Submitted:
18 August 2023
Posted:
21 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
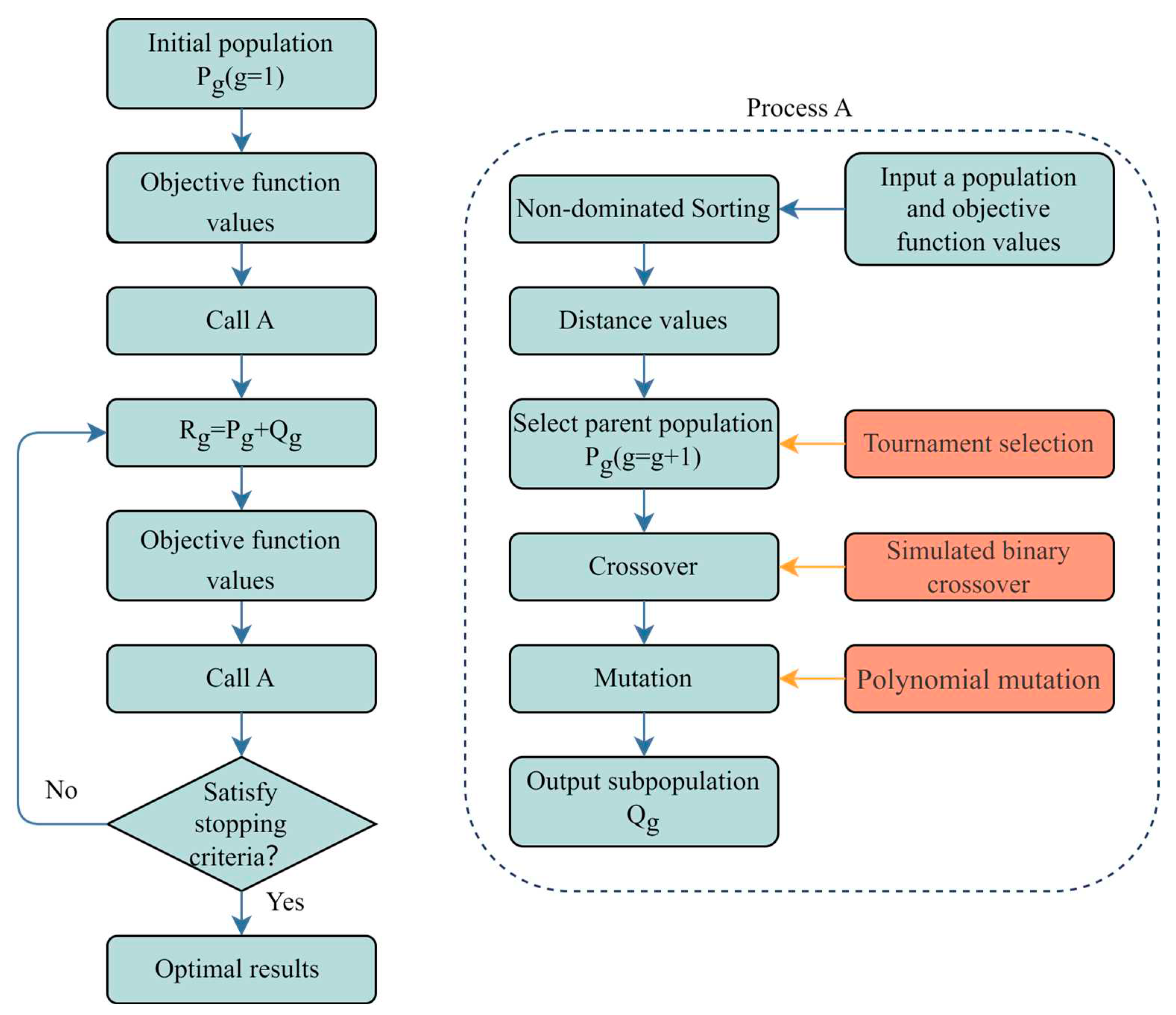
2. Method Description
2.1. NSGA-II
2.2. Ordinary Kriging (OK) Model
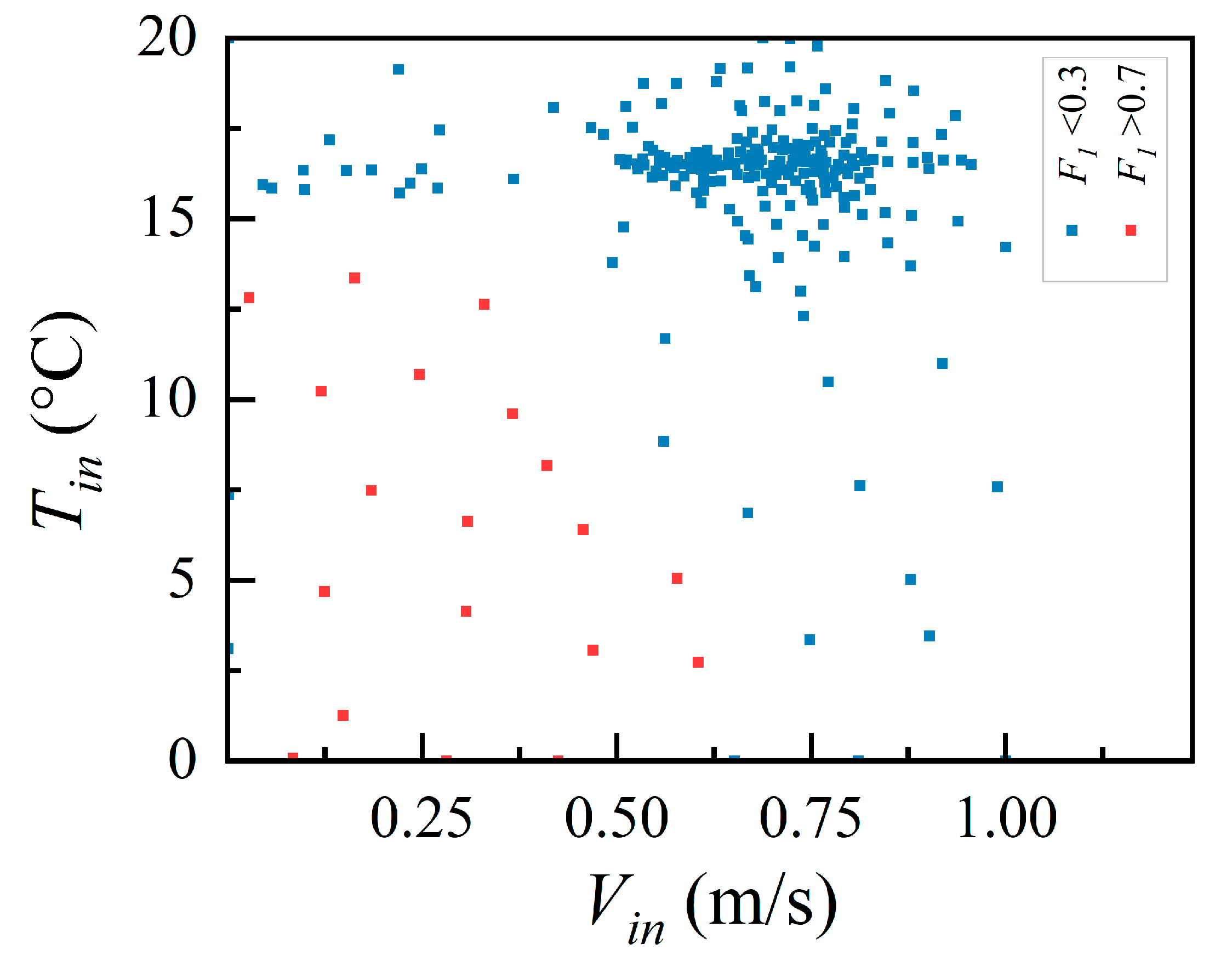
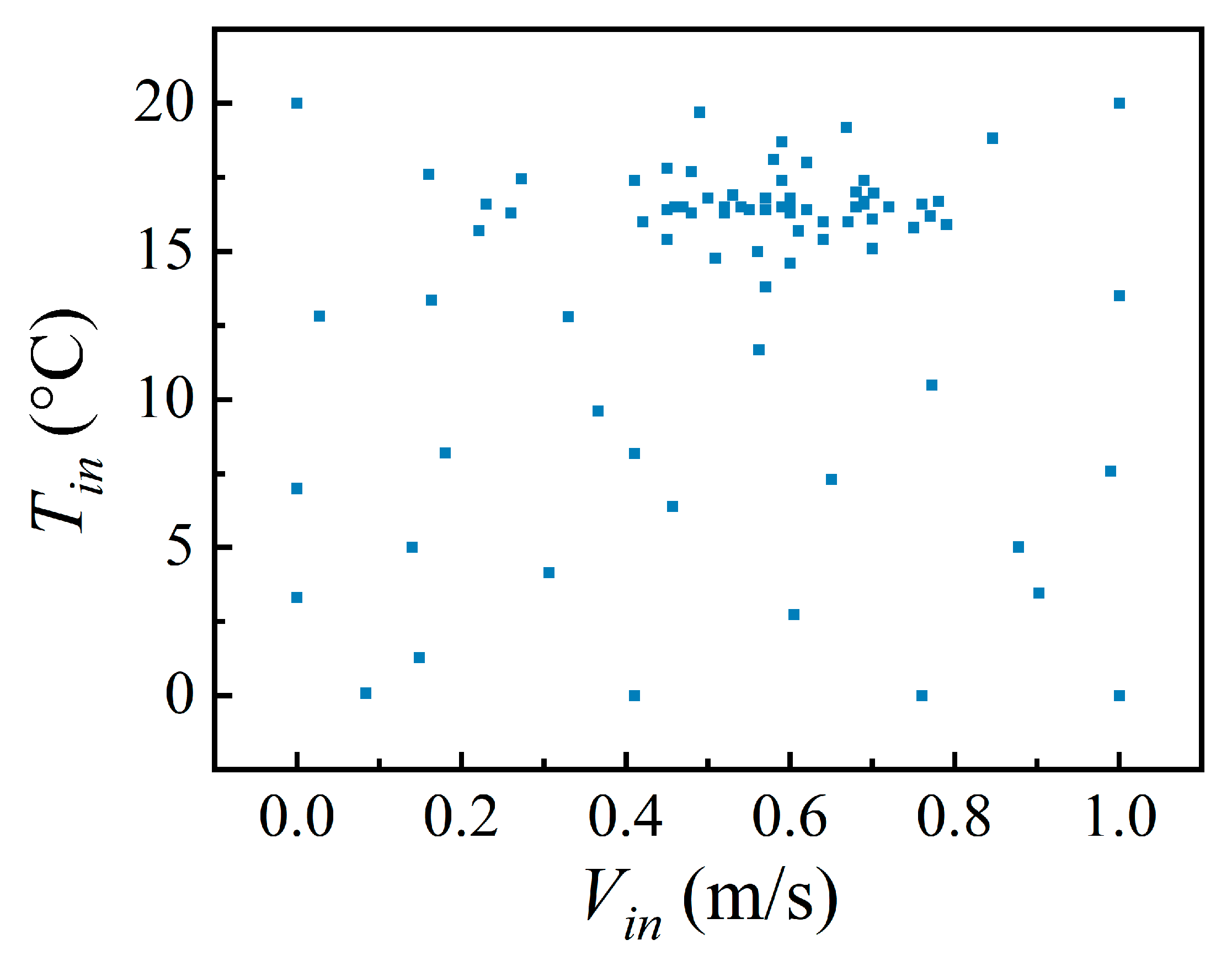
2.3. Latin Hypercube Sampling
2.4. Infill Criteria
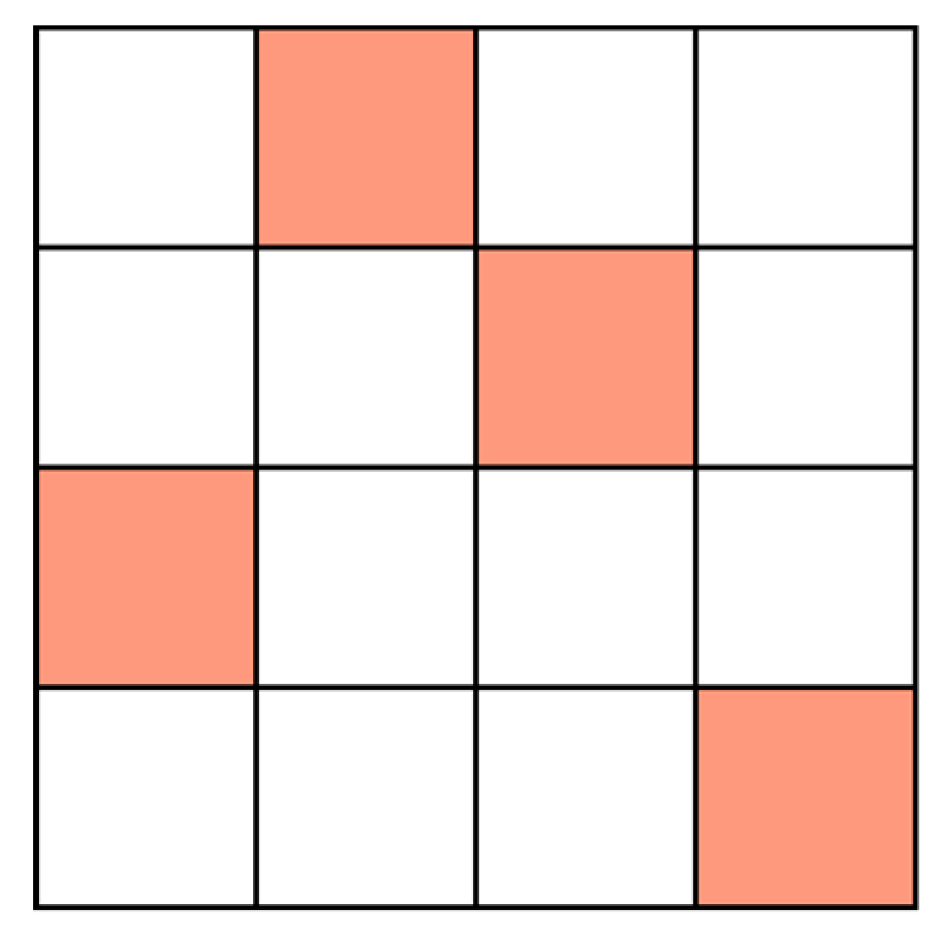
- All objects (points) in the initial set X are marked as 'unvisited';
- Select an unvisited object x randomly, mark x as 'visited', and check whether the neighborhood of x contains at least p objects;
- If not, then x is marked as a noise point. Otherwise, a new cluster C is created for x, and all objects in the neighborhood of x are placed in the candidate set N;
- Add objects that do not belong to other clusters from N to C iteratively. In this process, for an object u from N marked as 'unvisited', mark it as 'visited' and check its neighborhood, and if the neighborhood of u contains at least p objects, then all objects in the neighbourhood of u are added to N. Continue adding objects to C until C cannot be extended (N is empty). Then the generation of cluster C is complete;
- Select an unvisited object at random from the remaining objects and repeat steps 2 and 3 until all objects have been visited.
