Submitted:
18 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Multiple Environment Trials
2.2. Traits Measurement and Statistical Analysis
2.3. Marker Polymorphism and Analysis
2.4. Construction of Integrated Genetic Linkage Map
2.5. QTL Identification and Candidate Genes Prediction for QTL Hotspot
3. Results
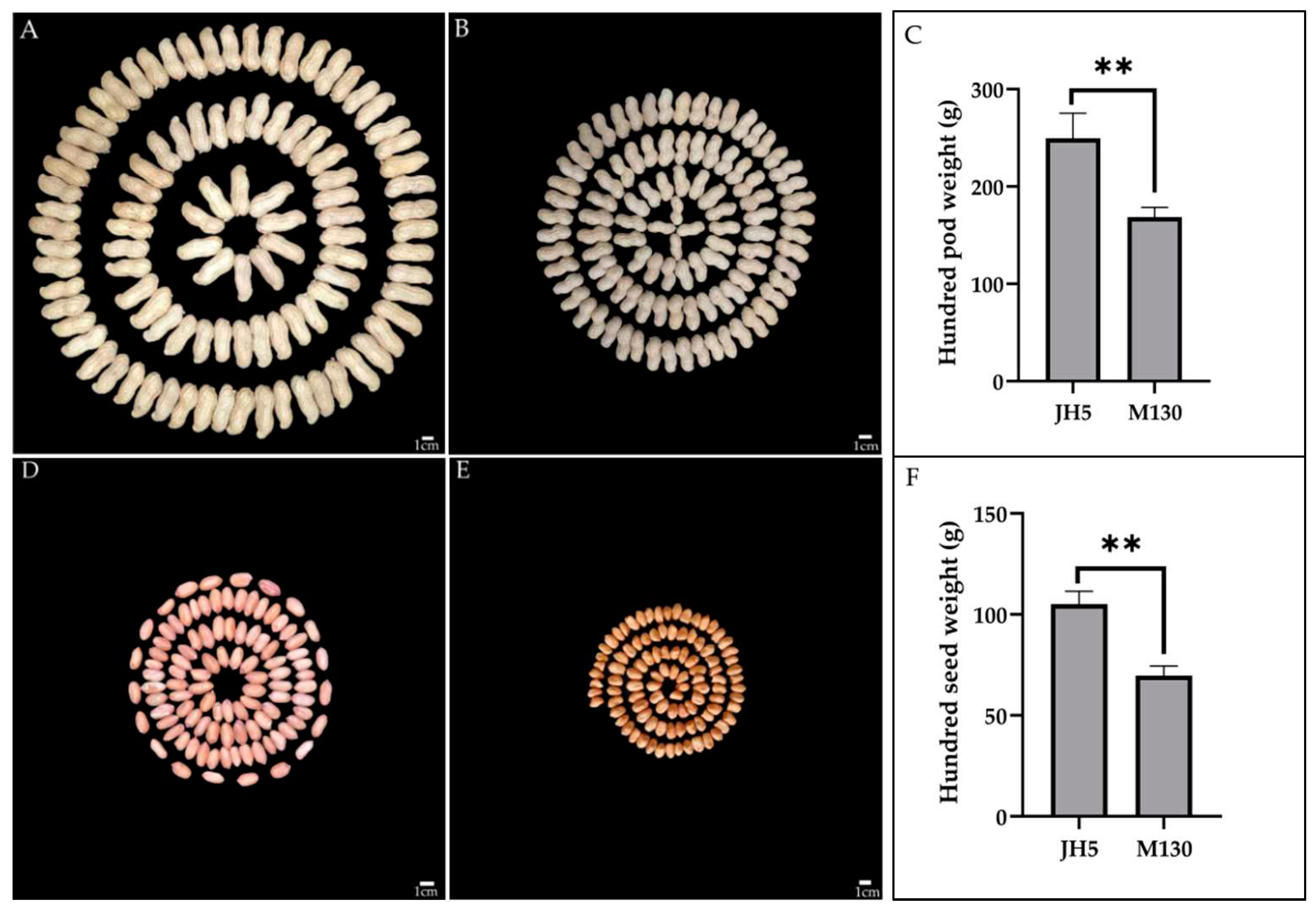
3.1. Phenotypic Variation of Parents and RIL Population
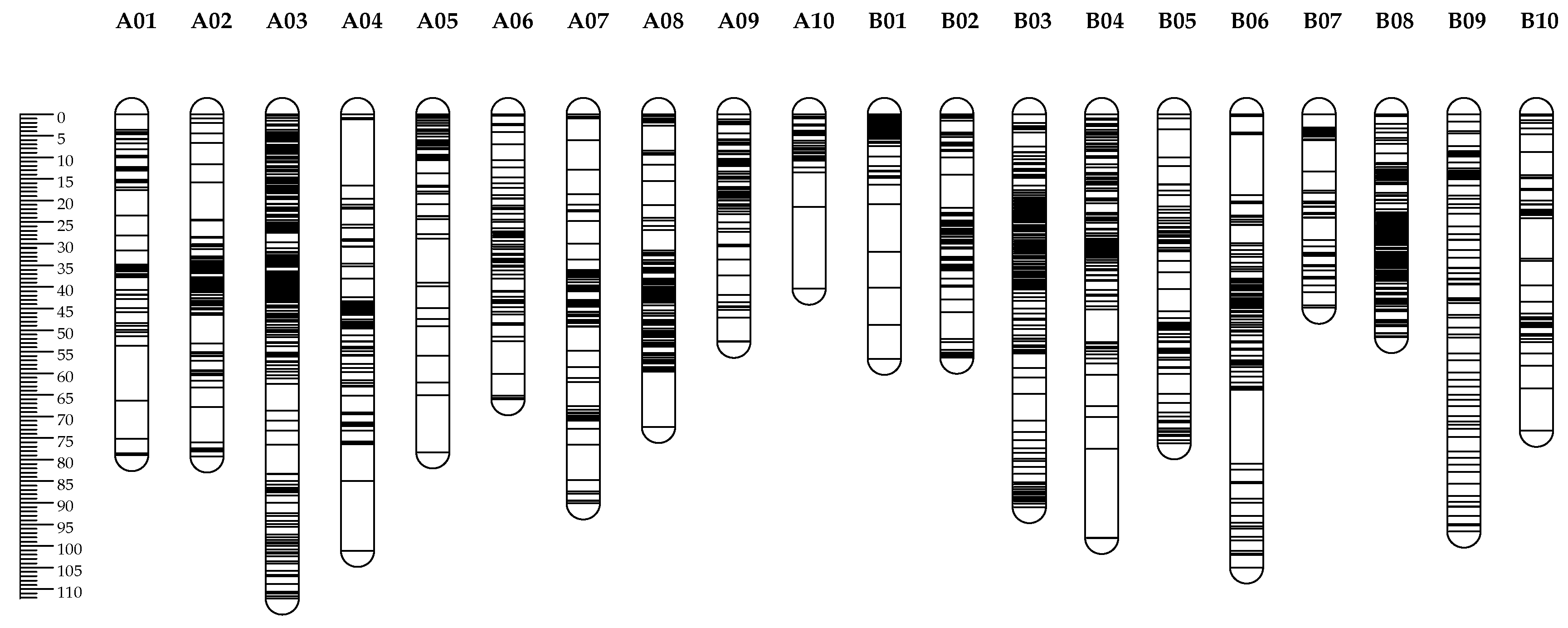
3.2. Integrated Genetic Map Construction and Marker Distribution
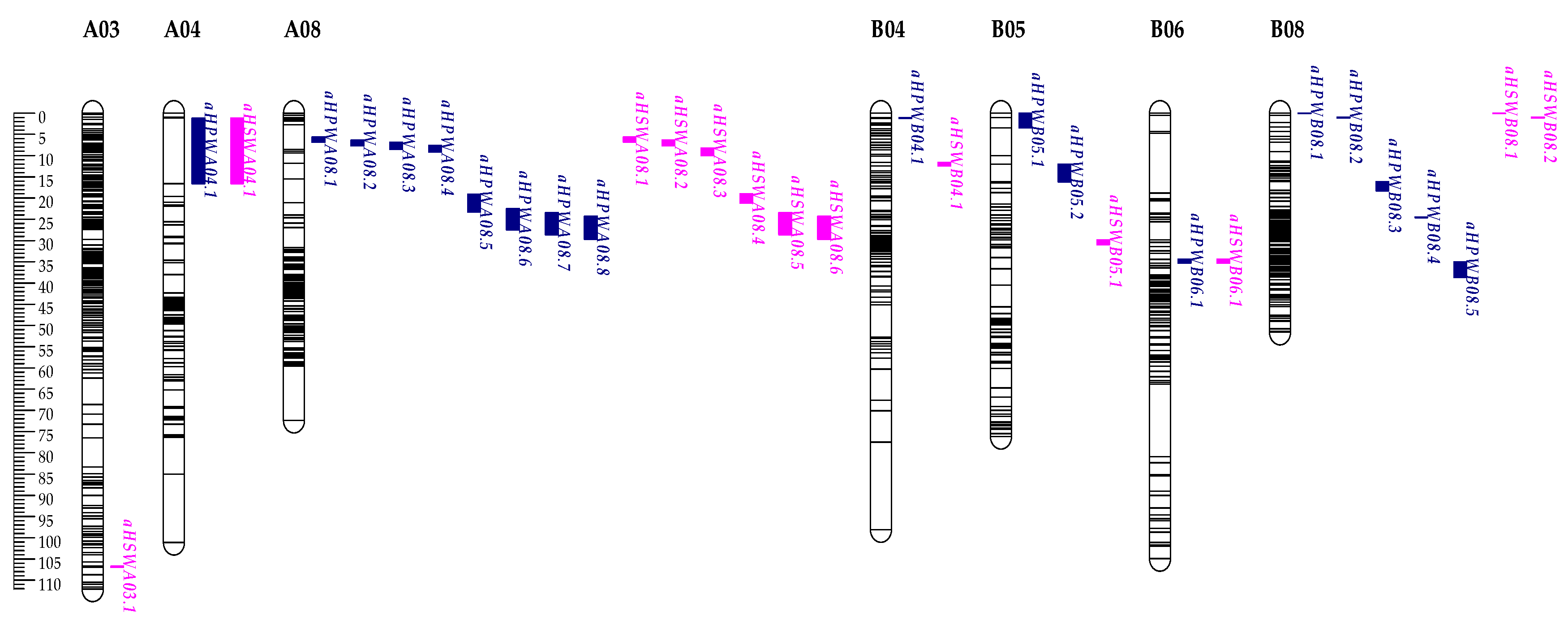
3.3. QTL Identification
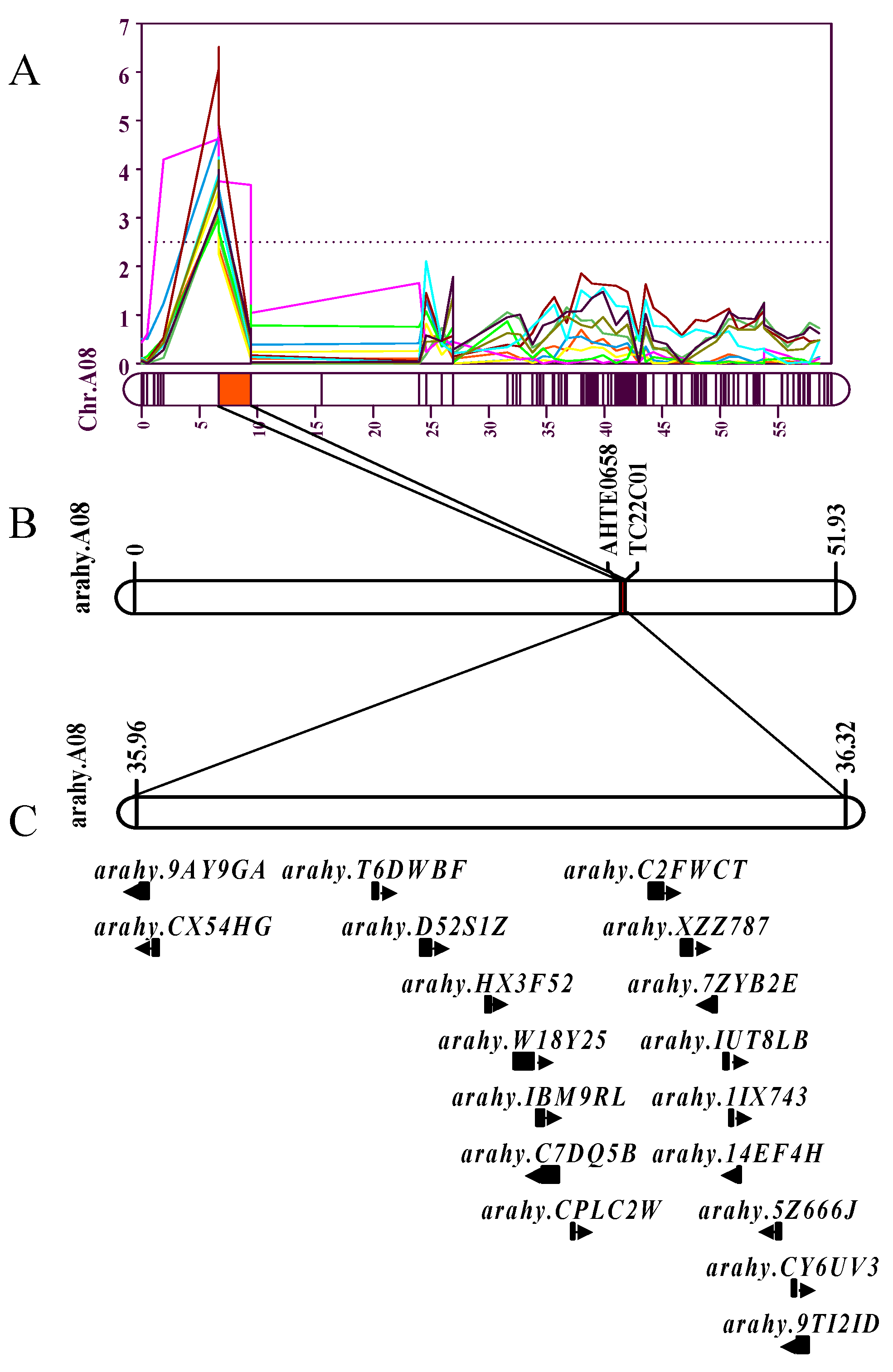
3.4. Co-localized Intervals and Putative Candidate Genes on Chromosomes A08
4. Discussion
5. Conclusions
Supplementary files
Author Contributions
Funding
Data Availability Statement
References
- Ding, Y.; Qiu, X.; Luo, H.; Huang, L.; Guo, J.; Yu, B.; Sudini, H.; Pandey, M.; Kang, Y.; Liu, N.; et al. Comprehensive evaluation of Chinese peanut mini-mini core collection and QTL mapping for aflatoxin resistance. Bmc Plant Biol 2022, 22, 207. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ren, X.; Zheng, Y.; Zhou, X.; Huang, L.; Yan, L.; Jiao, Y.; Chen, W.; Huang, S.; Wan, L. , et al. Genetic mapping of yield traits using RIL population derived from Fuchuan Dahuasheng and ICG6375 of peanut (Arachis hypogaea L.). Mol Breeding 2017, 37, 17. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Chen, T.; Wang, X.; Cao, J.; Li, X.; Xu, X.; Chen, L.; Xia, Q.; Dong, Y.; Huang, L. , et al. Physiological and Expressional Regulation on Photosynthesis, Starch and Sucrose Metabolism Response to Waterlogging Stress in Peanut. Front Plant Sci 2021, 12. [Google Scholar]
- Han, Y.; Dong, Q.; Zhang, K.; Sha, D.; Jiang, C.; Yang, X.; Liu, X.; Zhang, H.; Wang, X.; Guo, F. , et al. Maize-peanut rotational strip intercropping improves peanut growth and soil properties by optimizing microbial community diversity. Peerj 2022, 10, e13777. [Google Scholar] [CrossRef]
- Zhao, H.; Tian, R.; Xia, H.; Li, C.; Li, G.; Li, A.; Zhang, X.; Zhou, X.; Ma, J.; Huang, H. , et al. High-Density Genetic Variation Map Reveals Key Candidate Loci and Genes Associated With Important Agronomic Traits in Peanut. Front Genet 2022, 13, 845602. [Google Scholar] [CrossRef]
- Wan, L.; Ren, W.; Miao, H.; Zhang, J.; Fang, J. Genome-wide identification, expression, and association analysis of the monosaccharide transporter (MST) gene family in peanut (Arachis hypogaea L.). 3 Biotech 2020, 10, 130. [Google Scholar] [CrossRef]
- Hilu, K.W.; Stalker, H.T. Genetic relationships between peanut and wild species of Arachis sect. Arachis (Fabaceae): evidence from RAPDs. Plant Syst Evol 1995, 198, 167–178. [Google Scholar] [CrossRef]
