Submitted:
17 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
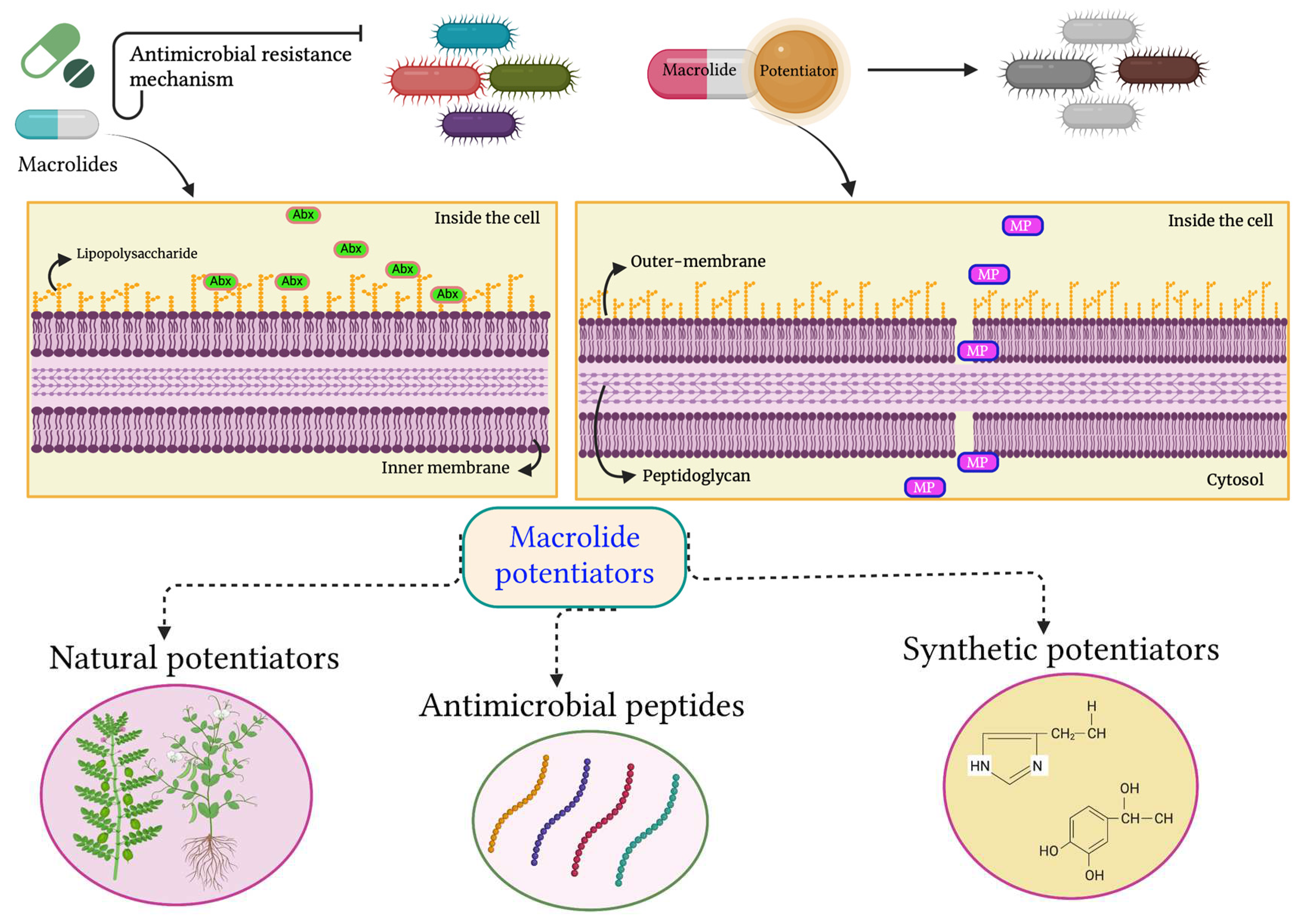
2. Resistance mechanisms to macrolide: Special emphasis on Gram-negative pathogens
2.1. Target modification
2.2. Bacterial efflux mechanism towards macrolide resistance
2.3. Enzymatic macrolide inactivation
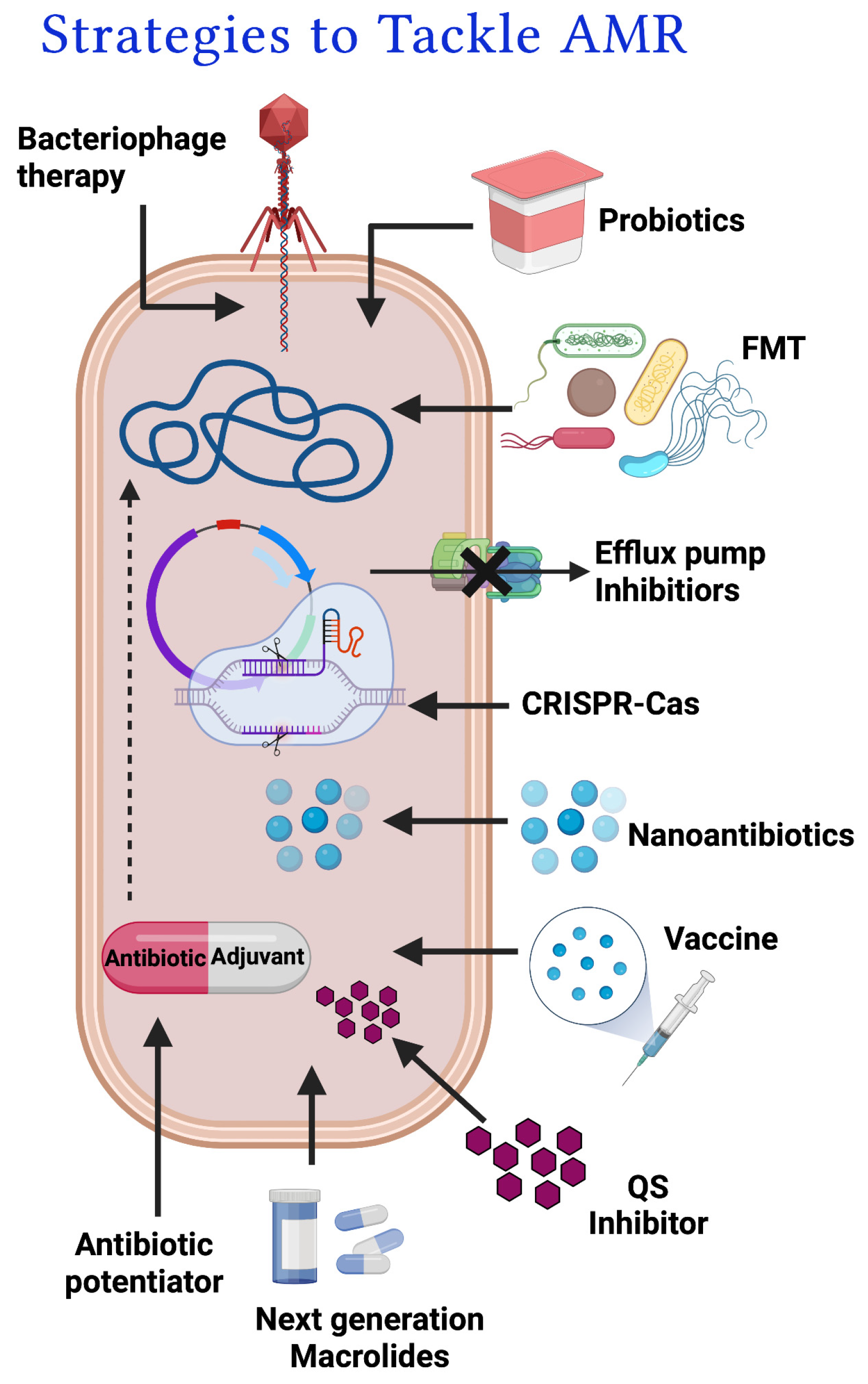
3. Approaches to tackle macrolide resistance
3.1. Antibiotic potentiators and exploration of their diverse mechanisms of action
3.1.1. Efflux pump inhibitors
3.1.2. Modifying enzyme inhibitors
3.1.3. Membrane permeabilizer
4. Macrolide Potentiators & their current status
4.1. Natural potentiators
4.2. Antimicrobial peptides as macrolide potentiators
| Sl. No. | Compound | Source | Antibiotic in combination | Organism tested | Current status | References |
|---|---|---|---|---|---|---|
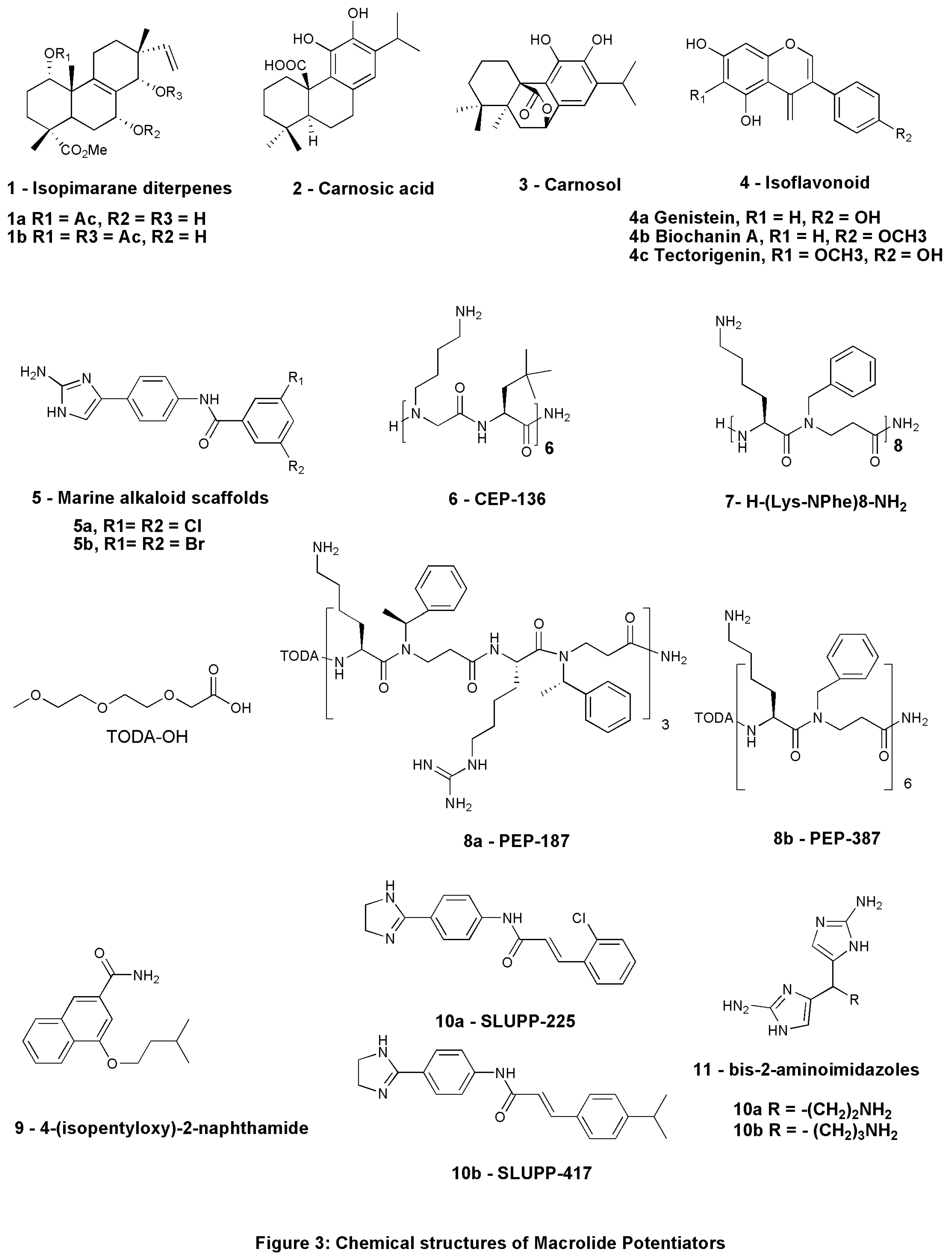
| 1 |
1a: Methyl-1a-acetoxy-7a-14a-dihydroxy-8,15-isopimaradien-18-oate 1b: Methyl-1a,14a-diacetoxy-7a-hydroxy-8,15-isopimaradien-18-oate |
Natural- L. europaeus |
Erythromycin | S. aureus isolates expressing msr(A) multidrug efflux pump |
Lab study- in vitro |
Gibbons et al., 2003 |
| 2 |
2: Carnosic acid 3: Carnosol |
Natural - Rosmarinus officinalis L | Erythromycin | msr(A) & NorA expressing S. aureus strain |
Lab study- in vitro |
Oluwatuyi et al., 2004 |
| 3 | Allyl sulphide | Natural - Allium sativum | Erythromycin | EmrD-3 expressing V. cholerae |
Lab study- in vitro |
Bruns et al., 2017 |
