Submitted:
18 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rubinoff, D.; Holland, B.S. Between two extremes: Mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference. Syst. Biol. 2005, 54, 952–961. https://doi.org/10.1080/10635150500234674. [CrossRef]
- Will, K.W.; Mishler, B.D.; Wheeler, Q.D. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 2005, 54, 844–851. https://doi.org/10.1080/10635150500354878. [CrossRef]
- Knowles, L. L.; Carstens, B. C. Delimiting species without monophyletic gene trees. Syst. Biol. 2007, 56, 887–895. https://doi.org/10.1080/10635150701701091. [CrossRef]
- Talavera, G.; Lukhtanov, V.A.; Pierce, N.E.; Vila, R. Establishing criteria for higher-level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae). Cladistics 2013, 29, 166–192. https://doi.org/10.1111/j.1096-0031.2012.00421.x. [CrossRef]
- Hernández-Roldán, J.L.; Dapporto, L.; Dincă, V.; Vicente, J.C.; Hornett, E.A.; Šíchová, J.; Lukhtanov, V.A.; Talavera, G.; Vila, R. Integrative analyses unveil speciation linked to host plant shift in Spialia butterflies. Mol. Ecol. 2016, 25, 4267–4284. https://doi.org/10.1111/mec.13756. [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Royal Soc. B: Biol. Sci. 2003, 270, 313–321. https://doi.org/10.1098/rspb.2002.2218. [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Nat. Acad. Sci. USA 2004, 101, 14812–14817. https://doi.org/10.1073/pnas.0406166101. [CrossRef]
- Talavera, G.; Lukhtanov, V.A.; Pierce, N.; Vila, R. DNA barcodes combined with multilocus data of representative taxa can generate reliable higher-level phylogenies. Syst. Biol. 2022, 71, 382–395. https://doi.org/10.1093/sysbio/syab038. [CrossRef]
- Kandul, N.P.; Lukhtanov, V.A.; Dantchenko, A.V.; Coleman, J.W.S.; Sekercioglu, C.H.; Haig, D.; Pierce, N.E. Phylogeny of Agrodiaetus Hübner 1822 (Lepidoptera: Lycaenidae) inferred from mtDNA sequences of COI and COII and nuclear sequences of EF1-α: Karyotype diversification and species radiation. Syst. Biol. 2004, 53, 278–298. https://doi.org/10.1080/10635150490423692. [CrossRef]
- Kandul, N.P.; Lukhtanov, V.A.; Pierce, N.E. Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 2007, 61, 546-559. https://doi.org/10.1111/j.1558-5646.2007.00046.x. [CrossRef]
- Eckweiler, W.; Bozano, G.C. Guide to the butterflies of the Palearctic region. Lycaenidae part IV. Omnes artes, Milano, Italy, 2016.
- de Lesse, H. Les nombres de chromosomes dans la classification du groupe d’Agrodiaetus ripartii Freyer (Lepidoptera, Lycaenidae). Rev. Fran. Entomol. 1960, 27, 240–264.
- de Lesse, H. Spéciation et variation chromosomique chez les Lépidoptères Rhopaloceres. Ann. Sci. Nat. Zool. Biol. Anim, 12e série. 1960, 2: 1–223.
- Eckweiler, W.; Häuser, C.L. An illustrated checklist of Agrodiaetus Hübner, 1822, a subgenus of Polyommatus Latreille, 1804 (Lepidoptera: Lycaenidae). Nachr. Entomol. Ver. Apollo 1997, Suppl. 16, 113–168.
- Olivier, A.; Puplesiene, J.; van der Pooten, D.; De Prins, W.; Wiemers, M. (1999) Revision of some taxa of Polyommatus (Agrodiaetus) transcaspicus group with description of a new species from Central Anatolia (Lepidoptera, Lycaenidae). Phegea 1999, 27, 1–24.
- Carbonell, F. Contribution a la conaissance du genre Agrodiaetus Hübner (1822), A. barmifiruze n. sp. et A. musa esfahensis n. ssp, en Iran méridional. Linneana Belgica 2000, 17, 211–217.
- Carbonell, F. (2001) Contribution a la connaissance du genre Agrodiaetus Hübner (1822), A. ahmadi et A. khorasanensis nouvelles espèces dans le Nord de l’Iran (Lepidoptera, Lycaenidae). Linneana Belgica 2001, 18, 105–110.