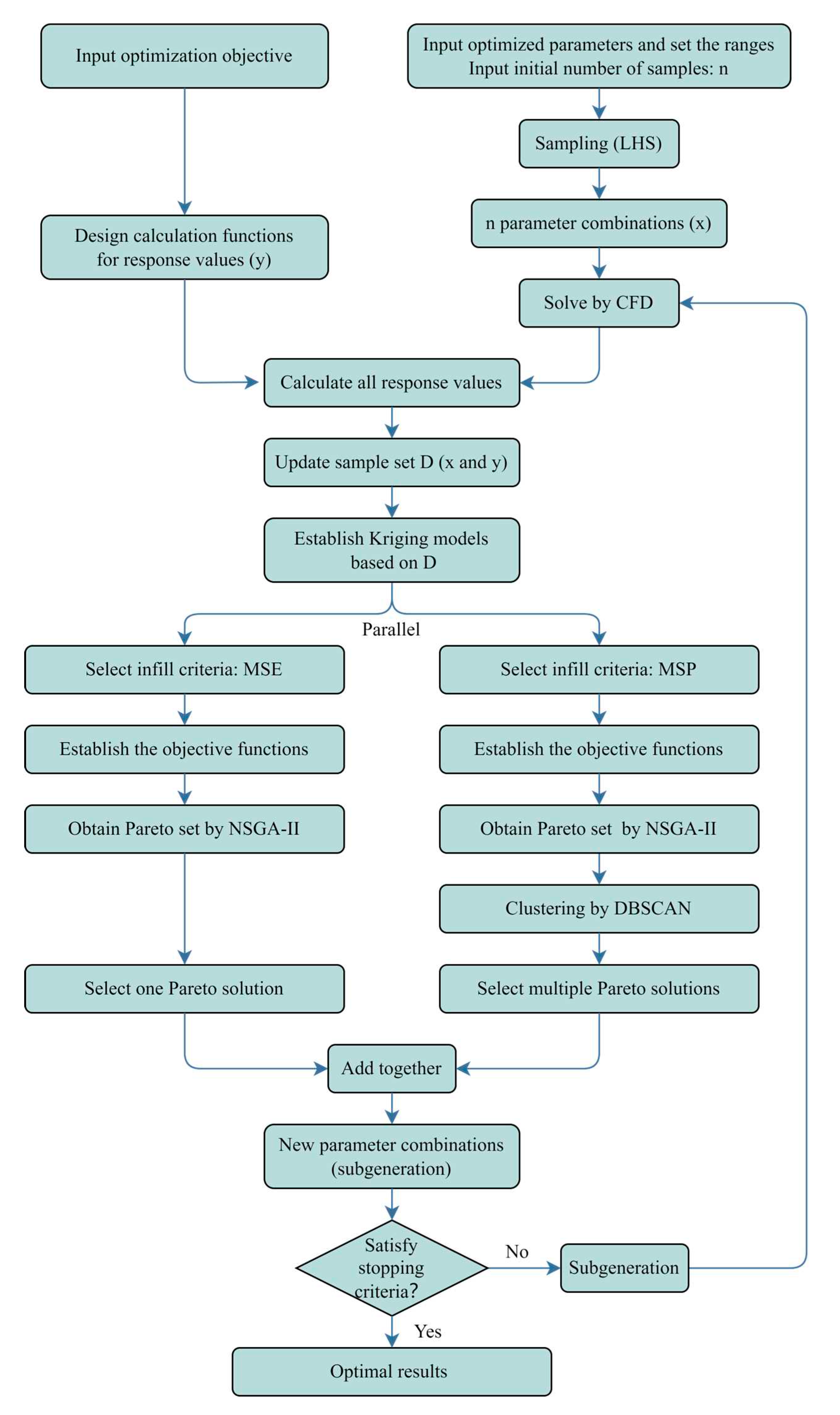
2.5. Process of Optimization
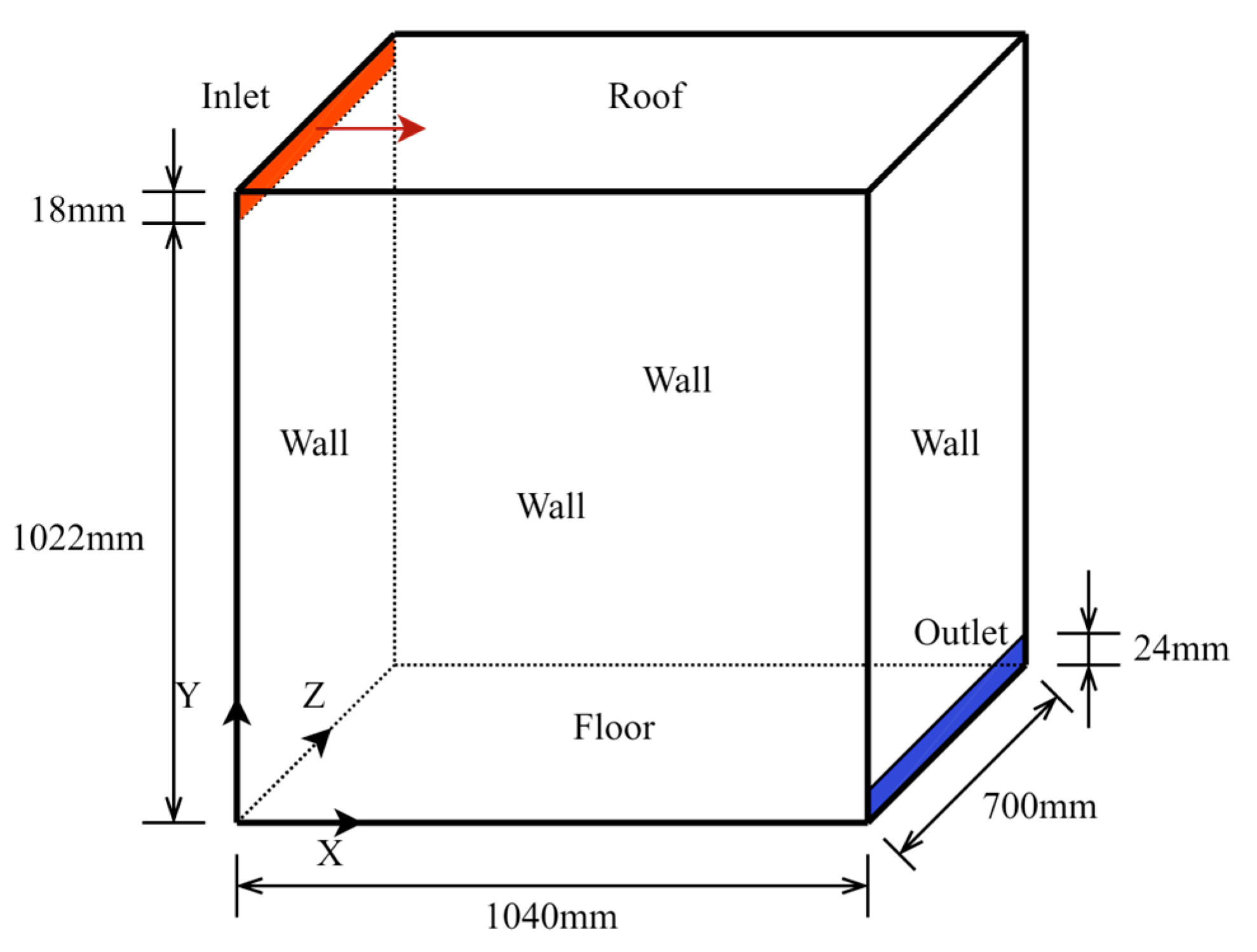
3. Testing with a Simple Room Case
3.1. Descriptions of the Case
3.2. Test Method
- Construct the objective/cost function as in Eq. (7) (minimization, which can be seen as a "distance");
- Set parameter range: 0 < Vin < 1 m/s, 0 < Tin < 20 ℃;
- Minimize the objective function by a single-objective genetic algorithm and output the corresponding parameters.
- Construct the objective function as in Eq. (8). Using MSP criteria as an example;
- 2.
- Set parameter range: 0 < Vin < 1 m/s, 0 < Tin <20 ℃.
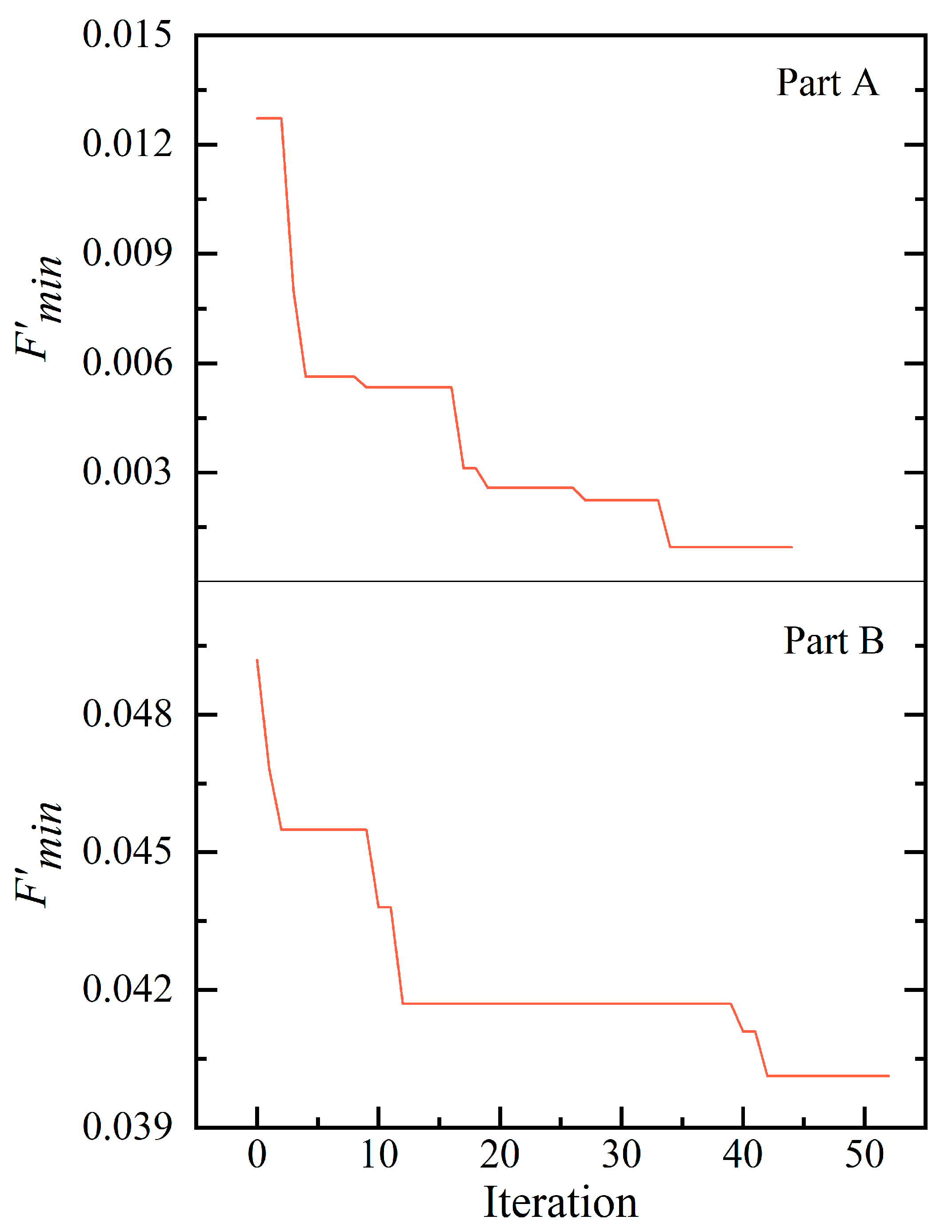
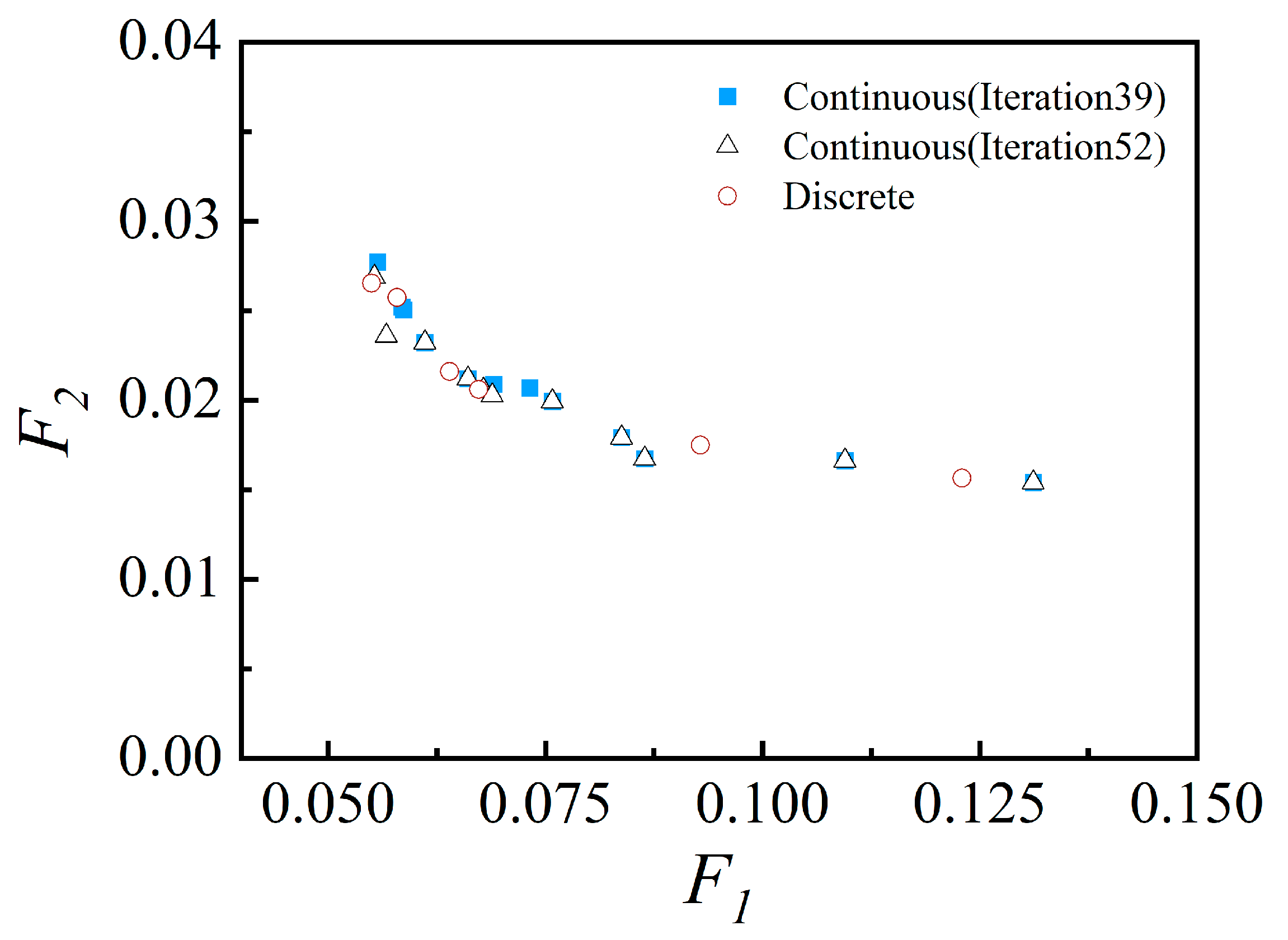
3.3. Inverse Prediction Results
3.3.1. Based on Simulation Data and 2D Simulations
3.3.2. Based on Experimental Data and 3D Simulations
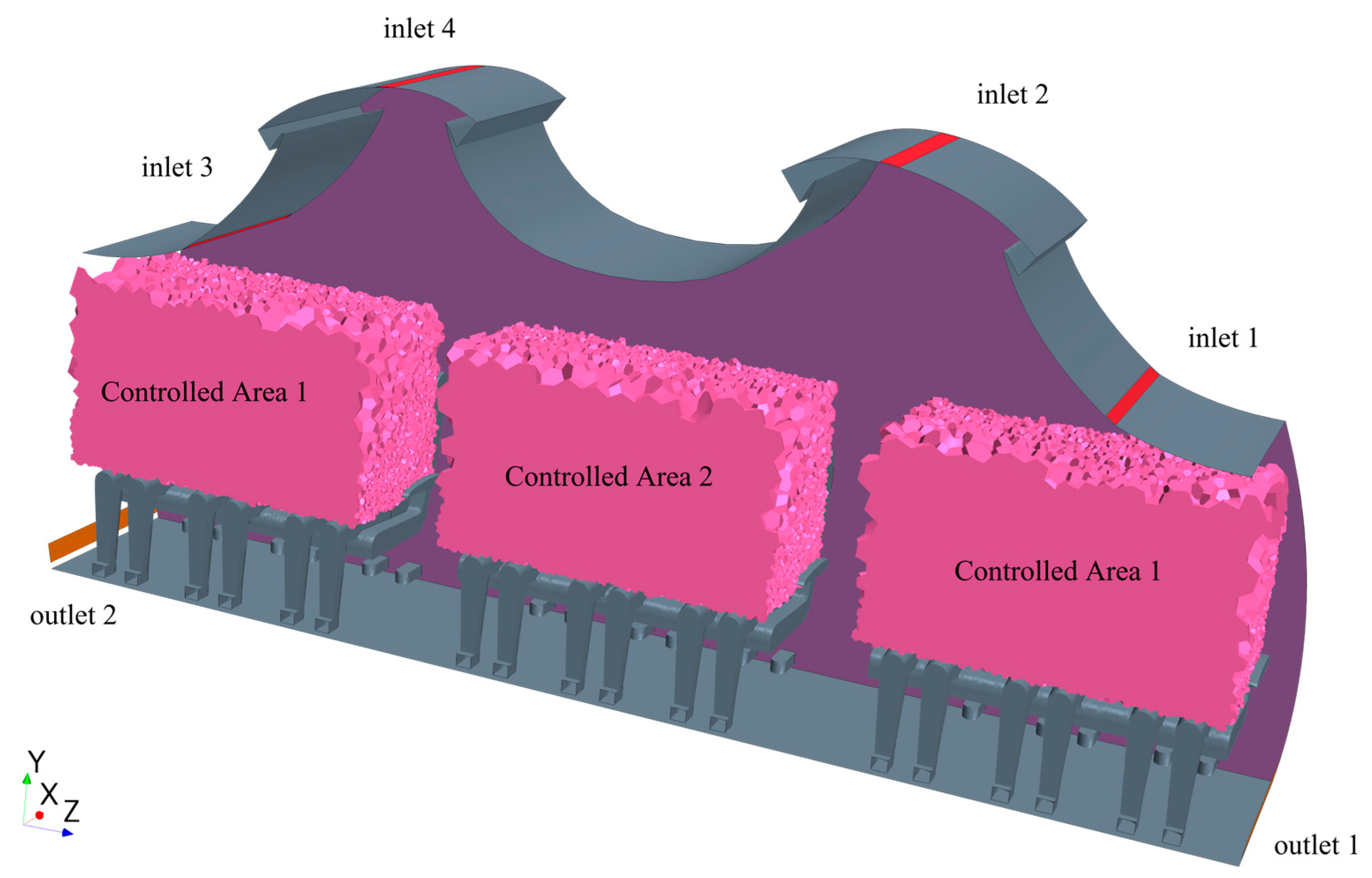
4. Optimizing the Air Supply Parameters of a Dual-Aisle Single-Row Cabin
4.1. Case Descriptions
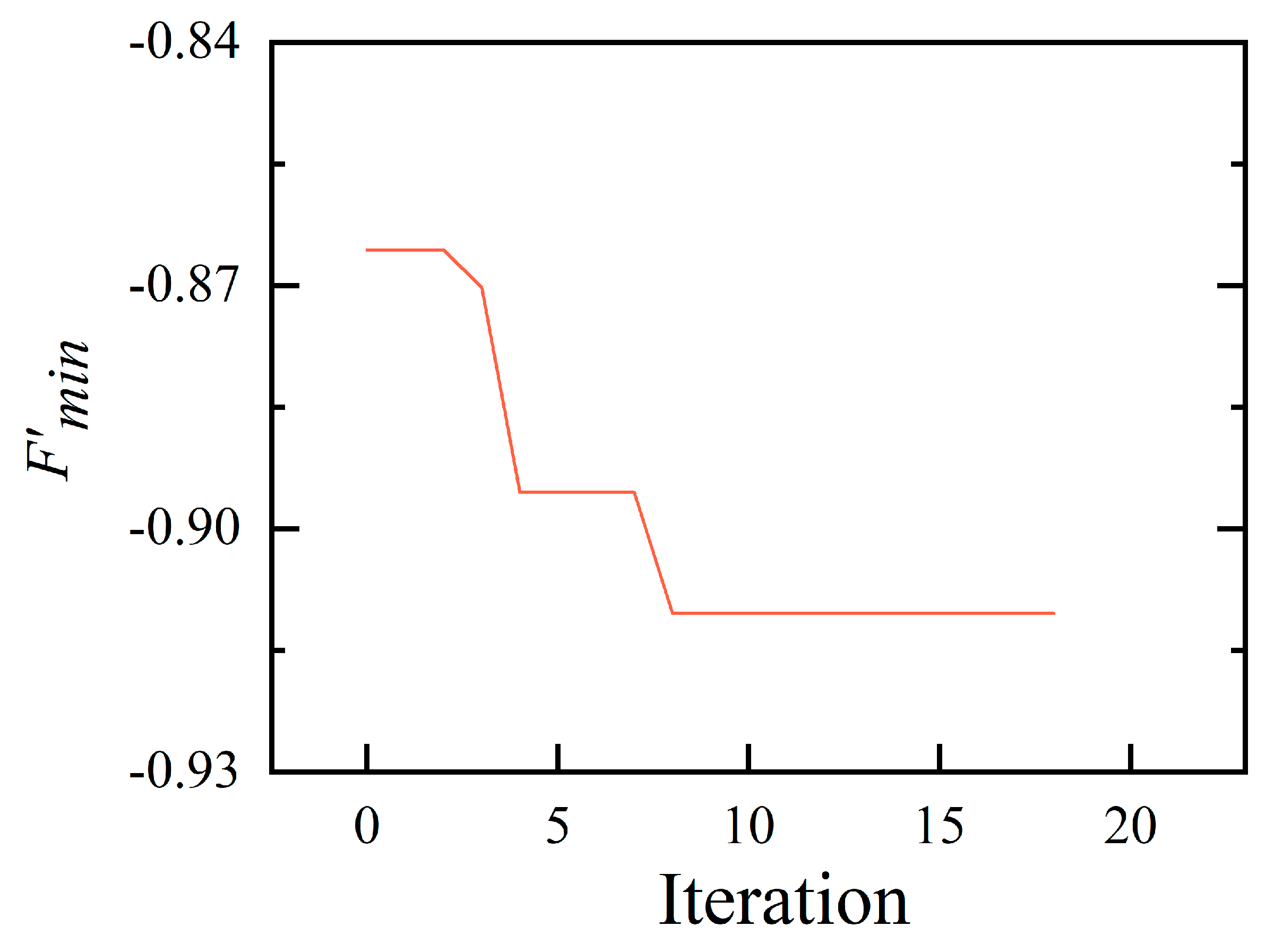
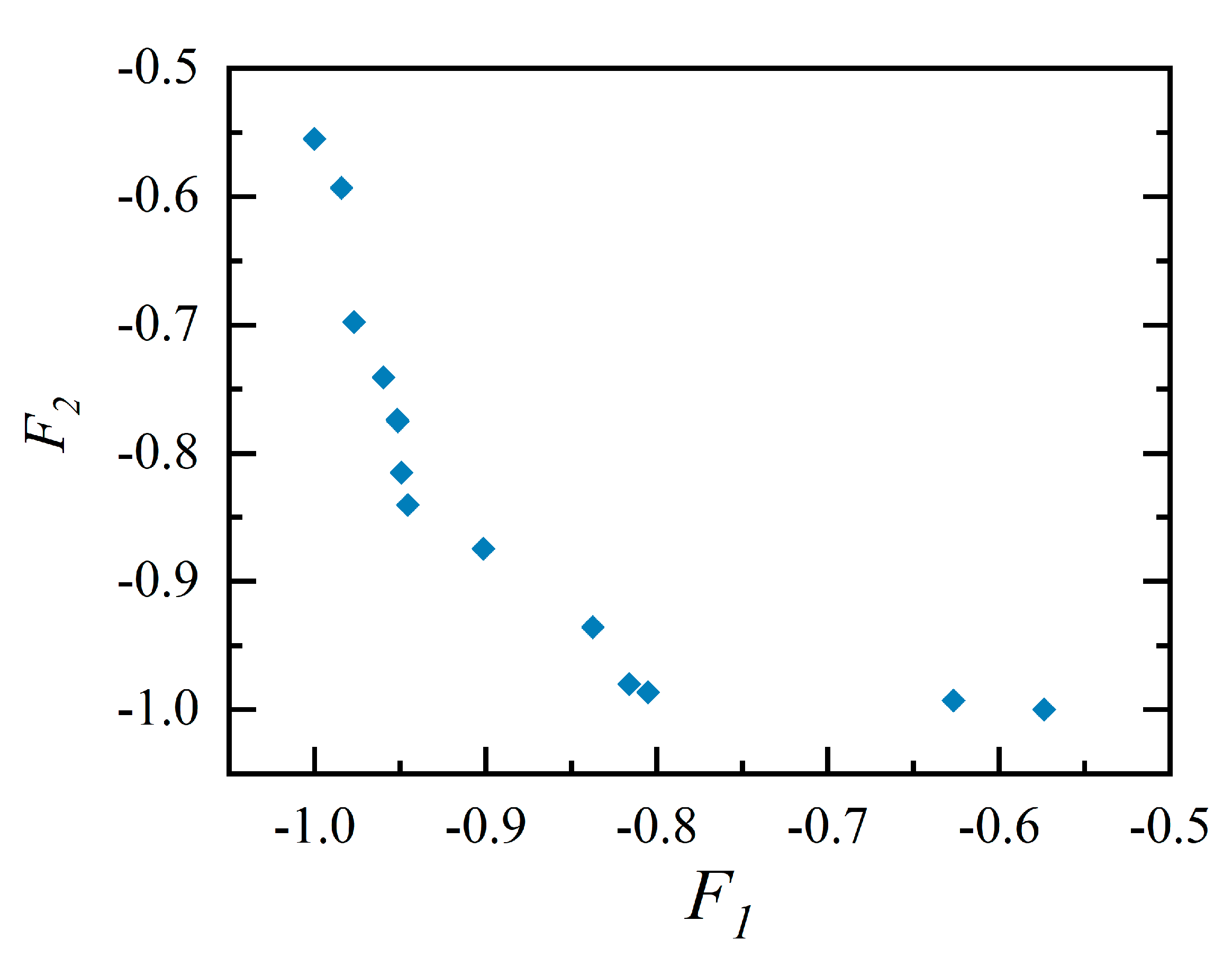
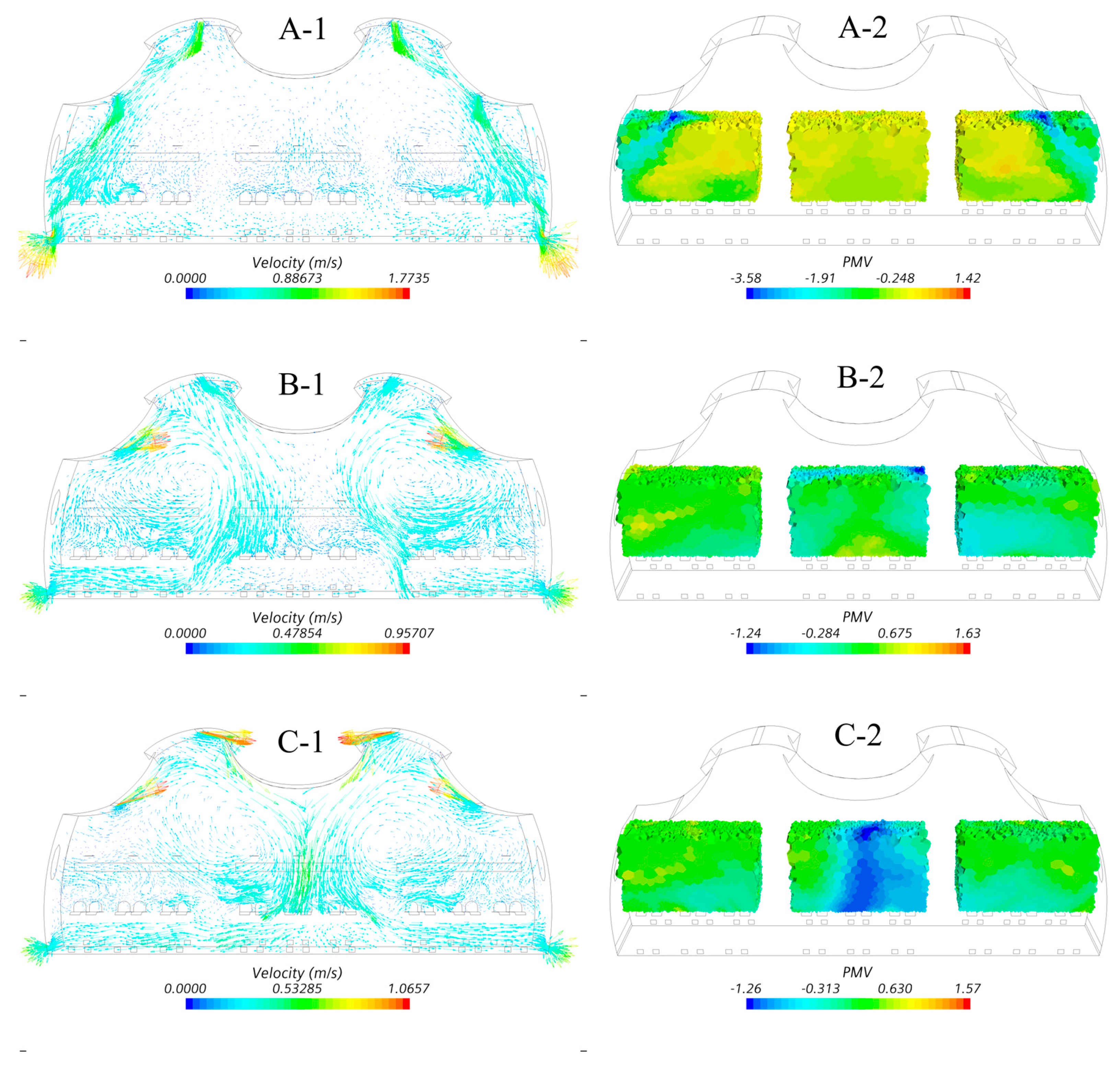
4.2. Optimization Results
5. Discussions
6. Conclusions
- In some common scenarios, the method in this paper can quickly provide an approximate Pareto Frontier. However, in the face of scenarios where the response values change rapidly, are locally non-differentiable, or are intermittent (surrogate models generate large gradients, and model errors make it challenging to optimize correctly), the optimization may stall. Because this paper assumes that the response values should be continuous and smooth, and then the Kriging model is used. Therefore, the method is more suitable for scenarios where the response values vary gently. In optimization based on 3D steady-state simulation, the use of discrete optimization with an appropriate discrete length can alleviate the above problems, speed up the optimization, and maintain accuracy.
