Submitted:
18 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
MSC: 6211; 68T09
1. Introduction
2. Related Work
3. Proposed Method
3.1. Overview
| Algorithm 1: ASISO |
|
Input: Data set ; hyperparameters k and
Output: Optimized data set

|
3.2. K-Space
| Algorithm 2: K-Space |
|
Input: Data set ; hyperparameter k
Output:
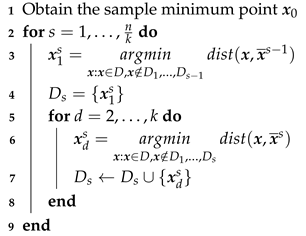
|
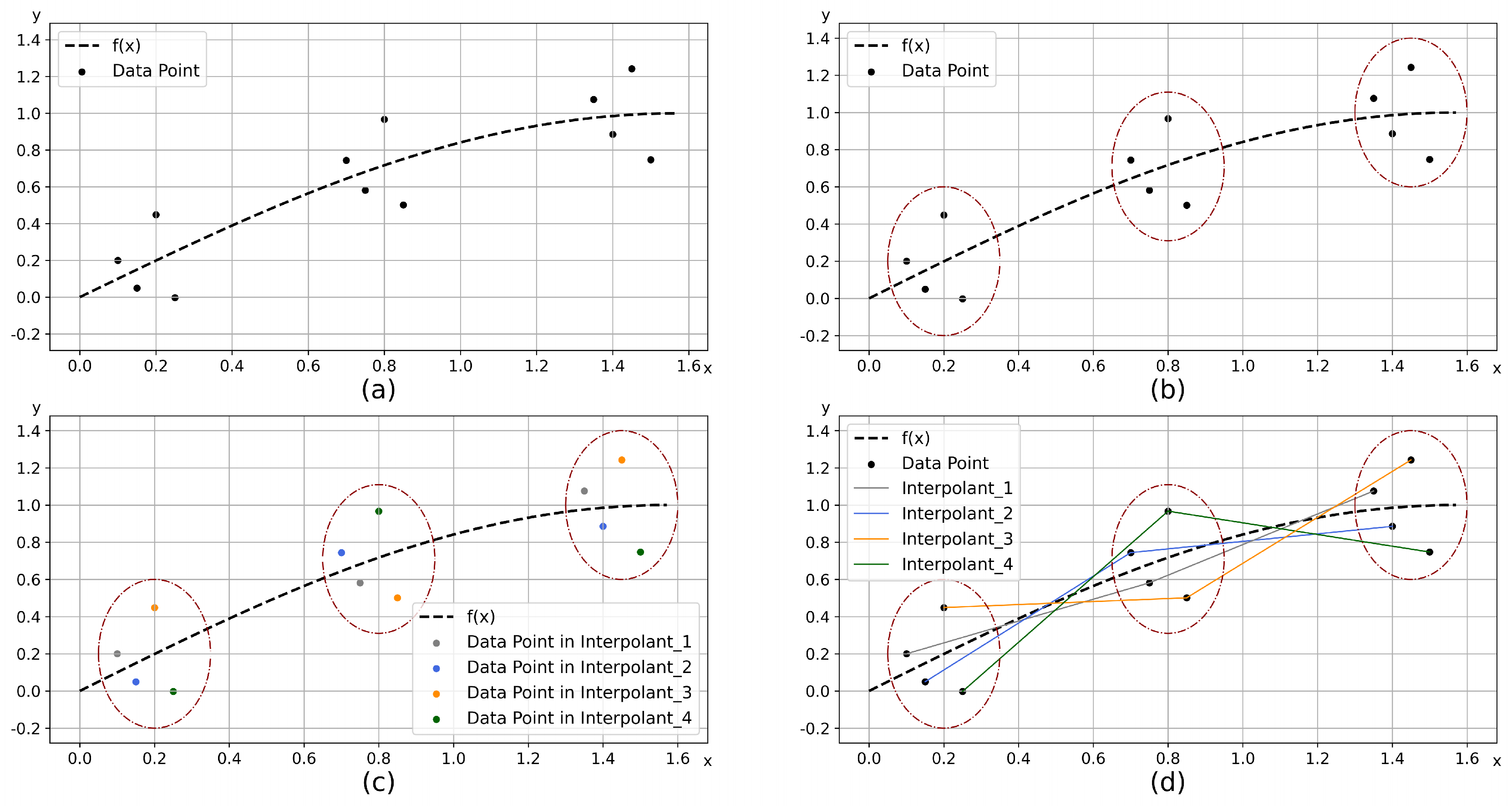
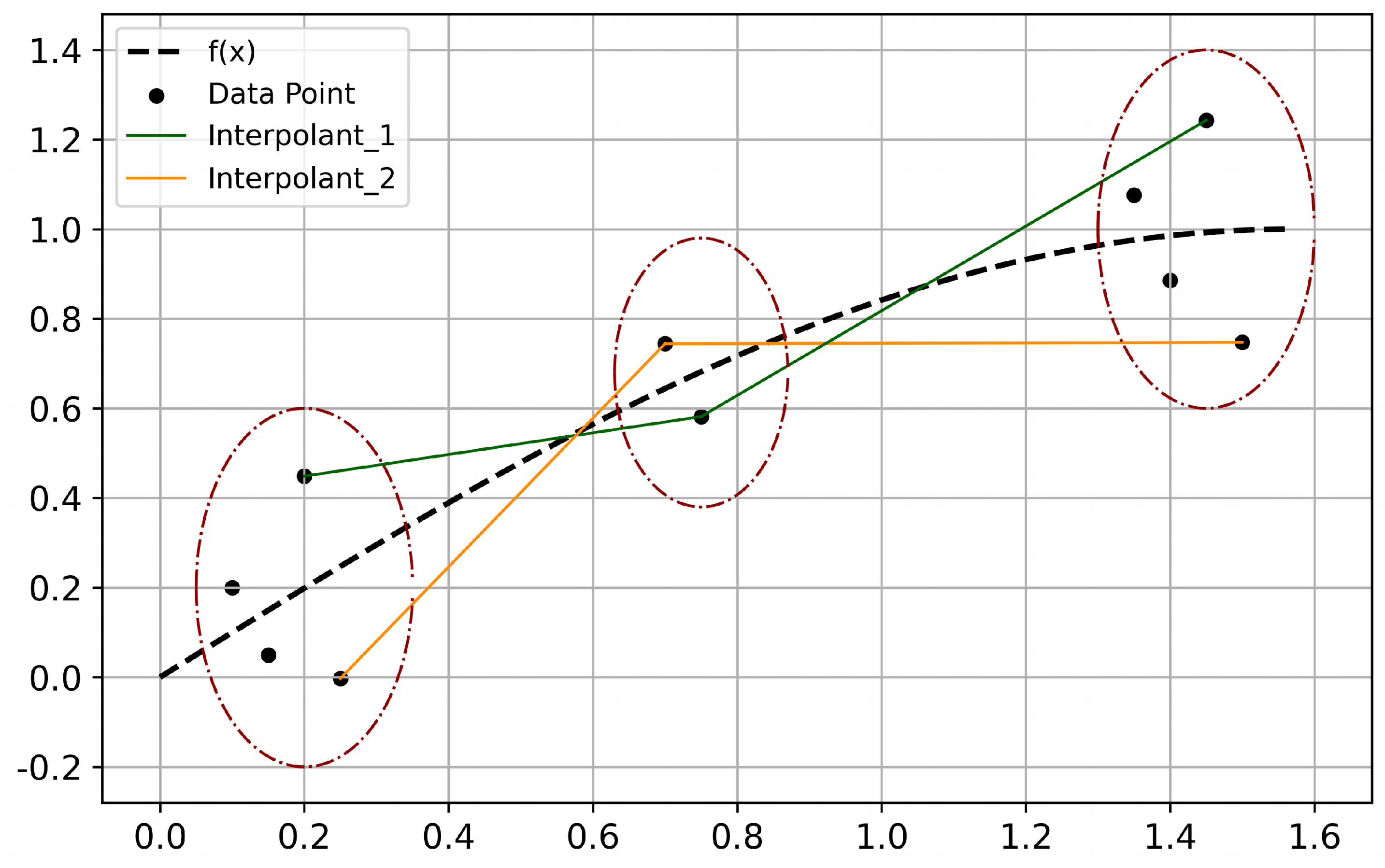
3.3. K-Match
| Algorithm 3: K-Match |
|
Input: Subset
Output: Matching scheme
1 Fitting the dataset and obtain
2 Obtain using (10)
3 Sorting the samples in and according to the value of , obtain and
4 Combine and into
|
3.4. Supplements
4. Experiments
4.1. Artificial Data Sets
4.1.1. Hyperparameter Selection
4.1.2. Comparison of Optimization Performance
4.2. Benchmark Data Sets
5. Conclusions
References
- ALRikabi H T S, Hazim H T. Enhanced data security of communication system using combined encryption and steganography. iJIM, 2021, 15(16): 145. [CrossRef]
- Kollias D. ABAW: learning from synthetic data & multi-task learning challenges. European Conference on Computer Vision. Cham: Springer Nature Switzerland, 2022: 157-172. [CrossRef]
- Mahesh B. Machine learning algorithms-a review. International Journal of Science and Research (IJSR).[Internet], 2020, 9(1): 381-386.