- Dantchenko, A.V. Genus Agrodiaetus. In: Tuzov, V.K. (ed.) Guide to the butterflies of Russia and adjacent territories, Vol. 2. Pensoft, Sofia & Moscow, 2000, 196–214.
- ten Hagen, W.; Eckweiler, W. Eine neue Art von Polyommatus (Agrodiaetus) aus Zentraliran (Lepidoptera, Lycaenidae). Nachr. Entomol. Ver. Apollo 2001, 22, 53–56.
- Skala, P. New taxa of the subgenus Agrodiaetus Hübner, 1822 from Iran: Polyommatus (Agrodiaetus) faramarzii sp. n., P. (A.) shahrami sp. n. and P. (A.) pfeifferi astyages ssp. n. (Lepidoptera, Lycaenidae). Nachr. Entomol. Ver. Apollo 2001, 22, 101–108.
- Wiemers, M. Chromosome differentiation and the radiation of the butterfly subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus) a molecular phylogenetic approach. Ph.D. Dissertation, University of Bonn, Bonn, Germany, 2003. 203 pp. http://nbn-resolving.de/urn:nbn:de:hbz:5n-02787.
- Schurian, K.G., ten Hagen W. Polyommatus (Agrodiaetus) urmiaensis sp. n. aus Nordwestiran (Lepidoptera: Lycaenidae). Nachr. Entomol. Ver. Apollo 2003, 24, 1–5.
- Vila, R.; Lukhtanov, V.A.; Talavera, G.; Gil-T.F.; & Pierce, N. E. (2010). How common are dot-like distribution ranges? Taxonomical oversplitting in Western European Agrodiaetus (Lepidoptera, Lycaenidae) revealed by chromosomal and molecular markers. Biol. J. Linn. Soc. 2010, 101, 130–154. https://doi.org/10.1111/j.1095-8312.2010.01481.x. [CrossRef]
- Lukhtanov, V.A.; Kandul, N.P.; Plotkin, J.B.; Dantchenko, A.V.; Haig, D.; Pierce, N.E. Reinforcement of pre-zygotic isolation and karyotype evolution in Agrodiaetus butterflies. Nature 2005, 436, 385–389. https://doi.org/10.1038/nature/03704. [CrossRef]
- Wiemers, M.; Keller, A.; Wolf, M. ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evol. Biol. 2009, 9, 300. https://doi.org/10.1186/1471-2148-9-300. [CrossRef]
- Przybyłowicz, Ł.; Lukhtanov, V.A.; Lachowska-Cierlik, D. Towards the understanding of the origin of the Polish remote population of Polyommatus (Agrodiaetus) ripartii (Lepidoptera: Lycaenidae) based on karyology and molecular phylogeny. J. Zool. Syst. Evol. Res. 2014, 52, 44–51. https://doi.org/10.1111/jzs.12040. [CrossRef]
- Vishnevskaya, M.S.; Saifitdinova, A.F.; Lukhtanov, V.A. Karyosystematics and molecular taxonomy of the anomalous blue butterflies (Lepidoptera, Lycaenidae) from the Balkan Peninsula. Comp. Cytogen. 2016, 10, 1–85. https://doi.org/10.3897/CompCytogen.v10i5.10944. [CrossRef]
- Vershinina, A.O.; Lukhtanov, V.A. Evolutionary mechanisms of runaway chromosome number change in Agrodiaetus butterflies. Sci. Rep. 2017, 7, 8199. https://doi.org/10.1038/s41598-017-08525-6. [CrossRef]
- Lukhtanov, V.A.; Shapoval, N.A.; Dantchenko, A.V. Taxonomic position of several enigmatic Polyommatus (Agrodiaetus) species (Lepidoptera, Lycaenidae) from Central and Eastern Iran: insights from molecular and chromosomal data. Comp.Cytogen. 2014, 8, 313–322. https://doi.org/10.3897/CompCytogen.v8i4.8939. [CrossRef]
- Lukhtanov, V.A.; Dantchenko, A.V. 2017. A new butterfly species from south Russia revealed through chromosomal and molecular analysis of the Polyommatus (Agrodiaetus) damonides complex (Lepidoptera, Lycaenidae). Comp. Cytogen. 2017, 11, 769–795, doi:10.3897/CompCytogen.v11i4.20072. [CrossRef]
- Lukhtanov, V.A.; Sourakov, A.; Zakharov, E.V.; Hebert, P.D.N. DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification. Mol. Ecol. Resour. 2009, 9, 1302–1310. https://doi.org/10.1111/j.1755-0998.2009.02577.x. [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299.