- The method in this paper, as a development of meta-heuristic optimization approaches, also has global search capability when a sufficient number of initial samples are provided. The difference is that the method generates a subgeneration with few individuals based on the prediction of the surrogate model, which improves the utilization of the samples, makes the subgeneration more likely to be generated in the right direction, greatly reduces the total number of samples, and reduces the computational cost. However, compared with the meta-heuristic optimization approaches that use selection operator, crossover operator and mutation operator to generate a subgeneration with many individuals directly, the diversity of subgeneration and the adaptability of method decrease.
- After dividing the dual-aisle cabin into two zones, the air supply parameters were optimized using this paper's method. 118 samples (cases) were calculated to obtain an approximate Pareto Frontier. For such a 5-parameter optimization process, thousands of samples may be required based on meta-heuristic optimization approaches. When using the POD method, if each parameter is divided into 4 intervals, uniform sampling requires 3125 cases to compose the initial database. The generated Pareto set suggests that an airflow organization with a left-right symmetric circulation structure may be optimal.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Nomenclature
| c | Coefficient of OK model (-) | y(x) | The distribution of predicted value |
| C | Cluster (-) | z(x) | Error function of the Kriging model |
| d | Neighborhood radius (-) | ||
| D | Sample set (-) | Superscripts/Subscripts | |
| m | Dimensionality (-) | - | Average |
| n | Initial sample size (-) | ^ | Predicted value |
| N | Candidate set in clustering (-) | 1 / 2 | A number |
| p | Minimum number of points in clustering (-) | i / j | A number |
| s | Number of iteration steps (-) | in | Inlet |