- Lepot M, Aubin J B, Clemens F H L R. Interpolation in time series: An introductive overview of existing methods, their performance criteria and uncertainty assessment. Water, 2017, 9(10): 796. [CrossRef]
- Chlap P, Min H, Vandenberg N, et al. A review of medical image data augmentation techniques for deep learning applications. Journal of Medical Imaging and Radiation Oncology, 2021, 65(5): 545-563. [CrossRef]
- Shorten C, Khoshgoftaar T M. A survey on image data augmentation for deep learning. Journal of big data, 2019, 6(1): 1-48. [CrossRef]
- Chawla N V, Bowyer K W, Hall L O, et al. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research, 2002, 16: 321-357. [CrossRef]
- Dablain D, Krawczyk B, Chawla N V. DeepSMOTE: Fusing deep learning and SMOTE for imbalanced data. IEEE Transactions on Neural Networks and Learning Systems, 2022. [CrossRef]
- Han H, Wang W Y, Mao B H. Borderline-SMOTE: a new over-sampling method in imbalanced data sets learning.International conference on intelligent computing. Berlin, Heidelberg: Springer Berlin Heidelberg, 2005: 878-887. [CrossRef]
- Bunkhumpornpat C, Sinapiromsaran K, Lursinsap C. Safe-level-smote: Safe-level-synthetic minority over-sampling technique for handling the class imbalanced problem. Advances in Knowledge Discovery and Data Mining: 13th Pacific-Asia Conference, PAKDD 2009 Bangkok, Thailand, April 27-30, 2009 Proceedings 13. Springer Berlin Heidelberg, 2009: 475-482. [CrossRef]
- Ha T, Dang T K, Dang T T, et al. Differential privacy in deep learning: an overview. 2019 International Conference on Advanced Computing and Applications (ACOMP). IEEE, 2019: 97-102. [CrossRef]
- Meng D, De La Torre F. Robust matrix factorization with unknown noise. Proceedings of the IEEE international conference on computer vision. 2013: 1337-1344. [CrossRef]
- Raghunathan T E. Synthetic data. Annual review of statistics and its application, 2021, 8: 129-140. [CrossRef]
- Sibson R. A brief description of natural neighbour interpolation. Interpreting multivariate data, 1981: 21-36.