- Halward, T.M.; Stalker, H.T.; Larue, E.A.; Kochert, G. Genetic variation detectable with molecular markers among unadapted germ-plasm resources of cultivated peanut and related wild species. Genome 1991, 34, 1013–1020. [Google Scholar] [CrossRef]
- Halward, T.G.U.A.; Stalker, H.T.; Kochert, G. Development of an RFLP linkage map in diploid peanut species. Theor Appl Genet 1993, 87, 379–384. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bertioli, D.J.; Moretzsohn, M.C.; Vadez, V.; Krishnamurthy, L.; Aruna, R.; Nigam, S.N.; Moss, B.J.; Seetha, K.; Ravi, K. , et al. The first SSR-based genetic linkage map for cultivated groundnut (Arachis hypogaea L.). Theor Appl Genet 2009, 118, 729–739. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, X.; Liang, X.; Liu, H.; Zhou, G.; Li, S.; Wen, S.; Holbrook, C.C.; Guo, B. SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. Bmc Plant Biol 2010, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Koilkonda, P.; Aoki, K.; Hirakawa, H.; Tabata, S.; Watanabe, M.; Hasegawa, M.; Kiyoshima, H.; Suzuki, S.; Kuwata, C. , et al. In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. Bmc Plant Biol 2012, 12, 80. [Google Scholar] [CrossRef]
- Huang, L.; Ren, X.; Wu, B.; Li, X.; Chen, W.; Zhou, X.; Chen, Y.; Pandey, M.K.; Jiao, Y.; Luo, H. , et al. Development and deployment of a high-density linkage map identified quantitative trait loci for plant height in peanut (Arachis hypogaea L.). Sci Rep-Uk 2016, 6, 39478. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, X.; Miao, H.; Chu, Y.; Cui, F.; Yang, W.; Wang, C.; Shen, Y.; Xu, T.; Zhao, L. , et al. QTL identification for seed weight and size based on a high-density SLAF-seq genetic map in peanut (Arachis hypogaea L.). Bmc Plant Biol 2019, 19, 537. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, M.P.; Gangurde, S.S.; Hake, A.A.; Yadawad, A.; Mahadevaiah, S.S.; Pattanashetti, S.K.; Gowda, M.; Shirasawa, K.; Varshney, R.K.; Pandey, M.K. , et al. Genotyping-by-Sequencing Based Genetic Mapping Identified Major and Consistent Genomic Regions for Productivity and Quality Traits in Peanut. Front Plant Sci 2021, 12, 668020. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.V.P.R.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P. , et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol Adv 2013, 31, 1120–1134. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S. , et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K. , et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Chen, W.; Jiao, Y.; Cheng, L.; Huang, L.; Liao, B.; Tang, M.; Ren, X.; Zhou, X.; Chen, Y.; Jiang, H. Quantitative trait locus analysis for pod- and kernel-related traits in the cultivated peanut (Arachis hypogaea L.). Bmc Genet 2016, 17, 25. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S. , et al. Sequencing of Cultivated Peanut, Arachis hypogaea, Yields Insights into Genome Evolution and Oil Improvement. Mol Plant 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Yin, D.; Ji, C.; Ma, X.; Li, H.; Zhang, W.; Li, S.; Liu, F.; Zhao, K.; Li, F.; Li, K. , et al. Genome of an allotetraploid wild peanut Arachis monticola: a de novo assembly. Gigascience 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.; Zhang, L.; Zhang, X.; Tang, R. , et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat Genet 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Huai, D.; Zhang, Z.; Cheng, K.; Kang, Y.; Wan, L.; Yan, L.; Jiang, H.; Lei, Y.; Liao, B. Development of a High-Density Genetic Map Based on Specific Length Amplified Fragment Sequencing and Its Application in Quantitative Trait Loci Analysis for Yield-Related Traits in Cultivated Peanut. Front Plant Sci 2018, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Kunta, S.; Agmon, S.; Chedvat, I.; Levy, Y.; Chu, Y.; Ozias-Akins, P.; Hovav, R. Identification of consistent QTL for time to maturation in Virginia-type Peanut (Arachis hypogaea L.). Bmc Plant Biol 2021, 21, 186. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.; Cui, S.; Meng, X.; Mu, G.; Hou, M.; He, M.; Zhang, H.; Liu, L.; Chen, C.Y. Construction of High-Density Genetic Map and Mapping Quantitative Trait Loci for Growth Habit-Related Traits of Peanut (Arachis hypogaea L.). Front Plant Sci 2019, 10, 745. [Google Scholar] [CrossRef]
- Hu, X.H.; Zhang, S.Z.; Miao, H.R.; Cui, F.G.; Shen, Y.; Yang, W.Q.; Xu, T.T.; Chen, N.; Chi, X.Y.; Zhang, Z.M. , et al. High-Density Genetic Map Construction and Identification of QTLs Controlling Oleic and Linoleic Acid in Peanut using SLAF-seq and SSRs. Sci Rep-Uk 2018, 8, 5479. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Cui, S.; Zhao, N.; Li, L.; Hou, M.; Mu, G.; Liu, L.; Li, Z. High-density genetic map development and QTL mapping for concentration degree of floret flowering date in cultivated peanut (Arachis hypogaea L.). Mol Breeding 2020, 40. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, X.; Ren, X.; Huang, L.; Luo, H.; Chen, Y.; Chen, W.; Liu, N.; Liao, B.; Lei, Y. , et al. A Major and Stable QTL for Bacterial Wilt Resistance on Chromosome B02 Identified Using a High-Density SNP-Based Genetic Linkage Map in Cultivated Peanut Yuanza 9102 Derived Population. Front Genet 2018, 9, 652. [Google Scholar] [CrossRef]
- Zhou, X.; Xia, Y.; Liao, J.; Liu, K.; Li, Q.; Dong, Y.; Ren, X.; Chen, Y.; Huang, L.; Liao, B. , et al. Quantitative Trait Locus Analysis of Late Leaf Spot Resistance and Plant-Type-Related Traits in Cultivated Peanut (Arachis hypogaea L.) under Multi-Environments. Plos One 2016, 11, e166873. [Google Scholar] [CrossRef]