| 4 | Conessine | Natural - Holarrhena antidysenterica | Erythromycin | P. aeruginosa PAO1 strain K767, MexAB-OprM overexpressed strain K1455, & MexB deleted strain K1523 |
Lab study- in vitro |
Siriyong et al., 2017 |
| 5 |
4a: Genistein, 4b: Biochanin A, 4c: Tectorigenin |
Cytisus striatus | Erythromycin | MRSA strains |
Lab study- in vitro |
Abreu et al., 2017 |
| 6 | Compound 5a & 5b | Nitrogen dense marine alkaloid scaffolds | Azithromycin, Erythromycin, Clarithromycin | A. baumannii AB5075 | Lab study - in vivo using a AB5075 infection model of Galleria mellonella | Martin et al., 2019 |
| 7 |
6: CEP-136 H-[NLys-tBuAla] 6-NH2 |
Peptide-based | Azithromycin, Clarithromycin | MDR strains including ESBL-producing isolates | Lab study - in vivo in mouse peritonitis model | Mood et al., 2021 |
| 8 | KLWKKWKKWLK-NH2 & GKWKKILGKLIR-NH2 | Peptide-based | Azithromycin, Erythromycin, Clarithromycin | K. pneumoniae, E. coli, & A. baumannii strains |
Lab study- in vitro |
Baker et al., 2019b |
| 9 | 7: H-(Lys-NPhe)8-NH2 | Peptide-based | Azithromycin, Erythromycin, Clindamycin | MDR strain of E. coli ST131 & K. pneumoniae ST258 |
Lab study- in vitro |
Baker et al., 2019a |
| 10 | 8: 4-isopentyloxy-2-naphthamide | Synthetic-2-naphthamide core | Erythromycin | AcrAB-TolC efflux pump expressing strains |
Lab study- in vitro |
Wang et al., 2017 |
| 11 |
9a: SLUPP-225 9b: SLUPP-417 |
Synthetic | Erythromycin | E. coli |
Lab study- in vitro |
Haynes et al., 2017 |
