of the link, calculated as described in [
23], and
is the Euclidean distance between the nodes (
i,j);
- ii)
an elementary capacitance to represent polarization:
where
is the dielectric constant of the nucleobase couple [
23].
The elements described by Equations (1,2) are connected in parallel and the global impedance is calculated: in this way it depends on the specific topology of the macromolecule, and constitutes a probe sensitive to the change in the structure. Finally, the global impedance of the network is calculated using the first and last node [
13,
24,
25,
30] as ideal electrical input/output contact.
Polarization is considered to be relevant only in the AC regime, so in the DC regime, impedance reduces to simple resistance. Furthermore, with increasing values of the applied voltage, deviation from linearity are commonly observed in macromolecule-based nano-devices [
24,
25,
30,
31]. This has been describes as due to a mechanism of sequential tunneling between the network nodes [
24,
25,
30,
31]. Thus, the resistivity is made dependent on the applied bias as follows:
where
is the potential drop across the couple of nodes (i,j),
is the elementary electric charge and
the potential barrier to be overcome for the transfer of electric charge to take place. We assume as a reference value
=0.22 eV [
24,
25].
The asymptotic values,
and
may be tuned on experimental data ( here still not present) as in [apl] and a stochastic procedure is implemented which assigns the final resistivity value according to the probability:
with
and
m the electron mass. The process is not deterministic and the final current oscillates, for each value of bias, around a mean value with fluctuations that testify to the coexistence of both tunneling probabilities of Equation (4). As the applied potential increases, probability described in Equation 4b becomes higher and the fluctuations smooth out.
Using Equations (1,2) the network impedance can be calculated in the AC regime.. This kind of impedance response partially reproduces a typical measurement performed using the electrochemical impedance spectroscopy technique [
22,
23]. In particular, neither the high-frequency resistance solution nor the low-frequency Warburg impedance are accounted for. On the other side, both of them do not give insights about the sensing action, which, usually is limited to the analysis of the Nyquist plots, i.e. the plots of the imaginary vs the real part of the impedance. In an ideal RC parallel circuit, this plot appears semicircular, with the maximal value of the imaginary part reached at the inverse of the characteristic time τ= RC. Often, in real systems, different characteristic times compete producing deviations from the ideal behavior. The origin of multiple characteristic times is not unique although it signals that the sample is not responding as a single object but contains multiple domains each with a specific characteristic [
22,
23].
3.2. Materials
The structures compared here concern the free aptamer TBA in its G-quadruplex form in the presence of different cations (
Table 1). In particular, we focused on TBA stabilized in the presence of : Pb
2+, Sr
2+, Mg
2+ and potassium in the stoichiometry 2:1 (2 cations:1 aptamer) and 1:1. The 3D structures were taken from the Public Data Bank (PDB) [
11] and are shown in
Table 1:
Some of these PDB entries refer to the same structure, although measured/calculated in different specific conditions. In particular 1c34,1c38,148d for TBA in the presence of K+, stoichiometry (1:1) ; 1c32, 1c35 for TBA in the presence of K+, stoichiometry (2:1) ;1qdh, 1qdf for TBA in the presence of Mg2+; 7d31,7d32, in the presence of Pb2+ . 7d33 is a mutant of 7d31 (G8C).
After some preliminary analyses, we conclude that the methods given here are unable to resolve significant differences within each group and for the subsequent analyzes contained in this article, only one of them was used, specifically 1c34 for K+, 1c32 for K+ 2:1, 1qdh for Mg2+ ,7d31 for Pb2+.