- deWaard, J.R.; Ivanova, N.V.; Hajibabaei, M.; Hebert, P.D.N. Assembling DNA barcodes: analytical protocols. In Environmental Genomics, Methods in Molecular Biology; Martin, C.C., Ed.; Humana Press: Totowa, N.J., United States, 2008; Volume 410, pp. 275–283, ISBN-13: 978-1588297778.
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61.
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. https://doi.org/10.1038/nmeth.2109. [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. https://doi.org/10.1093/sysbio/sys029. [CrossRef]
- Balint, Z. Reformation of the Polyommatus section with a taxonomic and biogeographic overview (Lepidoptera, Lycaenidae, Polyommatini). Neue Entomol. Nachricht. 1997, 40, 1–67.
- Coutsis, J.G. Notes concerning the taxonomic status of Agrodiaetus tankeri de Lesse (Lepidoptera: Lycaenidae). Nota Lep. 1985, 8, 8-14.
- Coutsis, J.G. The blue butterflies of the genus Agrodiaetus Hübner (Lep., Lycaenidae): symptoms of taxonomic confusion. Nota Lep. 1986, 9, 159-169.
- Parmentier, L.; Vila, R.; Lukhtanov, V.A. Integrative analysis reveals cryptic speciation linked to habitat differentiation within Albanian populations of the anomalous blues (Lepidoptera, Lycaenidae, Polyommatus Latreille, 1804). Comp. Cytogen. 2022, 16, 211–242. https://doi.org/10.3897/compcytogen.v16.i4.90558. [CrossRef]