| 12 | 10: Bis-2-aminoimidazoles (bis-2-AIs) | Synthetic- nitrogen-dense heterocycles | Azithromycin, Clarithromycin | P. aeruginosa |
Lab study- in vivo |
Hubble et al., 2019 |
4.3. Synthetic potentiators
5. Hindrances in taking macrolide potentiators from bench to bedside
5.1. Lack of comprehensive research and toxicity studies
5.2. Sensitivity and Specificity
5.3. Spectrum of activity
5.4. Macrolide potentiator resistance
6. Future perspective
7. Concluding remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lenz KD, Klosterman KE, Mukundan H, et al. Macrolides: From Toxins to Therapeutics. Toxins 2021; 13. Available at:. [CrossRef]
- Manesh A, Varghese GM. Rising antimicrobial resistance: an evolving epidemic in a pandemic. The Lancet Microbe 2021; 2: e419–20. Available at:. [CrossRef]
- Knight GM, Glover RE, McQuaid CF, et al. Antimicrobial resistance and COVID-19: Intersections and implications. Elife 2021; 10. Available at:. [CrossRef]
- Sultana J, Cutroneo PM, Crisafulli S, Puglisi G, Caramori G, Trifiro G. Azithromycin in COVID-19 Patients: Pharmacological Mechanism, Clinical Evidence and Prescribing Guidelines. Drug Saf 2020; 43: 691–8. [CrossRef]
- Doan T, Worden L, Hinterwirth A, et al. Macrolide and Non-macrolide Resistance with Mass Azithromycin Distribution. N Engl J Med 2020; 383: 1941–50. [CrossRef]
- World Health Organization: WHO (2017). WHO publishes list of bacteria for which new antibiotics are urgently needed. World Health Organization: WHO. Available at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed. /.