- Chen, X.; Li, H.; Pandey, M.K.; Yang, Q.; Wang, X.; Garg, V.; Li, H.; Chi, X.; Doddamani, D.; Hong, Y. , et al. Draft genome of the peanut A-genome progenitor (Arachis duranensis) provides insights into geocarpy, oil biosynthesis, and allergens. P Natl Acad Sci Usa 2016, 113, 6785–6790. [Google Scholar] [CrossRef]
- Gangurde, S.S.; Khan, A.W.; Janila, P.; Variath, M.T.; Manohar, S.S.; Singam, P.; Chitikineni, A.; Varshney, R.K.; Pandey, M.K. Whole-genome sequencing based discovery of candidate genes and diagnostic markers for seed weight in groundnut. Plant Genome-Us 2022, e20265. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Sun, Z.; Liu, H.; Zheng, Z.; Qin, L.; Shi, L.; Chen, Q.; Liu, H.; Lin, X.; Miao, L. , et al. QTL identification, fine mapping, and marker development for breeding peanut (Arachis hypogaea L.) resistant to bacterial wilt. Theor Appl Genet 2022, 135, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Ren, X.; Li, Z.; Xu, Z.; Li, X.; Huang, L.; Zhou, X.; Chen, Y.; Chen, W.; Lei, Y. , et al. Co-localization of major quantitative trait loci for pod size and weight to a 3.7 cM interval on chromosome A05 in cultivated peanut (Arachis hypogaea L.). Bmc Genomics 2017, 18, 58. [Google Scholar] [CrossRef]
- Mondal, S.; Badigannavar, A.M. Identification of major consensus QTLs for seed size and minor QTLs for pod traits in cultivated groundnut (Arachis hypogaea L.). 3 Biotech 2019, 9, 347. [Google Scholar] [CrossRef]
- Gangurde, S.S.; Pasupuleti, J.; Parmar, S.; Variath, M.T.; Bomireddy, D.; Manohar, S.S.; Varshney, R.K.; Singam, P.; Guo, B.; Pandey, M.K. Genetic mapping identifies genomic regions and candidate genes for seed weight and shelling percentage in groundnut. Front Genet 2023, 14, 1128182. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Qi, F.; Qin, L.; Zhang, M.; Sun, Z.; Li, H.; Cui, M.; Zhang, M.; Li, C.; Li, X. , et al. Mapping of a QTL associated with sucrose content in peanut kernels using BSA-seq. Front Genet 2022, 13, 1089389. [Google Scholar] [CrossRef]
- Quiros, G.L.C.F. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 2001. [Google Scholar]
- Hu, J.; Vick, B.A. Target region amplification polymorphism: A novel marker technique for plant genotyping. Plant Mol Biol Rep 2003, 21, 289–294. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, X.; Wang, X.; Li, Z.; Zhang, Y.; Liu, H.; Wu, L.; Zhang, G.; Yan, G.; Ma, Z. Mapping QTL for cotton fiber quality traits using simple sequence repeat markers, conserved intron-scanning primers, and transcript-derived fragments. Euphytica 2015, 201, 215–230. [Google Scholar] [CrossRef]
- Ooijen, J. W. van, J. W. van Ooijen, JW van ’t Verlaat, J. W. van Ooijen, Jolien Tol, Johan Dalén, Jessie B Van Buren, J.W.M. van der Meer, J. Han van Krieken, J. W. van Ooijen, J. S. Van Kessel, Ooijen Van, Roeland E. Voorrips and Lambert P. Heuvel. “JoinMap® 4, Software for the calculation of genetic linkage maps in experimental populations.” (2006).