| T | Temperature (℃) | n | A number |
| u | Object in clustering (-) | ||
| V | Velocity (m/s) | Greek symbols | |
| x | Variable (-) | σ | Standard deviation (-) |
| X | Initial set in clustering (-) | θ | Angle (°) |
| Functions | Abbreviations | ||
| average(x) | Average function | ANN | Artificial neural networks |
| Cov(xi,xj) | Covariance function | LHS | Latin hypercube sampling |
| f(x) | Basis function of the Kriging model | MSE | Mean squared error |
| F(x) | Objective (cost) function | MSP | Minimizing surrogate model prediction |
| F'(x) | Combinatorial function of the objective function | NSGA | Non-dominated Sorting Genetic Algorithms |
| g(x) | Constraint function | OK | Ordinary Kriging model |
| IF | Logic function | PMV | Predicted Mean Vote |
| max(x) | Find the maximum | POD | Proper orthogonal decomposition |
| R(xi,xj) | Function of correlation coefficient | PSO | Particle swarm optimization |
Appendix A
| y=0.52m, z=0.35m | ||||||||||
| i | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| xi (m) | 0.024 | 0.063 | 0.102 | 0.205 | 0.354 | 0.709 | 0.859 | 0.961 | 0.985 | 1.020 |
| uyi (m/s) | 0.220 | 0.240 | 0.222 | 0.140 | 0.068 | -0.06 | -0.125 | -0.204 | -0.268 | -0.270 |
| Notes: Data from Blay's experimental study [55]. | ||||||||||
Appendix B
| y=0.52m, z=0.35m | |||||||||||
| j | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| xj (m) | 0.00 | 0.02 | 0.038 | 0.084 | 0.229 | 0.520 | 0.811 | 0.964 | 1.00 | 1.01 | 1.04 |
| tj (K) | 288 | 292 | 293 | 293 | 292 | 292 | 292 | 291 | 291 | 291 | 288 |
| Notes: Data from Blay's experimental study [55]. | |||||||||||
References
- Pironneau, O. (1974). On optimum design in fluid mechanics. Journal of fluid mechanics, 64(1), 97-110. [CrossRef]
- Liu, W., & Chen, Q. (2015). Optimal air distribution design in enclosed spaces using an adjoint method. Inverse Problems in Science and Engineering, 23(5), 760-779. [CrossRef]