- Pazhenkova, E.A.; Lukhtanov, V.A. Сhromosomal conservatism vs chromosomal megaevolution: enigma of karyotypic evolution in Lepidoptera. Chromosome Research 2023, 31, 16. https://doi.org/10.1007/s10577-023-09725-9. [CrossRef]
- Hesselbarth, G.; Van Oorschot, H.; Wagener, S. Die Tagfalter der Türkei; Bocholt: Selbstverlag Sigbert Wagener, 1995; vol. 1, 754 pp.
- Koçak, A.O.; Kemal, M. 2001. A study on the biodiversity, zoogeography and taxonomy of the section Agrodiaetus Hbn. in the genus Polyommatus Latr. (Lycaenidae, Lepidoptera). Centre for Entomological Studies Miscellaneous Papers 2001, 78-79, 1-11.
- International Commission on Zoological Nomenclature. International Code of Zoological Nomenclature. The International Trust for Zoological Nomenclature, 1999. https://www.iczn.org/the-code/the-code-online/.

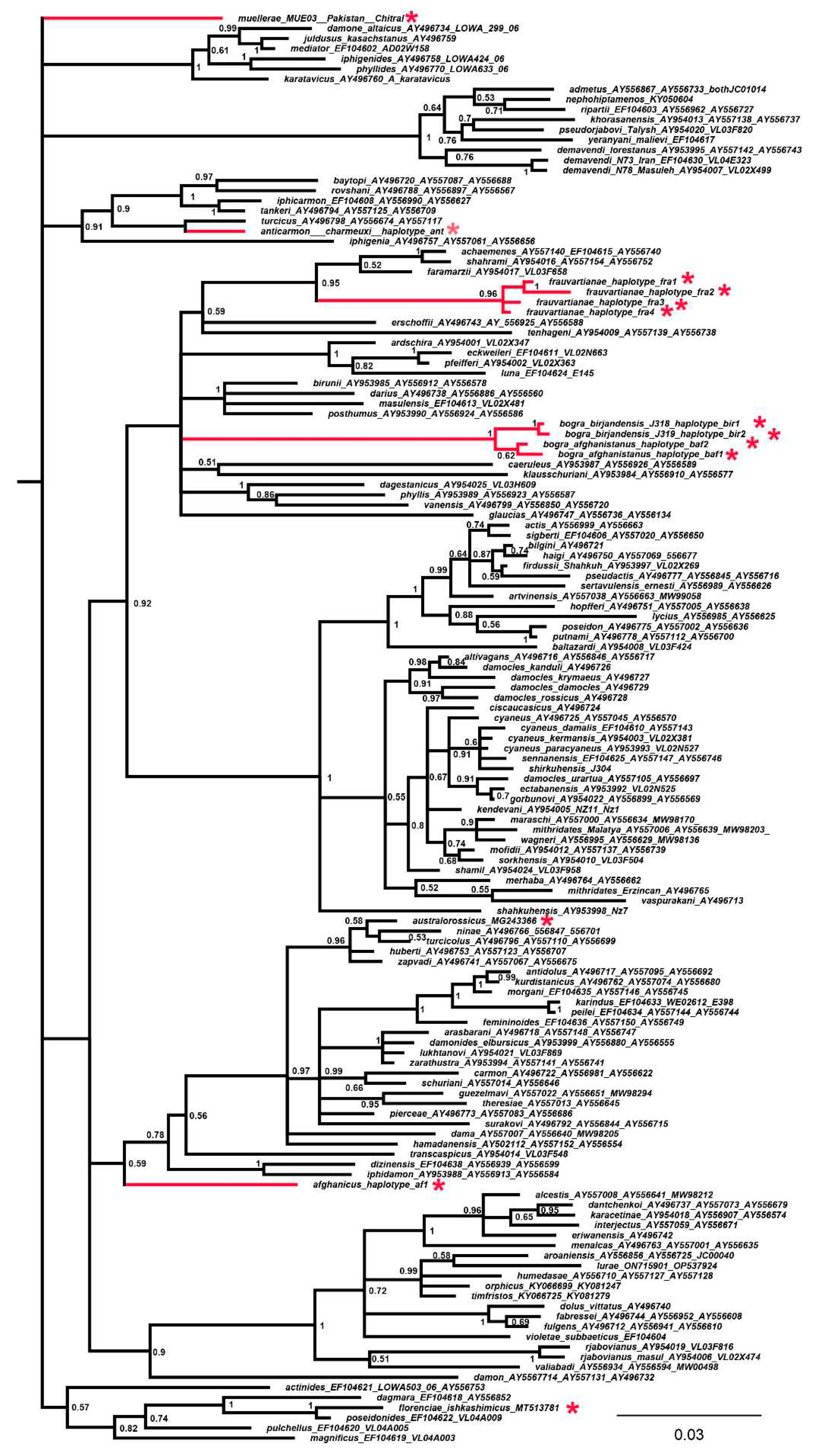
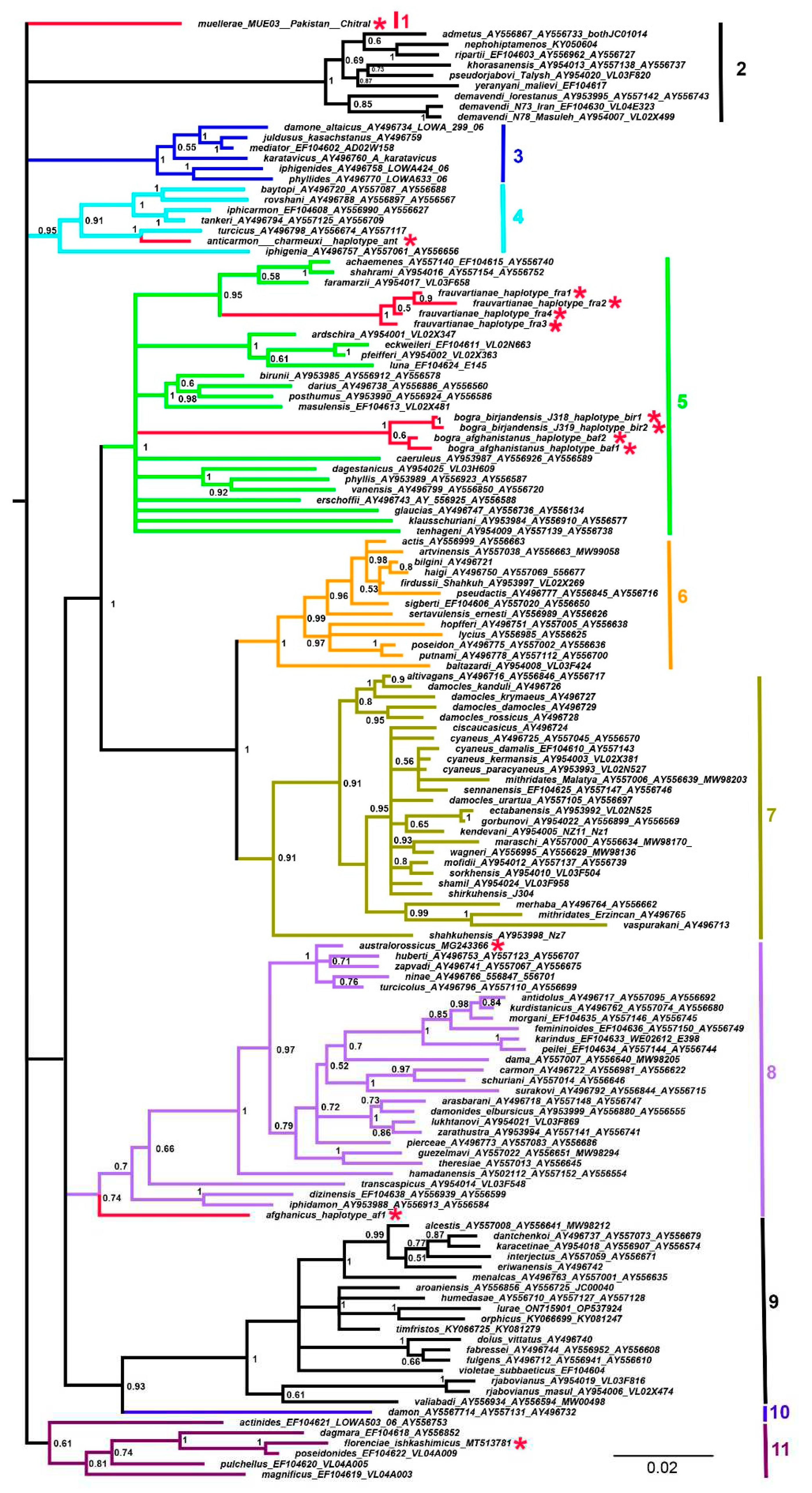

| Species | Laboratory ID | GeneBank | Haplotype | Country | Locality |
|---|---|---|---|---|---|
| P. (A.) afghanicus | AF05 | OR413713 | af1 | Afghanistan | Koh-i-Baba, Band-i-Amir, 34.8294° N, 67.1805° E, 2900-3000m., 2.07.2009, Yu. Skrylnik leg. |
| BPAL2125 | OR413714 | af1 | Afghanistan | near Kabul, July 2010, I. Pljushch leg. | |
| BPAL2126 | OR413715 | af1 | Afghanistan | the same locality | |
| BPAL2127 | OR413716 | af1 | Afghanistan | the same locality | |
| BPAL2128 | OR413717 | af1 | Afghanistan | the same locality | |
| P. (A.) anticarmon (=charmeuxi) | CHAR01 | OR424389 | ant | Turkey | Hakkari Prov., vic. Üzümcū 1300m, 28.06-04.07.1976 |