- Kosambi, D.D. The estimation of map distance from recombination values. Annals of Eugenics 1944. [Google Scholar]
- Voorrips, R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping:Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. The Crop Journal 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Liu, H.; Hong, Y.; Li, H.; Liu, H.; Li, X.; Wen, S.; Zhou, G.; Li, S.; Chen, X. , et al. Consensus map integration and QTL meta-analysis narrowed a locus for yield traits to 0.7 cM and refined a region for late leaf spot resistance traits to 0.38 cM on linkage group A05 in peanut (Arachis hypogaea L.). Bmc Genomics 2018, 19. [Google Scholar] [CrossRef]
- Sujay, V.; Gowda, M.V.; Pandey, M.K.; Bhat, R.S.; Khedikar, Y.P.; Nadaf, H.L.; Gautami, B.; Sarvamangala, C.; Lingaraju, S.; Radhakrishan, T. , et al. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.). Mol Breeding 2012, 30, 773–788. [Google Scholar] [CrossRef]
- Gautami, B.; Pandey, M.K.; Vadez, V.; Nigam, S.N.; Ratnakumar, P.; Krishnamurthy, L.; Radhakrishnan, T.; Gowda, M.V.; Narasu, M.L.; Hoisington, D.A. , et al. Quantitative trait locus analysis and construction of consensus genetic map for drought tolerance traits based on three recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.). Mol Breeding 2012, 30, 757–772. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, L.; Zhou, J.; Li, R.; Pandey, M.K.; Han, Y.; Cui, F.; Zhang, J.; Guo, F.; Chen, J. , et al. Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. J Adv Res 2022, 42, 237–248. [Google Scholar] [CrossRef]
- Sun, C.; Wang, Y.; Yang, X.; Tang, L.; Wan, C.; Liu, J.; Chen, C.; Zhang, H.; He, C.; Liu, C. , et al. MATE transporter GFD1 cooperates with sugar transporters, mediates carbohydrate partitioning and controls grain-filling duration, grain size and number in rice. Plant Biotechnol J 2022. [Google Scholar]
- Luo, S.; Jia, J.; Liu, R.; Wei, R.; Guo, Z.; Cai, Z.; Chen, B.; Liang, F.; Xia, Q.; Nian, H. , et al. Identification of major QTLs for soybean seed size and seed weight traits using a RIL population in different environments. Front Plant Sci 2022, 13, 1094112. [Google Scholar] [CrossRef]
- Zhang, M.; Zheng, H.; Jin, L.; Xing, L.; Zou, J.; Zhang, L.; Liu, C.; Chu, J.; Xu, M.; Wang, L. miR169o and ZmNF-YA13 act in concert to coordinate the expression of ZmYUC1 that determines seed size and weight in maize kernels. New Phytol 2022, 235, 2270–2284. [Google Scholar] [CrossRef]
- Alyr, M.H.; Pallu, J.; Sambou, A.; Nguepjop, J.R.; Seye, M.; Tossim, H.A.; Djiboune, Y.R.; Sane, D.; Rami, J.F.; Fonceka, D. Fine-Mapping of a Wild Genomic Region Involved in Pod and Seed Size Reduction on Chromosome A07 in Peanut (Arachis hypogaea L.). Genes-Basel 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, Z.; Qi, F.; Tian, M.; Wang, J.; Zhao, R.; Wang, X.; Wu, X.; Shi, X.; Liu, H. , et al. Comparative transcriptomics analysis of developing peanut (Arachis hypogaea L.) pods reveals candidate genes affecting peanut seed size. Front Plant Sci 2022, 13, 958808. [Google Scholar] [CrossRef] [PubMed]
- Xiao-jing, Z.; Yong, L.; You-lin, X.; Yan, Q.I.; Li-ying, Y.; Xiao-ping, R.; Li, H.; Huai-yong, L.; Nian, L.; Wei-gang, C. , et al. QTL mapping for traits of pod size and weight in cultivated peanut (Arachis hypogaea L.). Chinese Journal of Oil Crop Sciences 2019, 41, 869–877. [Google Scholar]