- Tachev G T. Piecewise linear interpolation with nonequidistant nodes. Numerical Functional Analysis and Optimization, 2000, 21(7-8): 945-953. [CrossRef]
- Blu T, Thévenaz P, Unser M. Linear interpolation revitalized. IEEE Transactions on Image Processing, 2004, 13(5): 710-719. [CrossRef]
- Berrut J P, Trefethen L N. Barycentric lagrange interpolation. SIAM review, 2004, 46(3): 501-517. [CrossRef]
- Musial J P, Verstraete M M, Gobron N. Comparing the effectiveness of recent algorithms to fill and smooth incomplete and noisy time series. Atmospheric chemistry and physics, 2011, 11(15): 7905-7923. [CrossRef]
- Fornberg B, Zuev J. The Runge phenomenon and spatially variable shape parameters in RBF interpolation. Computers & Mathematics with Applications, 2007, 54(3): 379-398. [CrossRef]
- Rabbath C A, Corriveau D. A comparison of piecewise cubic Hermite interpolating polynomials, cubic splines and piecewise linear functions for the approximation of projectile aerodynamics. Defence Technology, 2019, 15(5): 741-757. [CrossRef]
- Habermann C, Kindermann F. Multidimensional spline interpolation: Theory and applications. Computational Economics, bm2007, 30: 153-169. [CrossRef]
- Ganzburg M I. The Bernstein constant and polynomial interpolation at the Chebyshev nodes. Journal of Approximation Theory, 2002, 119(2): 193-213. [CrossRef]
- Cleveland W S. Robust locally weighted regression and smoothing scatterplots. Journal of the American statistical association, 1979, 74(368): 829-836. [CrossRef]
- Lichti D D, Chan T O, Belton D. Linear regression with an observation distribution model. Journal of geodesy, 2021, 95: 1-14. [CrossRef]