- Malkawi, A. M., Srinivasan, R. S., Yi, Y. K., & Choudhary, R. (2003). Performance-based design evolution: The use of genetic algorithms and CFD. Eighth International IBPSA. Eindhoven, Netherlands, 793-798.
- Xue, Y., Zhai, Z. J., & Chen, Q. (2013). Inverse prediction and optimization of flow control conditions for confined spaces using a CFD-based genetic algorithm. Building and Environment,64, 77-84. [CrossRef]
- Wang, J., Zhou, H., Zhang, T., & Wang, S. (2014). Inverse design of aircraft cabin environment based on proper decomposition of thermo-flow fields. In Proceedings of the 13th International Conference on Indoor Air Quality and Climate (Indoor Air 2014).
- Liu, W., Zhang, T., Xue, Y., Zhai, Z. J., Wang, J., Wei, Y., & Chen, Q. (2015). State-of-the-art methods for inverse design of an enclosed environment. Building and Environment, 91, 91-100. [CrossRef]
- Jie Luo, Yanhui Duan, Jinsheng Cai. (2014). Research on fast prediction method of flow field based on Proper orthogonal decomposition. Advances in Aeronautical Science and Engineering, 5(3), 350-357. (In Chinese).
- Wetter, M., & Wright, J. (2003, August). Comparison of a generalized pattern search and a genetic algorithm optimization method. In Proc. of the 8-th IBPSA Conference(Vol. 3, pp. 1401-1408).
- Wang, S., & Jin, X. (2000). Model-based optimal control of VAV air-conditioning system using genetic algorithm. Building and Environment, 35(6), 471-487. [CrossRef]
- Holland, J. H. (1992). Adaptation in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence. MIT press.
- Price, K. V. (2013). Differential evolution. Handbook of Optimization: From Classical to Modern Approach, 187-214.
- Kirkpatrick, S., Gelatt Jr, C. D., & Vecchi, M. P. (1983). Optimization by simulated annealing. science, 220(4598), 671-680.
- Kennedy, J., & Eberhart, R. (1995, November). Particle swarm optimization. In Proceedings of ICNN'95-international conference on neural networks (Vol. 4, pp. 1942-1948). IEEE.