- Luo, H.; Guo, J.; Ren, X.; Chen, W.; Huang, L.; Zhou, X.; Chen, Y.; Liu, N.; Xiong, F.; Lei, Y. , et al. Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.). Theor Appl Genet 2018, 131, 267–282. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, L.; Chen, Y.; Wang, X.; Huai, D.; Kang, Y.; Jiang, H.; Liu, K.; Lei, Y.; Liao, B. Detection of a major QTL and development of KASP markers for seed weight by combining QTL-seq, QTL-mapping and RNA-seq in peanut. Theor Appl Genet 2022, 135, 1779–1795. [Google Scholar] [CrossRef]
- Li, H.; Yu-Ning, C.; Huai-Yong, L.; Xiao-Jing, Z.; Nian, L.; Wei-Gang, C.; Yong, L.; Bo-Shou, L.; Hui-Fang, J. Advances of QTL mapping for seed size related traits in peanut. Acta Agronomica.
- Farrow, S.C.; Facchini, P.J. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front Plant Sci 2014, 5, 524. [Google Scholar] [CrossRef]
- Zeng, J.; Ding, Q.; Fukuda, H.; He, X.Q. Fertilization Independent Endosperm genes repress NbGH3.6 and regulate the auxin level during shoot development in Nicotiana benthamiana. J Exp Bot 2016, 67, 2207–2217. [Google Scholar] [CrossRef]
- Chen, R.; Wei, Q.; Liu, Y.; Li, J.; Du, X.; Chen, Y.; Wang, J.; Liu, Y. The pentatricopeptide repeat protein EMP601 functions in maize seed development by affecting RNA editing of mitochondrial transcript ccmC. The Crop Journal 2023. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu Rev Plant Biol 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.; Yu, M.; Zhang, M.; Zhong, Y.; Wang, T.; Liu, P.; Song, W.; Zhao, H.; Fastner, A. , et al. The sugar transporter ZmSUGCAR1 of the nitrate transporter 1/peptide transporter family is critical for maize grain filling. Plant Cell 2022, 34, 4232–4254. [Google Scholar] [CrossRef] [PubMed]
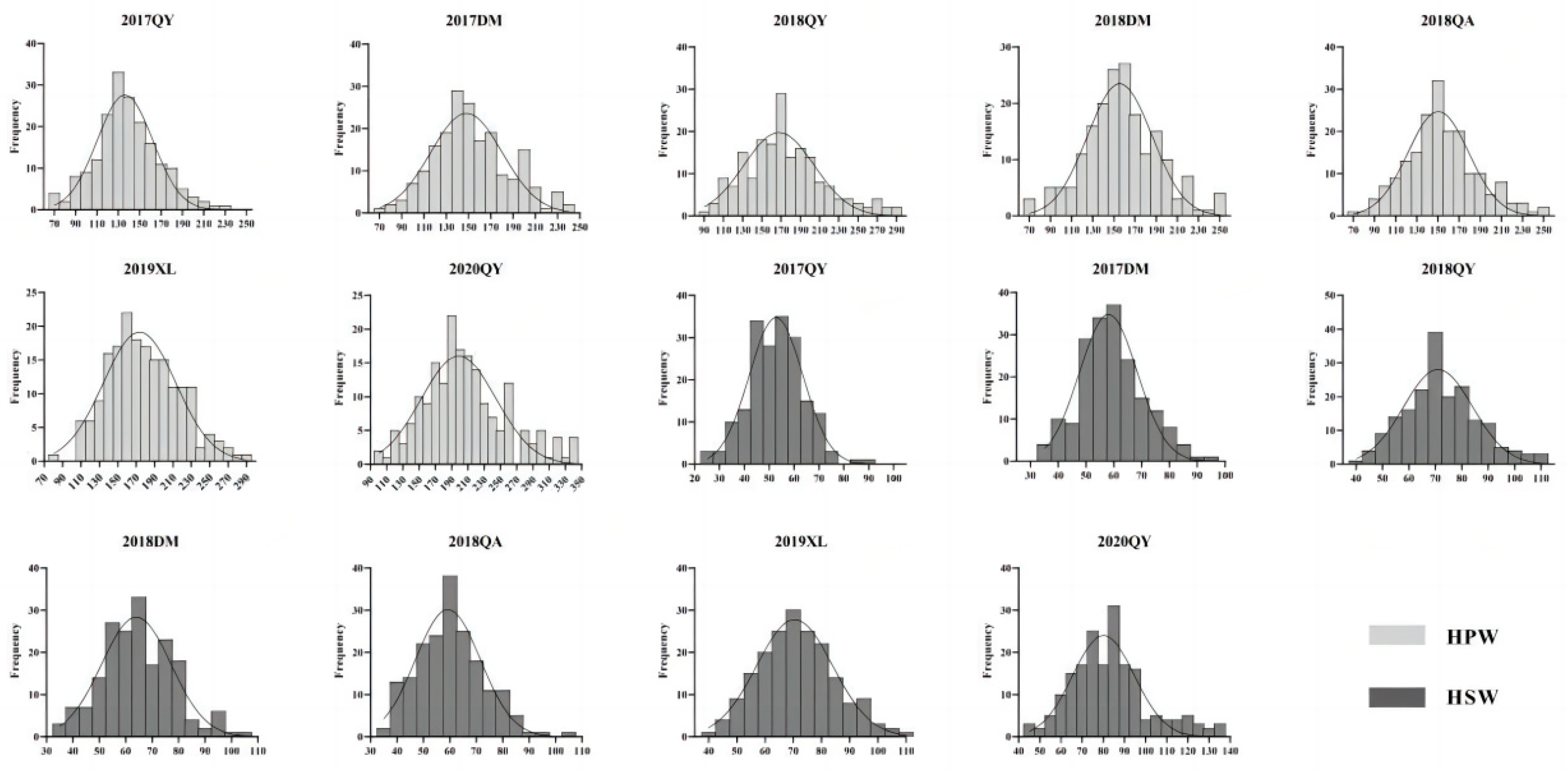
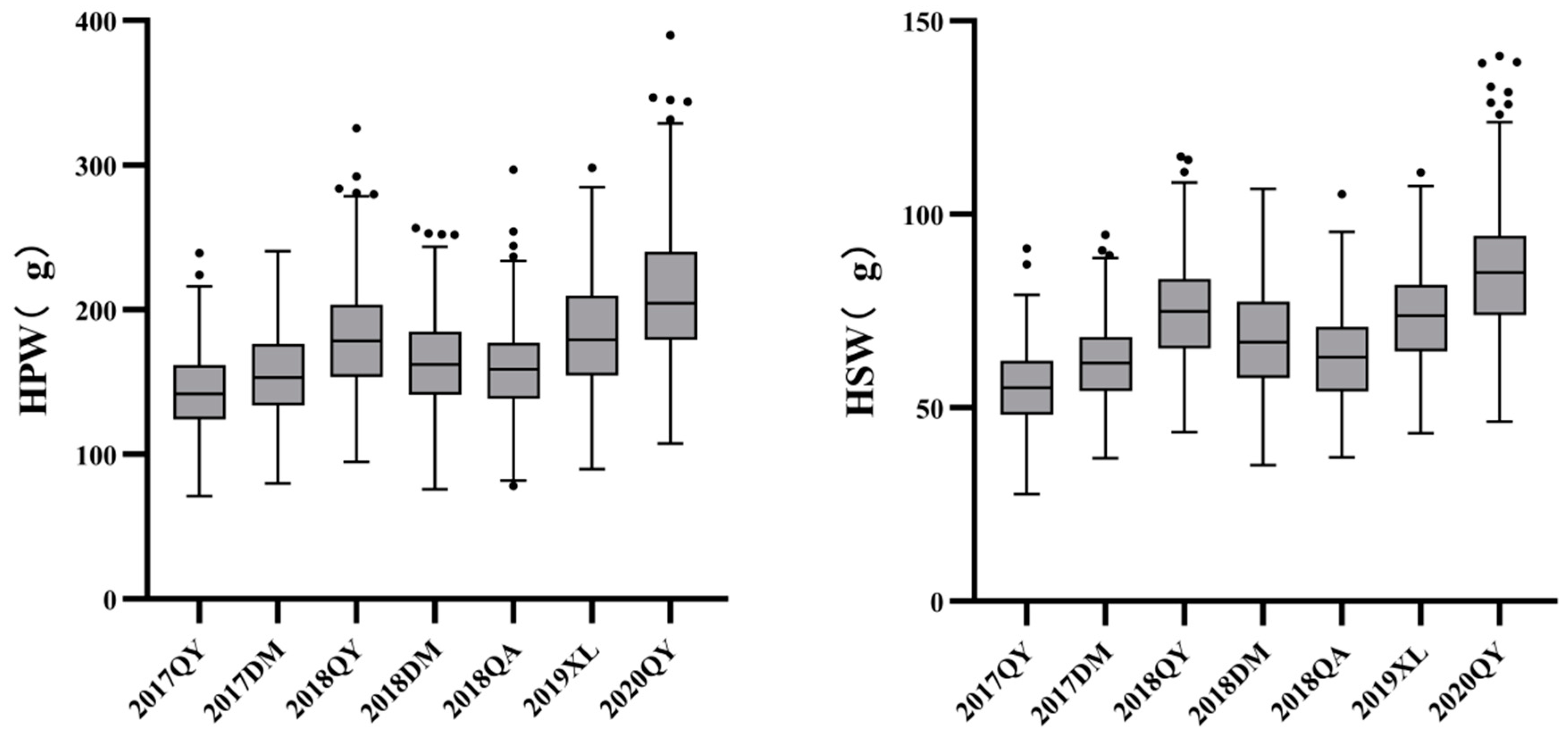
| Traits | Env. | Parents | RIL population | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| JH5 | M130 | Min | Max | Mean | SD | CV (%) | Shapiro–Wilk (w-test) | Skew | Kurt | ||
| HPW(g) | 17QY | 254.03±6.84** | 170.13±0.91 | 71.04 | 239.22 | 143.55 | 30.37 | 21.16 | 0.99 | 0.23 | 0.21 |
| 17DM | 247.18±2.30** | 164.64±3.78 | 79.86 | 240.61 | 156.36 | 33.06 | 21.14 | 0.99 * | 0.36 | -0.11 | |
| 18QY | 271.28±8.80** | 184.48±2.45 | 94.85 | 325.53 | 179.42 | 42.91 | 23.91 | 0.98 ** | 0.62 | 0.36 | |
| 18DM | 236.77±6.46** | 155.79±3.57 | 75.99 | 256.43 | 162.68 | 35.68 | 21.93 | 0.99 | 0.18 | 0.09 | |
| 18QA | 233.38±1.89** | 159.70±5.95 | 78.28 | 296.87 | 159.36 | 35.14 | 22.05 | 0.98 * | 0.51 | 0.86 | |
| 19XL | 213.20±1.74** | 167.76±10.46 | 89.71 | 298.00 | 183.19 | 38.75 | 21.15 | 0.99 | 0.39 | -0.17 | |
| 20QY | 291.31±1.41** | 177.85±0.92 | 107.61 | 389.97 | 212.66 | 52.48 | 24.68 | 0.97 ** | 0.66 | 0.36 | |
| HSW(g) | 17QY | 105.22±2.17** | 70.04±0.69 | 27.73 | 91.24 | 55.42 | 11.17 | 20.16 | 0.99 | 0.19 | 0.30 |
| 17DM | 100.82±1.49** | 68.83±1.14 | 36.93 | 94.67 | 62.00 | 11.60 | 18.71 | 0.99 | 0.32 | -0.10 | |
| 18QY | 114.32±2.73** | 76.20±3.11 | 43.73 | 114.88 | 74.99 | 14.47 | 19.29 | 0.99 | 0.33 | -0.14 | |