- Liu C, Li B, Vorobeychik Y, et al. Robust linear regression against training data poisoning. Proceedings of the 10th ACM workshop on artificial intelligence and security. 2017: 91-102. [CrossRef]
- Breunig M M, Kriegel H P, Ng R T, et al. LOF: identifying density-based local outliers. Proceedings of the 2000 ACM SIGMOD international conference on Management of data. 2000: 93-104. [CrossRef]
- Guo Y, Wang W, Wang X. A robust linear regression feature selection method for data sets with unknown noise. IEEE Transactions on Knowledge and Data Engineering, 2021, 35(1): 31-44. [CrossRef]
- Cukierski W. Bike Sharing Demand. Kaggle, https://kaggle.com/competitions/bike-sharing-demand, 2014.
- Dua D, Craff C. UCI Machine Learning Repository, http://archive.ics.uci.edu/ml, 2017.
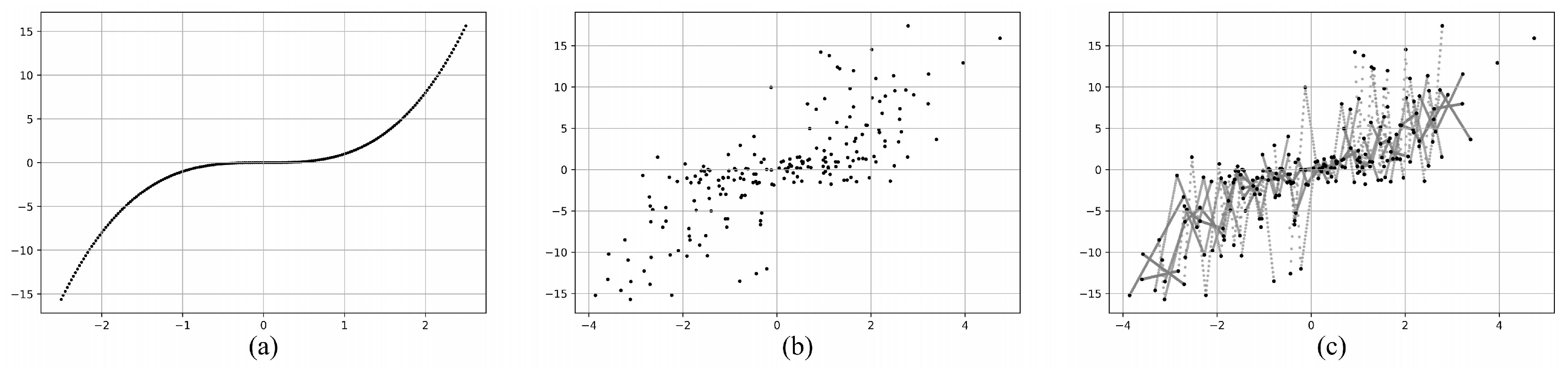
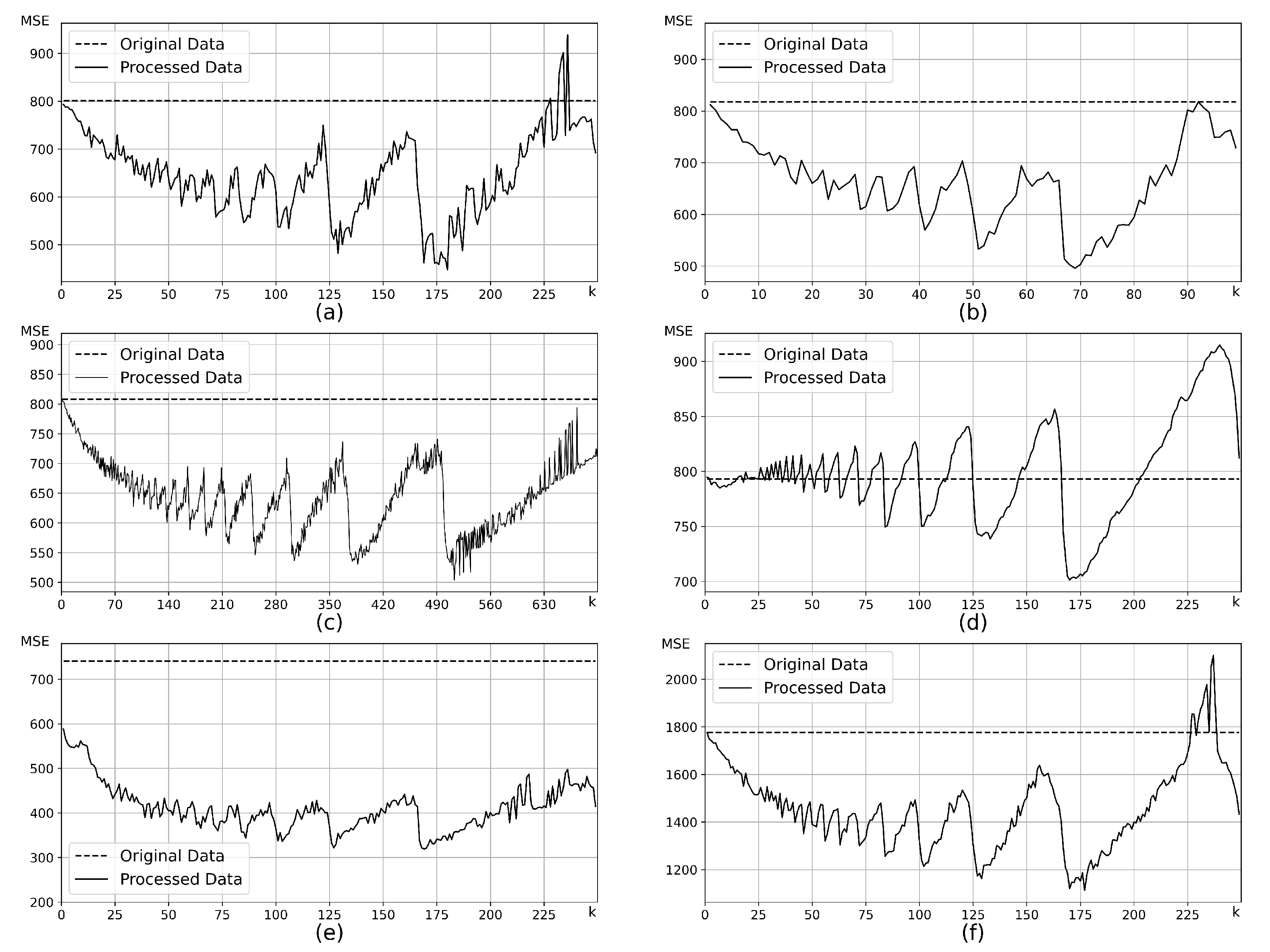