- Yu B, Roy Choudhury M, Yang X, et al. Restoring and Enhancing the Potency of Existing Antibiotics against Drug-Resistant Gram-Negative Bacteria through the Development of Potent Small-Molecule Adjuvants. ACS Infect Dis 2022; 8: 1491–508. [CrossRef]
- Narendrakumar L, Chakraborty M, Kumari S et al. β-Lactam potentiators to re-sensitize resistant pathogens: Discovery, development, clinical use and the way forward. Front Microbiol. 2023 Mar 10;13:1092556. [CrossRef]
- Chawla M, Verma J, Gupta R, et al. Antibiotic Potentiators Against Multidrug-Resistant Bacteria: Discovery, Development, and Clinical Relevance. Front Microbiol. 2022 Jul 1;13:887251. [CrossRef]
- Koya SF, Ganesh S, Selvaraj S, et al. Consumption of systemic antibiotics in India in 2019. The Lancet Regional Health - Southeast Asia; 2022; 4: 100025. [CrossRef]
- Hicks LA, Bartoces MG, Roberts RM, et al. US outpatient antibiotic prescribing variation according to geography, patient population, and provider speciality in 2011. Clin. Infect. Dis. 2015; 601308–1316. [CrossRef]
- Kumar M, Rao M, Mathur T, et al. Azithromycin Exhibits Activity Against Pseudomonas aeruginosa in Chronic Rat Lung Infection Model. Front. Microbiol. 2021; 12:603151. [CrossRef]
- Saini H, Chhibber S, Harjai K. Azithromycin and cipro- floxacin: a possible synergistic combination against Pseudomonas aeruginosa biofilm-associated urinary tract infections. Int J Antimicrob Agents 2015; 45:359–7. [CrossRef]
- Putnam SD, Castanheira M, Moet GJ, et al. CEM-101, a novel fluoroketolide: antimicrobial activity against a diverse collection of Gram-positive and Gram-negative bacteria. Diagn Microbiol Infect Dis 2010; 66:393–401.
- Gomes C, Martínez-Puchol S, Palma N, et al. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit Rev Microbiol. 2017; 43:1-30. [CrossRef]
- O’Brien KS, Emerson P, Hooper PJ et al. Antimicrobial resistance following mass azithromycin distribution for trachoma: a systematic review. Lancet Infect Dis 2019; 19: e14–25.
- Dinos, GP. The macrolide antibiotic renaissance. Br J Pharmacol 2017; 174: 2967-83. [CrossRef]
- Golkar T, Zielinski M, Berghuis AM. Look and Outlook on Enzyme-Mediated Macrolide Resistance. Front Microbiol 2018; 9: 1942.
- Gomes C, Martinez-Puchol S, Durand D, et al. Which mechanisms of azithromycin resistance are selected when efflux pumps are inhibited? Int J Antimicrob Agents 2013a; 4:307–11. [CrossRef]
- Gutierrez-Castrellon P, Mayorga-Buitron JL, Bosch-Canto V, et al. Efficacy and safety of clarithromycin in pediatric patients with upper respiratory infections: a systematic review with meta-analysis. Rev Invest Clin 2012; 64: 126–35.
- Bohnert JA, Schuster S, Feahnrich E, et al. Altered spectrum of multidrug resistance associated with a single point mutation in the Escherichia coli RND-type MDR efflux pump YhiV (MdtF). J Antimicrob Chemother 2007; 59:1216–22. [CrossRef]
- Pawlowski AC, Stogios PJ, Koteva K, et al. The evolution of substrate discrimination in macrolide antibiotic resistance enzymes. Nat Commun 2018; 9: 112. [CrossRef]
- Andersen JL, He GX, Kakarla P, et al. Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int J Environ Res Public Health. 2015 Jan 28;12(2):1487-547. [CrossRef]
- Xiang Y, Wu F, Chai Y, et al. A new plasmid carrying mphA causes prevalence of azithromycin resistance in enterotoxigenic Escherichia coli serogroup O6. BMC Microbiol. 2020 Aug 11;20(1):247. [CrossRef]
- Micoli F, Bagnoli F, Rappuoli R et al. The role of vaccines in combatting antimicrobial resistance. Nat Rev Microbiol. 2021;19:287-302.