- Lee, K. D., & Kim, K. Y. (2011). Surrogate based optimization of a laidback fan-shaped hole for film-cooling. International Journal of Heat and Fluid Flow, 32(1), 226-238. [CrossRef]
- Fontes, D. B., Homayouni, S. M., & Gonçalves, J. F. (2023). A hybrid particle swarm optimization and simulated annealing algorithm for the job shop scheduling problem with transport resources. European Journal of Operational Research, 306(3), 1140-1157. [CrossRef]
- Zhao, X., Liu, W., Lai, D., & Chen, Q. (2018). Optimal design of an indoor environment by the CFD-based adjoint method with area-constrained topology and cluster analysis. Building and Environment, 138, 171-180. [CrossRef]
- Li, L., He, Y., Zhang, H., Fung, J. C., & Lau, A. K. (2023). Enhancing IAQ, thermal comfort, and energy efficiency through an adaptive multi-objective particle swarm optimizer-grey wolf optimization algorithm for smart environmental control. Building and Environment, 235, 110235. [CrossRef]
- Lin, C. J., Wang, K. J., Dagne, T. B., & Woldegiorgis, B. H. (2022). Balancing thermal comfort and energy conservation–a multi-objective optimization model for controlling air-condition and mechanical ventilation systems. Building and Environment, 219, 109237. [CrossRef]
- Hou, F., Ma, J., Kwok, H. H., & Cheng, J. C. (2022). Prediction and optimization of thermal comfort, IAQ and energy consumption of typical air-conditioned rooms based on a hybrid prediction model. Building and Environment, 225, 109576. [CrossRef]
- Ye, X., Qi, H., Kang, Y., & Zhong, K. (2022). Optimization study of heating performance for an impinging jet ventilation system based on data-driven model coupled with TOPSIS method. Building and Environment, 223, 109465. [CrossRef]
- Li, L., Zhang, Y., Fung, J. C., Qu, H., & Lau, A. K. (2022). A coupled computational fluid dynamics and back-propagation neural network-based particle swarm optimizer algorithm for predicting and optimizing indoor air quality. Building and Environment, 207, 108533. [CrossRef]
- Aruta, G., Ascione, F., Bianco, N., Mauro, G. M., & Vanoli, G. P. (2023). Optimizing heating operation via GA-and ANN-based model predictive control: Concept for a real nearly-zero energy building. Energy and Buildings, 292, 113139. [CrossRef]
- Wang, X., Zhao, J., Wang, F., Song, B., & Zhang, Q. (2021). Air supply parameter optimization of a custom nonuniform temperature field based on the POD method. Building and Environment, 206, 108328. [CrossRef]
- Liu, Y., Pan, W., & Long, Z. (2021). Optimization of air supply parameters for stratum ventilation based on proper orthogonal decomposition. Sustainable Cities and Society, 75, 103291. [CrossRef]
- Shao, X., Liu, Y., Wang, B., Li, X., Chen, J., Zhu, Z., & Ma, X. (2023). Fast regulation of multi-position differentiated environment: Multi-step joint optimization of air supply parameters. Building and Environment, 239, 110425. [CrossRef]
- Baba, F. M., Ge, H., Zmeureanu, R., & Wang, L. L. (2023). Optimizing overheating, lighting, and heating energy performances in Canadian school for climate change adaptation: Sensitivity analysis and multi-objective optimization methodology. Building and Environment, 237, 110336. [CrossRef]
- Fan, Z., Liu, M., & Tang, S. (2022). A multi-objective optimization design method for gymnasium facade shading ratio integrating energy load and daylight comfort. Building and Environment, 207, 108527. [CrossRef]
- Rafati, N., Hazbei, M., & Eicker, U. (2023). Louver configuration comparison in three Canadian cities utilizing NSGA-II. Building and Environment, 229, 109939. [CrossRef]
- Wang, Y., Yang, W., & Wang, Q. (2022). Multi-objective parametric optimization of the composite external shading for the classroom based on lighting, energy consumption, and visual comfort. Energy and Buildings, 275, 112441. [CrossRef]
- Mostafazadeh, F., Eirdmousa, S. J., & Tavakolan, M. (2023). Energy, economic and comfort optimization of building retrofits considering climate change: A simulation-based NSGA-III approach. Energy and Buildings, 280, 112721. [CrossRef]
- Li, C., & Chen, Y. (2023). A multi-factor optimization method based on thermal comfort for building energy performance with natural ventilation. Energy and Buildings, 285, 112893. [CrossRef]
- Sun, R., Liu, J., Lai, D., & Liu, W. (2023). Building form and outdoor thermal comfort: Inverse design the microclimate of outdoor space for a kindergarten. Energy and Buildings, 284, 112824. [CrossRef]
- Yin, Y., Li, A., Wu, D., Li, J., & Guo, J. (2023). Low-resistance optimization and secondary flow analysis of elbows via a combination of orthogonal experiment design and simple comparison design. Building and Environment, 236, 110263. [CrossRef]
- Chen, M., Zhang, Z., Deng, Q., Feng, Y., & Wang, X. (2023). Optimization of underfloor air distribution systems for data centers based on orthogonal test method: A case study. Building and Environment, 232, 110071. [CrossRef]
- Yeh, W. C., Lin, Y. P., Liang, Y. C., Lai, C. M., & Huang, C. L. (2023). Simplified swarm optimization for hyperparameters of convolutional neural networks. Computers & Industrial Engineering, 177, 109076. [CrossRef]
- Ong, Y. S., Nair, P. B., Keane, A. J., & Wong, K. W. (2005). Surrogate-assisted evolutionary optimization frameworks for high-fidelity engineering design problems. Knowledge Incorporation in Evolutionary Computation, 307-331. [CrossRef]
- Wang, C., Zhang, J., & Zhou, J. (2016). Optimization of a fan-shaped hole to improve film cooling performance by RBF neural network and genetic algorithm. Aerospace Science and Technology, 58, 18-25. [CrossRef]
- Wang, C. H., Zhang, J. Z., & Zhou, J. H. (2017). Data mining optimization of laidback fan-shaped hole to improve film cooling performance. Journal of Central South University, 24(5), 1183-1189. [CrossRef]
- Regis, R. G. (2023, March). Hyperparameter Tuning of Random Forests Using Radial Basis Function Models. In Machine Learning, Optimization, and Data Science: 8th International Workshop, LOD 2022, Certosa di Pontignano, Italy, September 19–22, 2022, Revised Selected Papers, Part I (pp. 309-324). Cham: Springer Nature Switzerland. [CrossRef]
- Yuan, H., Pan, C., Song, L., Zhao, G., & Zheng, C. (2022). Modeling and optimization of laser shock hole-clinching using response surface methodology and genetic algorithm. The International Journal of Advanced Manufacturing Technology, 122(5-6), 2391-2406. [CrossRef]
- Forrester, A. I., & Keane, A. J. (2009). Recent advances in surrogate-based optimization. Progress in aerospace sciences, 45(1-3), 50-79. [CrossRef]
- Johnson, M. E., Moore, L. M., & Ylvisaker, D. (1990). Minimax and maximin distance designs. Journal of statistical planning and inference, 26(2), 131-148.
- McKay, M. D., Beckman, R. J., & Conover, W. J. (2000). A comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics, 42(1), 55-61.
- Booker, A. J., Dennis, J. E., Frank, P. D., Serafini, D. B., Torczon, V., & Trosset, M. W. (1999). A rigorous framework for optimization of expensive functions by surrogates. Structural optimization, 17, 1-13.
- Jones, D. R., Schonlau, M., & Welch, W. J. (1998). Efficient global optimization of expensive black-box functions. Journal of Global optimization, 13(4), 455. [CrossRef]
- Sasena, M. J., Papalambros, P., & Goovaerts, P. (2002). Exploration of metamodeling sampling criteria for constrained global optimization. Engineering optimization, 34(3), 263-278. 3. [CrossRef]
- Liu, W., Duan, R., Chen, C., Lin, C. H., & Chen, Q. (2015). Inverse design of the thermal environment in an airliner cabin by use of the CFD-based adjoint method. Energy and Buildings, 104, 147-155. [CrossRef]
- Chand, S., & Wagner, M. (2015). Evolutionary many-objective optimization: A quick-start guide. Surveys in Operations Research and Management Science, 20(2), 35-42. [CrossRef]
- Srinivas, N., & Deb, K. (1994). Muiltiobjective optimization using nondominated sorting in genetic algorithms. Evolutionary computation, 2(3), 221-248. [CrossRef]
- Deb, K., Pratap, A., Agarwal, S., & Meyarivan, T. A. M. T. (2002). A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE transactions on evolutionary computation, 6(2), 182-197. [CrossRef]
- Ishibuchi, H., Imada, R., Setoguchi, Y., Nojima, Y. (Jul 2016). Performance comparison of NSGA-V2 and NSGA-V3 on various many-objective test problems. In 2016 IEEE Congress on Evolutionary Computation (CEC), pp. 3045-3052. IEEE.