| P. (A.) bogra afghanistanus | XX21 | OR424390 | baf1 | Afghanistan | Bamyan prov., 8 km S Bamyan, 2700m, 31.05.2010, O. Pak leg. |
| AAF02 | OR424391 | baf2 | Afghanistan | the same locality | |
| AAF10 | OR424392 | baf1 | Afghanistan | Bamiyan prov., 34.2155° N, 66.8994° E, 2545m, 23.06.2016. O. Pak leg. |
|
| AAF11 | OR424393 | baf1 | Afghanistan | the same locality | |
| P. (A.) bogra birjandensis | J318 | OR413718 | bir1 | Iran | South Khorasan Prov., 26 km N of Birjand, 1900- 2000m, 14.07.2005, V. Lukhtanov leg. |
| J319 | OR413719 | bir2 | Iran | the same locality | |
| P. (A.) frauvartianae | AAF01 | OR424394 | fra1 | Afghanistan | Bamiyan prov., Yakawlang District, Bandi-Amir env., 3300m, 03.08.2011, O. Pak leg. |
| AAF03 | OR424395 | fra2 | Afghanistan | the same locality | |
| AAF04 | OR424396 | fra1 | Afghanistan | the same locality | |
| AAF05 | OR424397 | fra1 | Afghanistan | the same locality | |
| AAF07 | OR424398 | fra3 | Afghanistan | the same locality | |
| AAF08 | OR424399 | fra4 | Afghanistan | Bamiyan prov., Panjab District, Kohi-Baba Mts., Rashak Mts., Shatu Pass, 3490m, 06.08.2011, O. Pak leg. | |
| AAF09 | OR424400 | fra1 | Afghanistan | the same locality | |
| P. (A.) muellerae | MUE03 | OR413720 | mu1 | Pakistan | Chitral, Birmogh Lasht, 35.8981°N, 71.7712°E, 2600-3000m, 01.07.2001, leg. V. Tuzov |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