- Eleraky NE, Allam A, Hassan SB, et al. Nanomedicine Fight against Antibacterial Resistance: An Overview of the Recent Pharmaceutical Innovations. Pharmaceutics 2020; 12. Available at:. [CrossRef]
- Morakul B, Suksiriworapong J, Chomnawang MT, et al. Dissolution enhancement and in vitro performance of clarithromycin nanocrystals produced by precipitation-lyophilization-homogenization method. Eur J Pharm Biopharm. 2014; 88:886-96. [CrossRef]
- Azhdarzadeh M, Lotfipour F, Zakeri-Milani P, et al. Anti-bacterial performance of azithromycin nanoparticles as colloidal drug delivery system against different Gram-negative and Gram-positive bacteria. Adv Pharm Bull 2012; 2: 17–24.
- Mood EH, Goltermann L, Brolin C, et al. Antibiotic Potentiation in Multidrug-Resistant Gram-Negative Pathogenic Bacteria by a Synthetic Peptidomimetic. ACS Infect Dis 2021; 7: 2152–63. [CrossRef]
- Hyun S, Choi Y, Jo D, et al. Proline Hinged Amphipathic α-Helical Peptide Sensitizes Gram-Negative Bacteria to Various Gram-Positive Antibiotics. J Med Chem 2020; 63: 14937–50. [CrossRef]
- MacNair CR, Brown ED. Outer Membrane Disruption Overcomes Intrinsic, Acquired, and Spontaneous Antibiotic Resistance. MBio 2020; 11. Available at:. [CrossRef]
- Tyers M, Wright GD. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat Rev Microbiol. 2019;17:141-155. [CrossRef]
- Du D, Wang-Kan X, Neuberger A, et al. Author Correction: Multidrug efflux pumps: structure, function and regulation. Nat Rev Microbiol 2018; 16: 577.
- Jeon B, Zhang Q. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J Antimicrob Chemother 2009; 63: 946–8.
- Gill EE, Franco OL, Hancock RE. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des. 2015;85:56-78. [CrossRef]
- 36. Mojica MF, Rossi MA, Vila AJ, Bonomo RA. The urgent need for metallo-β-lactamase inhibitors: an unattended global threat. Lancet Infect Dis 2022; 22: e28–34. [CrossRef]
- 37. Laws M, Shaaban A, Rahman KM. Antibiotic resistance breakers: current approaches and future directions. FEMS Microbiol Rev 2019; 43: 490–516. [CrossRef]
- Klobucar K, Brown ED. New potentiators of ineffective antibiotics: Targeting the Gram-negative outer membrane to overcome intrinsic resistance. Curr Opin Chem Biol 2022; 66: 102099. [CrossRef]
- Pandey P, Sahoo R, Singh K, et al. Drug Resistance Reversal Potential of Nanoparticles/Nanocomposites via Antibiotic’s Potentiation in Multi Drug Resistant P. aeruginosa. Nanomaterials 2021; 12: 117. Available at:. [CrossRef]
- . Pruul H, McDonald PJ. Potentiation of antibacterial activity of azithromycin and other macrolides by normal human serum. Antimicrob Agents Chemother. 1992; 36(1):10-6.
- Liu Y, Li R, Xiao X, Wang Z. Antibiotic adjuvants: an alternative approach to overcome multi-drug resistant Gram-negative bacteria. Crit Rev Microbiol 2019; 45: 301–14.
- Khameneh B, Iranshahy M, Soheili V, et al. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob Resist Infect Control 2019; 8: 118. [CrossRef]
- Gibbons S, Oluwatuyi M, Veitch NC, Gray AI. Bacterial resistance modifying agents from Lycopus europaeus. Phytochemistry 2003; 62: 83–7. [CrossRef]
- Oluwatuyi M, Kaatz GW, Gibbons S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry 2004; 65: 3249–54. [CrossRef]
- Bruns MM, Kakarla P, Floyd JT, et al. Modulation of the multidrug efflux pump EmrD-3 from Vibrio cholerae by Allium sativum extract and the bioactive agent allyl sulfide plus synergistic enhancement of antimicrobial susceptibility by A. sativum extract. Arch Microbiol 2017; 199: 1103–12.
- Siriyong T, Srimanote P, Chusri S, et al. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement Altern Med 2017; 17: 405. [CrossRef]
- Abreu AC, Coqueiro A, Sultan AR, et al. Looking to nature for a new concept in antimicrobial treatments: isoflavonoids from Cytisus striatus as antibiotic adjuvants against MRSA. Sci Rep 2017; 7: 3777.
- Martin SE, Melander RJ, Brackett CM, et al. Small Molecule Potentiation of Gram-Positive Selective Antibiotics against. ACS Infect Dis 2019; 5: 1223–30. [CrossRef]
- Hubble VB, Bartholomew KR, Weig AW, et al. Augmenting the Activity of Macrolide Adjuvants against. ACS Med Chem Lett 2020; 11: 1723–31.
- Huan Y, Kong Q, Mou H, Yi H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front Microbiol 2020; 11: 582779. [CrossRef]
- Costa F, Teixeira C, Gomes P, et al. Clinical Application of AMPs. Adv Exp Med Biol 2019; 1117: 281–98.
- She P, Liu Y, Xu L, et al. SPR741, Double- or Triple-Combined With Erythromycin and Clarithromycin, Combats Drug-Resistant, Its Biofilms, and Persister Cells. Front Cell Infect Microbiol 2022; 12: 858606.