| 18DM | 96.12±2.48 ** | 63.88±1.06 | 35.13 | 106.51 | 67.30 | 13.70 | 20.35 | 0.99 | 0.25 | -0.06 | |
| 18QA | 100.24±1.44** | 63.90±1.65 | 37.21 | 105.15 | 63.18 | 13.04 | 20.64 | 0.98 | 0.39 | -0.05 | |
| 19XL | 109.07±0.61** | 74.07±2.7 | 43.44 | 110.75 | 73.99 | 13.40 | 18.10 | 1.00 | 0.26 | -0.14 | |
| 20QY | 109.57±1.05** | 71.01±1.34 | 46.39 | 140.89 | 85.87 | 18.73 | 21.81 | 0.96 ** | 0.68 | 0.53 | |
| Env. | Traits | HPW | HSW |
|---|---|---|---|
| 17QY | HPW | 1 | 0.844** |
| HSW | 0.844** | 1 | |
| 17DM | HPW | 1 | 0.848** |
| HSW | 0.848** | 1 | |
| 18QY | HPW | 1 | 0.881** |
| HSW | 0.881** | 1 | |
| 18DM | HPW | 1 | 0.908** |
| HSW | 0.908** | 1 | |
| 18QA | HPW | 1 | 0.858** |
| HSW | 0.858** | 1 | |
| 19XL | HPW | 1 | 0.881** |
| HSW | 0.881** | 1 | |
| 20QY | HPW | 1 | 0.962** |
| HSW | 0.962** | 1 |
| Traits | Variables | df | MS | F-value | P-value | hB2 |
|---|---|---|---|---|---|---|
| HPW | Geno. | 187 | 20956.938 | 314.2 | P<0.001 | 0.64 |
| Env. | 6 | 289675.957 | 4343.012 | P<0.001 | ||
| G×E | 1122 | 1828.708 | 27.417 | P<0.001 | ||
| HSW | Geno. | 187 | 2566.214 | 182.797 | P<0.001 | 0.52 |
| Env. | 6 | 56659.777 | 4035.995 | P<0.001 | ||
| G×E | 1122 | 249.054 | 17.741 | P<0.001 |
| Linkage | Marker_number | Length (cM) | Average interval (cM) | Maximum gap (cM) |
|---|---|---|---|---|
| A01 | 65 | 110.60 | 1.70 | 12.85 |
| A02 | 256 | 92.49 | 0.36 | 8.18 |
| A03 | 259 | 104.09 | 0.40 | 6.76 |
| A04 | 99 | 105.20 | 1.06 | 16.03 |
| A05 | 238 | 131.83 | 0.55 | 13.20 |
| A06 | 264 | 72.80 | 0.28 | 7.51 |
| A07 | 119 | 182.98 | 1.54 | 8.14 |
| A08 | 159 | 117.05 | 0.74 | 12.74 |
| A09 | 91 | 63.74 | 0.70 | 5.39 |
| A10 | 44 | 58.09 | 1.32 | 18.93 |
| A subgenome | 1594 | 1038.87 | 0.68 | |
| B01 | 203 | 50.20 | 0.25 | 11.12 |
| B02 | 84 | 90.38 | 1.08 | 6.21 |
| B03 | 153 | 76.53 | 0.50 | 6.21 |
| B04 | 196 | 65.14 | 0.33 | 20.59 |
| B05 | 96 | 72.62 | 0.76 | 6.58 |
| B06 | 247 | 192.61 | 0.78 | 17.14 |
| B07 | 48 | 59.51 | 1.24 | 7.25 |
| B8 | 313 | 138.82 | 0.44 | 2.23 |
| B09 | 104 | 54.89 | 0.53 | 3.63 |
| B10 | 92 | 159.35 | 1.73 | 9.87 |
| B subgenome | 1536 | 960.05 | 0.63 | |
| Whole genome | 3130 | 1998.92 | 0.64 |
| Traits | QTLs | Env.a | Chr.b | Position(cM) | Marker interval | LODc | PVEd (%) | Adde | Dirf |
|---|---|---|---|---|---|---|---|---|---|
| HPW | qHPWA04.1 | 18QY | A04 | 2.21 | SMK547-SMK549 | 3.42 | 8.11 | 12.27 | Jihua 5 |
| qHPWA08.1 | 20QY | A08 | 6.26 | AhTE0658-TC22C01 | 6.69 | 8.07 | 16.38 | Jihua 5 | |
| qHPWA08.2 | 17DM | A08 | 6.97 | AhTE0658-TC22C01 | 3.38 | 4.55 | 7.93 | Jihua 5 | |
| qHPWA08.3 | 17QY | A08 | 7.69 | AhTE0658-TC22C01 | 3.60 | 4.41 | 7.64 | Jihua 5 | |
| 18DM | A08 | 7.69 | AhTE0658-TC22C01 | 7.31 | 8.89 | 11.85 | Jihua 5 | ||
| 19XL | A08 | 7.69 | AhTE0658-TC22C01 | 5.44 | 6.79 | 11.23 | Jihua 5 | ||
| qHPWA08.4 | 18QA | A08 | 8.40 | AhTE0658-TC22C01 | 6.07 | 7.83 | 10.34 | Jihua 5 | |
| qHPWA08.5 | 18QY | A08 | 21.20 | me3em14-196-Ah4-4 | 4.32 | 6.21 | 12.02 | Jihua 5 | |
| qHPWA08.6 | 19XL | A08 | 25.00 | Ah4-4-Ah2TC09B08 | 4.92 | 5.54 | 10.09 | Jihua 5 | |
| qHPWA08.7 | 18QA | A08 | 26.00 | Ah4-4-Ah2TC09B08 | 3.60 | 4.55 | 7.85 | Jihua 5 | |
| qHPWA08.8 | 18DM | A08 | 27.00 | Ah4-4-Ah2TC09B08 | 4.60 | 5.32 | 9.13 | Jihua 5 | |