- Sacks, J., Welch, W. J., Mitchell, T. J., & Wynn, H. P. (1989). Design and analysis of computer experiments. Statistical science, 4(4), 409-423. [CrossRef]
- Bailey, R. A. (2008). Design of comparative experiments (Vol. 25). Cambridge University Press.
- Ester, M., Kriegel, H. P., Sander, J., & Xu, X. (1996, August). A density-based algorithm for discovering clusters in large spatial databases with noise. In kdd (Vol. 96, No. 34, pp. 226-231).
- Blay, D. (1992). Confined turbulent mixed convection in the presence of horizontal buoyant wall jet. HTD Vol. 213, Fundamentals of Mixed Convection.
- Wang, M., & Chen, Q. (2009). Assessment of various turbulence models for transitional flows in an enclosed environment (RP-1271). Hvac&r Research, 15(6), 1099-1119. [CrossRef]
- Yang Changwei, Zhang Xiwen, Yao Zhaowui, Cao, Xiaodong; Liu, Junjie, He Feng (2016). Numerical study of the instantaneous flow fields by large eddy simulation and stability analysis in a single aisle cabin model. Building and Environment, 96(2016), 1–11.
- Strøm-Tejsen, P., Wyon, D. P., Lagercrantz, L., & Fang, L. (2007). Passenger evaluation of the optimum balance between fresh air supply and humidity from 7-h exposures in a simulated aircraft cabin. Indoor Air, 17(2), 92-108. [CrossRef]
- Munoz, A. G., Ayala-Ramirez, V., Alfaro-Ayala, J. A., & Acosta, B. T. (2011). Optimization of the transition piece applying genetic algorithms. Applied thermal engineering, 31(16), 3214-3225. [CrossRef]
- Sun, Y., & Wang, X. (2010). Optimization of air flow field of the melt blowing slot die via numerical simulation and genetic algorithm. Journal of applied polymer science, 115(3), 1540-1545. 3. [CrossRef]
- Xu, Y., Wei, X., Yu, Y., Xia, Y., Zeng, L., Cao, G., & Gao, J. (2023). Inverse design of dynamic protective air supply based on proper orthogonal decomposition. Sustainable Cities and Society, 95, 104591. [CrossRef]

| Methods | Authors | Other tools | Situations |
|---|---|---|---|
| CFD-based adjoint | Zhao, X. et al. (2018) [16] | The centroid-based hierarchical cluster analysis | Steady-state and single-objective |
| Artificial neural networks (ANN) | Li, L. et al. (2023) [17] | Particle swarm optimizer-grey wolf optimization | Transient and multi-objective |
| Lin, C. J. et al. (2022) [18] | Whale optimization algorithm | Steady-state and multi-objective | |
| Hou, F. et al. (2022) [19] | Grey wolf optimization | Steady-state and multi-objective | |
| Ye, X. et al. (2022) [20] | Technique for order preference by similarity to an ideal solution (TOPSIS) | Steady-state and multi-objective | |
| Li, L. et al. (2022) [21] | PSO | Steady-state and single-objective | |
| Aruta, G. et al. (2023) [22] | Non-dominated sorting genetic algorithm-II (NSGA-II) | Multi-objective | |
| Proper orthogonal decomposition (POD) | Wang, X. et al. (2021) [23] | Radial basis functions | Steady-state and multi-objective |
| Liu, Y. et al. (2021) [24] | Steady-state and multi-objective | ||
| Multi-step joint optimization | Shao, X. et al. (2023) [25] | Three flow field characteristic indicators | Transient and multi-objective |
| Meta-heuristic optimization approaches | Baba, F. M. et al. (2023) [26] | NSGA-II | Steady-state and multi-objective |
| Fan, Z. et al. (2022) [27] | Improving the strength Pareto evolutionary algorithm-2 (SPEA-2) | Steady-state and multi-objective | |
| Rafati, N. et al. (2023) [28] | NSGA-II | Multi-objective | |
| Wang, Y. et al. (2022) [29] | NSGA-II and K-means | Multi-objective | |
| Mostafazadeh, F. et al. (2023) [30] | NSGA-III and TOPSIS | Multi-objective | |
| Li, C. et al. (2023) [31] | PSO | Steady-state and single-objective | |
| Sun, R. et al. (2023) [32] | Genetic algorithm | Steady-state and single-objective | |
| Orthogonal experiment designs | Yin, Y. et al. (2023) [33] | Steady-state and single-objective | |
| Chen, M. et al. (2023) [34] | Steady-state and single-objective |
| NO. |
Vin (m/s) (deviation) |
Tin (℃) (deviation) | F1 | F2 | F' |
|---|---|---|---|---|---|
| 1 | 0.57103 (0.18%) | 15.0359 (0.24%) | 0.000151 (min) |
0.001755 | 0.000953 (min) |
| 2 | 0.55203 (3.15%) | 14.9596 (0.29%) | 0.005745 | 0.000493 (min) |
0.003119 |
| NO. |
Vin (m/s) (deviation) |
Tin (℃) (deviation) | F1 | F2 | F' | |
|---|---|---|---|---|---|---|
| Iteration 39 (continuous) | 1 | 0.61033 (7.075%) | 16.43050 (9.53%) | 0.05569 (min) |
0.02771 | 0.04170 (min) |
| 2 | 0.72669 (27.5%) | 16.63695 (10.9%) | 0.13114 | 0.01539 (min) |
0.07327 | |
| Iteration 52 (continuous) | 1 | 0.61394 (7.71%) | 16.81935 (12.1%) | 0.05531 (min) |
0.02688 | 0.04109 |
| 2 | 0.55865 (1.99%) | 16.64436 (11.0%) | 0.05667 | 0.02359 | 0.04013 (min) |
|
| 3 | 0.72669 (27.5%) | 16.63695 (10.9%) | 0.13114 | 0.01539 (min) |
0.07327 | |
| Iteration 12 (discrete) | 1 | 0.59 (3.51%) |
16.5 (10.0%) |
0.05497 (min) |
0.02654 | 0.04075 (min) |
| 2 | 0.76 (33.3%) |
16.6 (10.7%) |
0.12294 | 0.01565 (min) |
0.06930 | |
| 1 | 0.61033 (7.075%) | 16.43050 (9.53%) | 0.05569 (min) |
0.02771 | 0.04170 (min) |
| Parameters | Lower limit | Upper limit | Precision |
|---|---|---|---|
| t (℃) | 8 | 20 | 0.1 |
| V1 (m/s) | 0.1 | 1.5 | 0.01 |
| V2 (m/s) | 0.1 | 1.5 | 0.01 |
| θ1 (°) | -80 | 80 | 1 |
| θ2 (°) | -80 | 80 | 1 |
| NO. | t (℃) | V1 (m/s) | V2 (m/s) | θ1 (°) | θ2 (°) | volume1 (m3) | volume2 (m3) | total (m3) |
|---|---|---|---|---|---|---|---|---|
| 1 | 8 | 0.76 | 0.98 | 19 | 12 | 0.786 | 0.709 (max) | 1.496 |
| 2 | 10.8 | 0.83 | 0.29 | -56 | -15 | 1.236 | 0.621 | 1.856 |
| 3 | 10.8 | 0.87 | 0.28 | -57 | -17 | 1.296 | 0.596 | 1.893 (max) |
| 4 | 10.6 | 0.9 | 0.27 | -55 | -20 | 1.301 | 0.578 | 1.880 |
| 5 | 11 | 0.87 | 0.31 | -60 | -14 | 1.304 | 0.550 | 1.854 |
| 6 | 10.9 | 0.86 | 0.29 | -58 | -17 | 1.305 | 0.549 | 1.854 |
| 7 | 10.9 | 0.87 | 0.29 | -59 | -15 | 1.316 | 0.526 | 1.841 |
| 8 | 10.8 | 0.86 | 0.31 | -59 | -17 | 1.340 | 0.495 | 1.834 |
| 9 | 9.8 | 1.01 | 0.97 | -72 | -68 | 1.371 (max) | 0.394 | 1.765 |
| CFD-based genetic algorithms | POD method | Surrogate-based (this paper) |
|
|---|---|---|---|
| Target number (achievable) | multiple | multiple | multiple |
| Target number (current study) |
multiple | single | double |
| Initial sample size | small | large | medium |
| Sampling method | random | uniform/orthogonal experimental design | Latin hypercube sampling technique |
| Randomness | existent | non-existent | existent |
| Continuity assumption | non-existent | existent | existent |
| Prediction process | non-existent | interpolation | interpolation |
| Prediction method | non-existent | spline/polynomial/ radial basis function |
Kriging |
| Gradient dependence | non-existent | existent | existent |
| Database update | existent | usually non-existent | existent |
| Subgeneration generation tool | three kinds of operators | usually non-existent | infill criteria and NSGA-II |
| Number of new individuals | usually equal to the initial sample size | usually non-existent | few |
| Validation times | many | one or few | many |
| Effect of outliers | non-existent | existent | existent |
| Costs | higher | medium | lower |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