- Sakoulas G, Okumura CY, Thienphrapa W, et al. Nafcillin enhances innate immune-mediated killing of methicillin-resistant Staphylococcus aureus. J Mol Med 2014; 92: 139–49. [CrossRef]
- Lin L, Nonejuie P, Munguia J, et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine. 2015 Jun 10;2(7):690-8. [CrossRef]
- Baker KR, Jana B, Hansen AM, et al. Repurposing azithromycin and rifampicin against Gram-negative pathogens by combination with peptide potentiators. Int J Antimicrob Agents 2019; 53: 868–72. [CrossRef]
- Lenci E, Trabocchi A. Peptidomimetic toolbox for drug discovery. Chem Soc Rev 2020; 49: 3262–77.
- Baker KR, Jana B, Hansen AM, et al. Repurposing Azithromycin and Rifampicin Against Gram-Negative Pathogens by Combination With Peptidomimetics. Front Cell Infect Microbiol 2019; 9: 236. [CrossRef]
- Haynes KM, Abdali N, Jhawar V, et al. Identification and Structure-Activity Relationships of Novel Compounds that Potentiate the Activities of Antibiotics in Escherichia coli. J Med Chem 2017; 60: 6205–19. [CrossRef]
- Wang Y, Mowla R, Guo L, et al. Evaluation of a series of 2-napthamide derivatives as inhibitors of the drug efflux pump AcrB for the reversal of antimicrobial resistance. Bioorg Med Chem Lett 2017; 27: 733–9.
- Hubble VB, Hubbard BA, Minrovic BM, et al. Using Small-Molecule Adjuvants to Repurpose Azithromycin for Use against Pseudomonas aeruginosa. ACS Infect Dis 2019; 5: 141–51.
- Cui ZH, He HL, Zheng ZJ et al. Phentolamine Significantly Enhances Macrolide Antibiotic Antibacterial Activity against MDR Gram-Negative Bacteria. Antibiotics 2023, 12, 760. [Google Scholar] [CrossRef]
- Ma TKW, Chow KM, Choy ASM, et al. Clinical manifestation of macrolide antibiotic toxicity in CKD and dialysis patients. Clin Kidney J 2014; 7: 507–12.
- Woodhead JL, Yang K, Oldach D, et al. Analyzing the Mechanisms Behind Macrolide Antibiotic-Induced Liver Injury Using Quantitative Systems Toxicology Modeling. Pharm Res 2019; 36: 48. [CrossRef]
- Fohner AE, Sparreboom A, Altman RB, et al. Pharm GKB summary: Macrolide antibiotic pathway, pharmacokinetics/pharmacodynamics. Pharmaco genet Genomics. 2017; 27:164-167.
- Pani A, Lauriola M, Romandini A, et al. Macrolides and viral infections: focus on azithromycin in COVID-19 pathology. Int J Antimicrob Agents 2020; 56: 106053.
- Mendez-Samperio, P. Peptidomimetics as a new generation of antimicrobial agents: current progress. Infect Drug Resist 2014; 7: 229–37.
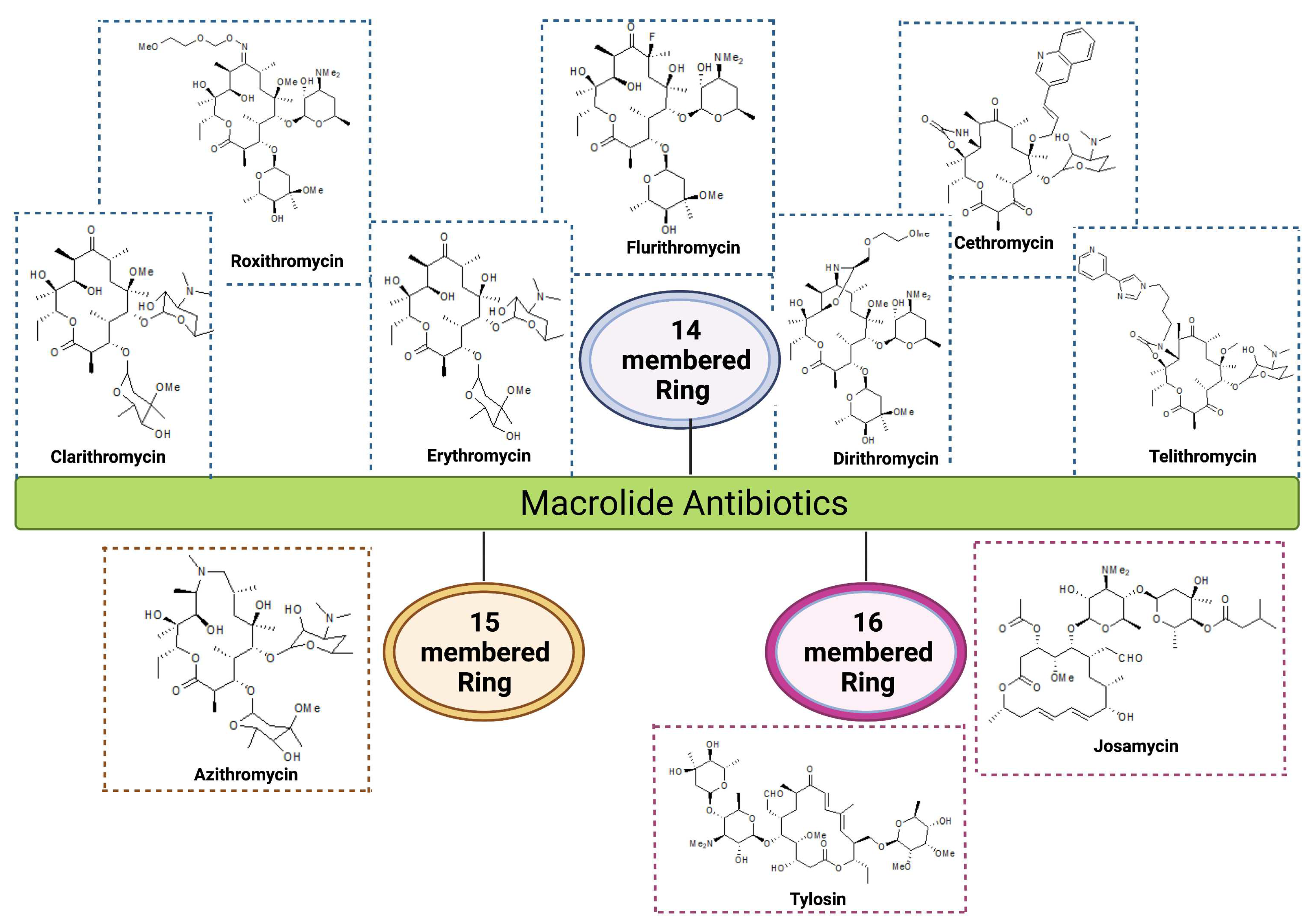
| Group | Ring Structure | Molecule | Origin | Target pathogens | Treatment | Reference |
|---|---|---|---|---|---|---|
| First generation | 14-membered | Erythromycin | Streptomyces erythreus | Gram-positive bacteria: Staphylococcus aureus, Streptococcus pneumoniae, & S. pyogenes Gram-negative bacteria: Neisseria meningitis, N. gonorrhoeae, & Bordetella pertussis |
RTI, Skin, soft tissues, Urogenital tract and Middle ear infections |
Kaneko et al., 2007; Farzam et al., 2021 |
| Second generation | 14-membered | Clarithromycin |
Semi-synthetic conversion of erythromycin | Gram-positive bacteria: S. aureus, S. pneumoniae, & S. pyogenes Gram-negative bacteria : Mycoplasma pneumoniae, Legionella pneumophila, & Chlamydia pneumoniae, Helicobacter pylori, Pseudomonas aeroginosa |