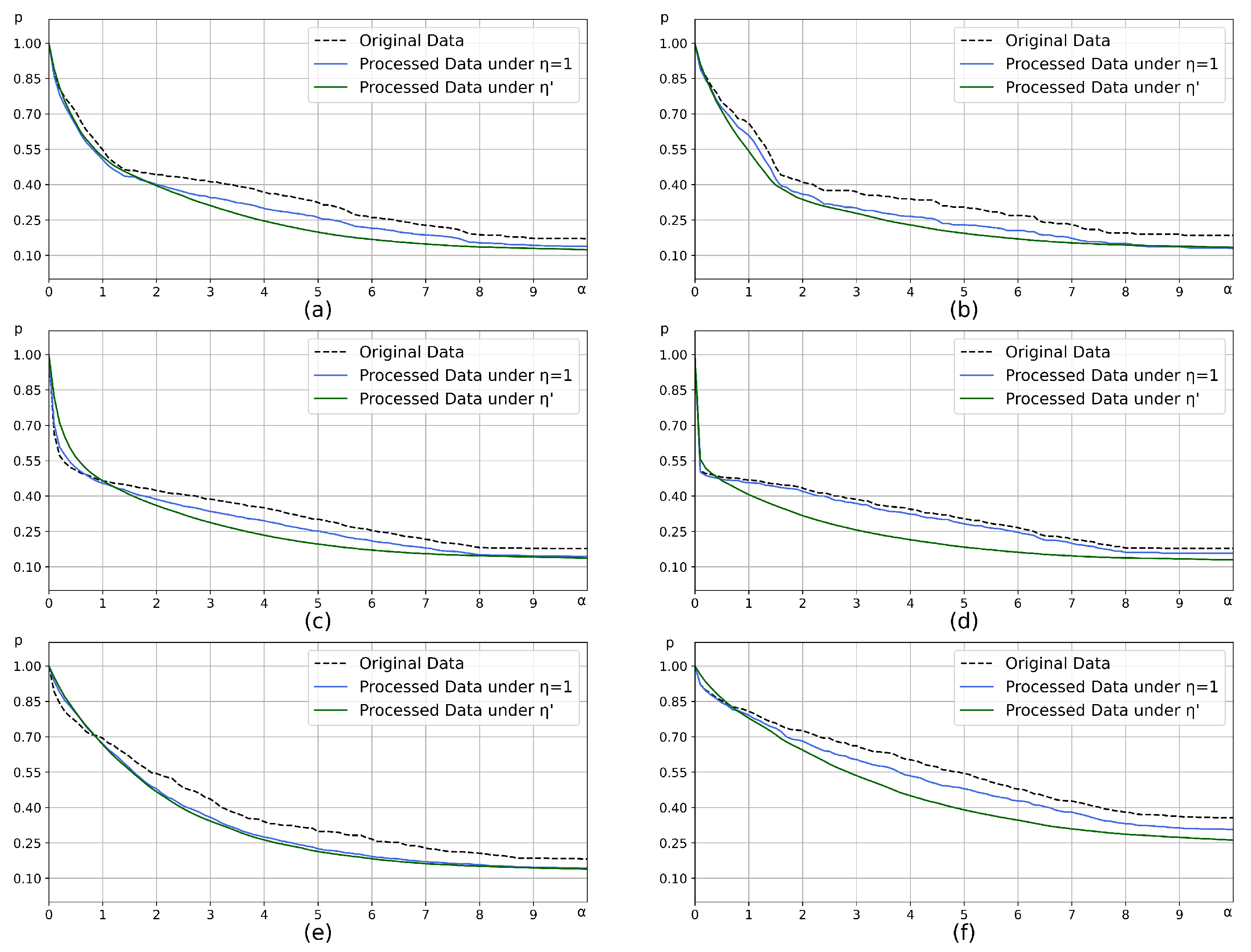
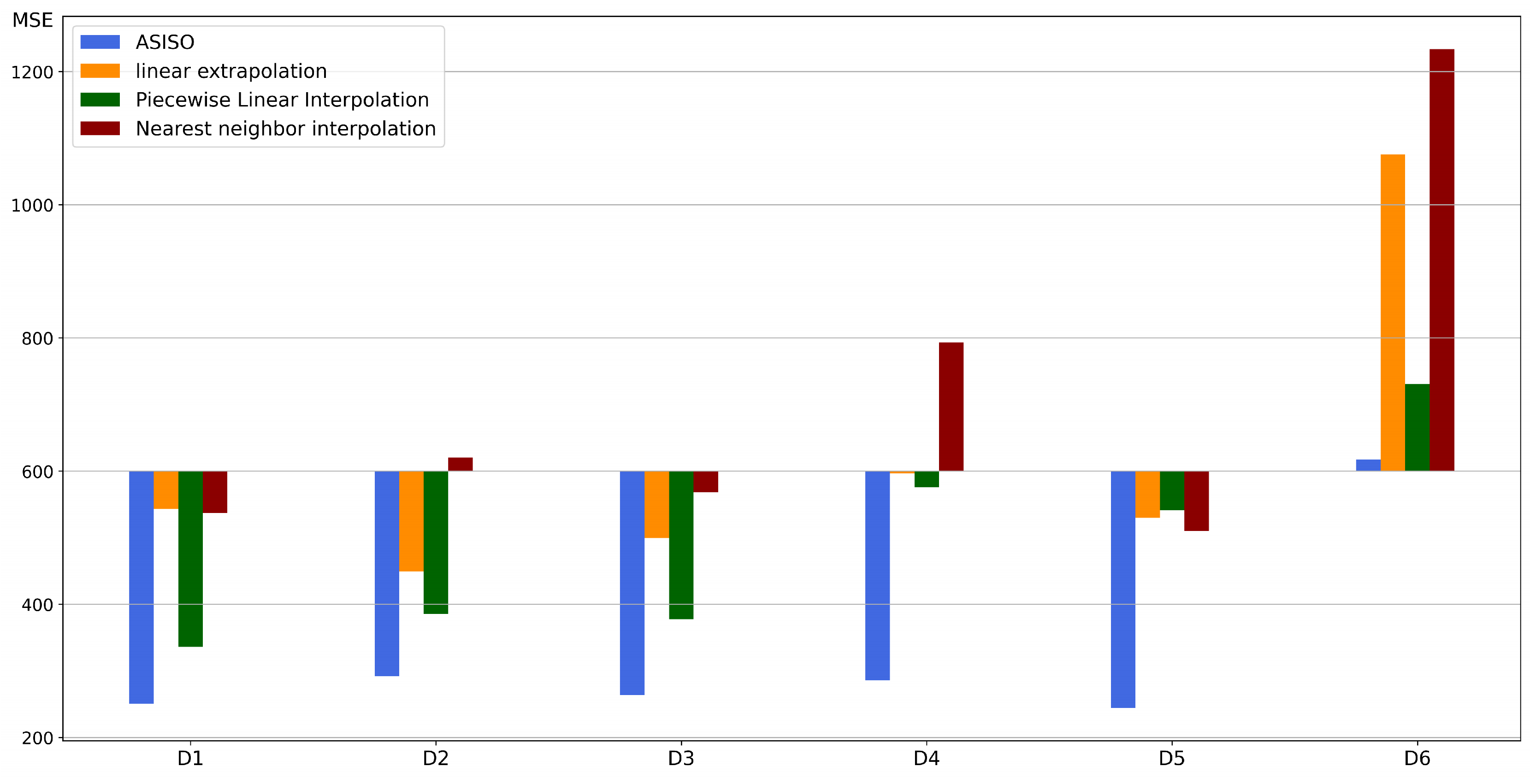
| Artificial Data Sets | Distribution | Samples | ) |
| 20%-N(0,64) 30%-U(-8,8) 50%-N(0,0.04) |
500 | (5,3) | |
| 20%-N(0,64) 30%-U(-8,8) 50%-N(0,0.04) |
200 | (5,3) | |
| 20%-N(0,64) 30%-U(-8,8) 50%-N(0,0.04) |
1500 | (5,3) | |
| 20%-N(0,64) 30%-U(-8,8) 50%-N(0,0.04) |
500 | (1,3) | |
| 20%-N(0,64) 30%-U(-8,8) 50%-N(0,0.04) |
500 | (20,10) | |
| 40%-N(0,64) 45%-U(-8,8) 15%-N(0,0.04) |
500 | (5,3) |
| Data sets | Processing | Hyperparameter | Testing MAE () | ||||
| KNN | RF | MLP | SVR | GBDT | |||
| Bike Sharing | - | - | |||||
| ASISO | |||||||
| - | - | ||||||
| ASISO | |||||||
| Air Quality | - | - | |||||
| ASISO | |||||||
| Forest Fires | - | - | |||||
| ASISO | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