RTI, Chronic inflammation of stomach ulcers, MAC infections in HIV patients |
Yanagihara et al., 2002; Lenz et al., 2021 |
| Roxithromycin | Semi-synthetic derivative of erythromycin | Gram-positive bacteria: S. aureus, S. pyogenes, S. pneumoniae, Listeria monocytogens Gram-negative bacteria: N. meningitidis, B. pertussis, Haemophilus influenzae |
RTI, skin and soft tissue infection and gastrointestinal infections |
Dinos et al., 2017 | ||
| Flurithromycin | Fluorinated derivative of erythromycin A |
H. pylori, Bacteroides forsythus |
Chronic gastritis, Periodontal disease | Fera et al., 2001; Blandino et al., 2013 |
||
| Dirithromycin | Semi-synthetic derivative of erythromycin | Gram-positive bacteria: S. aureus, S. pneumoniae, Gram-negative bacteria: H. influenzae, L. pneumophila, Moraxella catarrhalis, and M. pneumoniae |
Bronchitis, Pneumoniae, tonsillitis and skin infections | Dinos et al., 2017 | ||
| 15-membered | Azithromycin | Derivative of erythromycin | Gram-positive bacteria: S. aureus, S. pneumoniae Gram-negative bacteria: H. influenzae, M. catarrhalis, C. trachomatis, & H. pylori |
RTI, otitis media, skin and soft tissue infections, gastric and duodenal infections, trachoma eye infections & sexually transmitted diseases | Pawlowski et al., 2018 Leroy et al., 2021 |
|
| Third generation | 14-membered ketolides |
Telithromycin |
Semi-synthetic derivative of erythromycin | Gram-positive bacteria: S. pneumoniae Gram-negative bacteria: M. pneumoniae, C. pneumoniae, H. influenzae & L. pneumophilia |
Community-acquired respiratory tract infections |
Kaneko et al., 2007; Wolter et al 2008 |
| Cethromycin | Derivative of erythromycin | Gram-positive bacteria: Macrolide-resistant S. pneumoniae, S. pyogenes Gram-negative bacteria: H. influenzae |
Community acquired pneumonia | Mansour et al., 2016, Rafie et al., 2010 |
||
| 16-membered | Josamycin |
S. narbonensis var. josamyceticus | Gram-positive bacteria: S. aureus, S. pneumoniae, and S. pyogenes Gram-negative bacteria: H. influenzae, M. catarrhalis , M. genitalium, N. gonorrhea, N. meningitidis |
RTI, Urethritis |
Gupta et al., 2020 |
|
| Tylosin |
S. fradiae, H. influenzae |
H. influenzae, Gram-positive pathogens and mycoplasma | Respiratory diseases, mastitis, and dysentery in cattle and other farm animals |
Arsic et al., 2012 |
| Mechanism of resistance | Target/ Enzymes |
Genes | Organisms | Location/Associated MGEs/ Co-resistance determinants | Accession number | References |
|---|---|---|---|---|---|---|
| Efflux pumps | Mef proteins | mef(A) | Streptococcus pyogenes, S. pneumoniae, Staphylococcus aureus, Enterococcus spp. | Plasmid, Tn1207.1, Tn1207.2 |
U70055, AF227520, AF227521 |
Miklasinska-Majdanik 2021; Dinos 2017; Roberts et al., 1999 |
| mef(B) | Escherichia coli | Plasmid, Class 1 integron, IS440 | FJ196385 | |||
| mef(C) | Photobacterium damselae | Plasmid, tet(M), flo(R), Tet(C), Tet(D) | AB571865 | |||
| mef(D) | S. aureus | Chromosome | LR130509 | |||
| mef(E) | S. pneumoniae | Plasmid, Chromosome | U83667, AF274302 | |||
| mef(F) | Macrococcus canis | Plasmid | CP046364 | |||
| mef(G) | S. mitis | Chromosome | HG423652 | |||
| mef(H) | Clostridioides difficile | Chromosome | MW269960.1 | |||
| mef(I) | S. pneumoniae | Chromosome, Tn916, tet(M) | AJ971089 | |||
| mef(J) | S. pyogenes | Chromosome | CP065927.1 | |||
| mef(O) | S. pyogenes | Chromosome | DQ016305 | |||
| Msr proteins | msr(A) | S. epidermidis, S. aureus | Plasmids | X52085 | Fyfe et al., 2016; Roberts et al., 1999; Schwendener et al., 2020 |
|
| msr(B) | S. xylosus | Plasmid | M81802.1 | |||
| msr(C) | E. faecium | Chromosome | AF313494 | |||
| msr(D) | S. pyogenes | Tn1207.2 | AF227521 | |||
| msr(E) | Pasteurella multocida, S. pneumoniae, Klebsiella pneumoniae | Chromosome, Plasmid, Tn1207.1, Sul1, blaKPC-2, blaDHA-1, qnrB4, & armA |
FR751518, FJ628167.2, AF227520 | |||
| msr(F) | Macrococcus canis | Chromosome | MN728681 | |||
| msr(G) | M. canis | Plasmid | CP046364 | |||
| msr(H) | M. canis | Chromosome | BK011995 | |||
| msr(I) | S. pyogenes | Chromosome, tet(M) | CP065927.1 | |||
| Enzymatic degradation | Phosphotransferases | mph(A) | E. coli, | Plasmid, IS26 | D16251 |
Pawlowski et al., 2018; Golkar et al., 2018 |
| mph(B) | E. coli, | Plasmid | D85892 | |||
| mph(C) | S. aureus | Plasmid, msrA | AF167161 | |||
| mph(D) | Pseudomonas aeruginosa | Chromosome | AB048591 | |||
| mph(E) |
P. multocida, K. pneumoniae, Serratia marcescens, E. coli,Acinetobacter baumannii,Citrobacter freundii |
Chromosome, plasmid, IS26, ISCR1, ISEc28, ISEc29, IS26, ISAba125, Tn5393, IS18, ISAba3, Class 1 integron, ISEcp1, msr(E) blaKPC-2, blaDHA-1, qnrB4, armA, blaOXA-58, ant3/, linF, sul1, blaCTX-M, aac3, dhfr, aadA2, qac |
FR751518, FJ628167.2 FJ917355.1 FJ187822.1 EU294228.1 AY522431.4 AF550415.2 |
|||
| mph(F) | Uncultured bacterium | Plasmid | AM260957 | |||
| mph(G) |
Photobacterium damselae |
Plasmid, floR, tet (B), tet(C) tet (D), tet (M) | AB571865 | |||
| mph(H) |
Brachybacterium faecium |
Chromosome | NC_013172.1 | |||
| mph(I) | Paenibacillus sp | Chromosome | KX531056.1 | |||
| mph(J) |
Brevibacillus brevis |
Chromosome | KY753883.1 | |||
| mph(K) | Bacillus subtilis | Chromosome | NC_000964.3 | |||
| mph(L) |
B. cereus |
Plasmid, FosB, TetV, | ACMJ01000036.1 | |||
| mph(M) | B. cereus | Chromosome | AHFH01000066.1 | |||
| Macrolide esterases | ere(A) |
E. coli,Providencia stuartii, Enterobacter aerogenes |
Plasmids, Integrons, Tn7, sat, aadA1, dfr16, aac(6')-Ib |
M11277, AY183453 DQ157752. 1 AF512546.1 |
Zielinski et al., 2021 |
|
| ere(B) |
E. coli |
Plasmids | A15097, X03988 | |||
| ere(C) |
Riemerella anatipestifer |
Chromosome | CP004020 | |||
| ere(D) |
R. anatipestifer |
Chromosome | KP265721 | |||
| Ribosomal Modification | Mutation in 23S rRNA genes | erm(A) |
S. aureus |
Tn554 | KT803896.1 |
Gupta et al., 2013 |
| erm(B) |
S. pneumoniae |
Tn551, IS1216v | LC125351.1 |
|||
| erm(C) | ˇ S. aureus, S. cohnii | Tn917 | JQ219851.1 |
|||
| Mutation in ribosomal proteins | Mutation in ribosomal protein L4 & L22 |
S. pneumoniae, H. influenzae, and E. coli |
Chromosome | AF126059 |
Zaman et al,, 2007; Schroeder et al., 2016 |
|
| Mutation in ribosomal RNA | Mutation in A2058 & A2059 | S. pneumoniae, Helicobacter pyroli, E.coli | Chromosome, Plasmid | CP000936.1 |
Jiang et al., 2015; Dinos et al., 2017 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