| 20QY | A08 | 27.00 | Ah4-4-Ah2TC09B08 | 3.29 | 3.73 | 11.08 | Jihua 5 | ||
| qHPWB04.1 | 17QY | B04 | 1.11 | SMK1996-SMK1995 | 3.20 | 6.77 | -9.71 | M130 | |
| qHPWB05.1 | 19XL | B05 | 3.01 | SMK2087-SMK2088 | 2.60 | 5.72 | -10.30 | M130 | |
| qHPWB05.2 | 19XL | B05 | 16.01 | SMK2085-SMK2084 | 3.19 | 6.70 | -13.45 | M130 | |
| qHPWB06.1 | 17QY | B06 | 34.51 | SMK2106-SMK2107 | 2.84 | 5.99 | 7.51 | Jihua 5 | |
| 17DM | B06 | 34.51 | SMK2106-SMK2107 | 4.05 | 8.48 | 9.92 | Jihua 5 | ||
| qHPWB08.1 | 17DM | B08 | 0.00 | AHGS1286-Ah3TC20B05 | 5.47 | 6.20 | -9.30 | M130 | |
| 18QA | B08 | 0.00 | AHGS1286-Ah3TC20B05 | 3.22 | 3.66 | -7.07 | M130 | ||
| 19XL | B08 | 0.00 | AHGS1286-Ah3TC20B05 | 3.51 | 3.73 | -8.34 | M130 | ||
| qHPWB08.2 | 17QY | B08 | 1.00 | AHGS1286-Ah3TC20B05 | 3.97 | 4.42 | -7.66 | M130 | |
| 18DM | B08 | 1.00 | AHGS1286-Ah3TC20B05 | 5.90 | 6.57 | -10.21 | M130 | ||
| 20QY | B08 | 1.00 | AHGS1286-Ah3TC20B05 | 4.79 | 5.11 | -13.05 | M130 | ||
| qHPWB08.3 | 20QY | B08 | 17.21 | SMK2658-SMK2393 | 2.65 | 10.83 | 20.94 | Jihua 5 | |
| qHPWB08.4 | 20QY | B08 | 24.51 | SMK2406-SMK2423 | 2.57 | 5.56 | 16.27 | Jihua 5 | |
| qHPWB08.5 | 18QA | B08 | 36.81 | SMK2628-SMK2626 | 3.65 | 8.09 | -12.85 | M130 | |
| HSW | qHSWA03.1 | 17DM | A03 | 106.81 | SMK539-SMK540 | 4.04 | 8.54 | -3.88 | M130 |
| 18QY | A03 | 106.81 | SMK539-SMK540 | 2.67 | 5.87 | -3.74 | M130 | ||
| qHSWA04.1 | 17DM | A04 | 1.21 | SMK547-SMK549 | 2.75 | 5.79 | 2.84 | Jihua 5 | |
| qHSWA08.1 | 20QY | A08 | 6.23 | AhTE0658-TC22C01 | 5.65 | 6.93 | 5.32 | Jihua 5 | |
| qHSWA08.2 | 19XL | A08 | 6.97 | AhTE0658-TC22C01 | 3.75 | 4.47 | 3.30 | Jihua 5 | |
| qHSWA08.3 | 18DM | A08 | 9.12 | AhTE0658-TC22C01 | 5.00 | 6.00 | 3.52 | Jihua 5 | |
| qHSWA08.4 | 18QA | A08 | 19.00 | Ah1TC06H03-AhTE0477 | 3.91 | 6.37 | 3.59 | Jihua 5 | |
| qHSWA08.5 | 18QY | A08 | 26.00 | Ah4-4-Ah2TC09B08 | 6.66 | 9.20 | 4.52 | Jihua 5 | |
| 19XL | A08 | 26.00 | Ah4-4-Ah2TC09B08 | 5.74 | 6.59 | 3.99 | Jihua 5 | ||
| qHSWA08.6 | 17QY | A08 | 27.00 | Ah4-4-Ah2TC09B08 | 3.49 | 4.87 | 2.62 | Jihua 5 | |
| 18DM | A08 | 27.00 | Ah4-4-Ah2TC09B08 | 3.49 | 4.85 | 3.16 | Jihua 5 | ||
| 20QY | A08 | 27.00 | Ah4-4-Ah2TC09B08 | 4.00 | 4.63 | 4.32 | Jihua 5 | ||
| qHSWB04.1 | 18QA | B04 | 11.61 | SMK1978-SMK1848 | 4.39 | 10.43 | 5.84 | Jihua 5 | |
| qHSWB05.1 | 20QY | B05 | 29.91 | SMK2063-SMK2062 | 2.55 | 5.61 | 6.31 | Jihua 5 | |
| qHSWB06.1 | 17QY | B06 | 34.51 | SMK2106-SMK2107 | 3.89 | 8.52 | 3.26 | Jihua 5 | |
| qHSWB08.1 | 18DM | B08 | 0.00 | AHGS1286-Ah3TC20B08 | 4.73 | 5.50 | -3.38 | M130 | |
| qHSWB08.2 | 20QY | B08 | 1.00 | AHGS1286-Ah3TC20B08 | 3.86 | 4.14 | -4.11 | M130 |
| Chr. | Gene Names | Physical Position (bp) | Nr_annotation |
|---|---|---|---|
| A08 | Arahy.9AY9GA | 35,966,338~35,970,068 | DDRGK domain-containing protein 1-like [Glycine max] |
| A08 | Arahy.CX54HG | 35,973,257~35,975,499 | Translation initiation factor SUI1 family protein |
| A08 | Arahy.T6DWBF | 36,090,499~36,092,669 | Trafficking protein particle complex subunit-like protein |
| A08 | Arahy.D52S1Z | 36,116,001~36,121,083 | Probable 2-oxoglutarate/Fe(II)-dependent dioxygenase-like [Glycine max] |
| A08 | Arahy.HX3F52 | 36,151,497~36,153,080 | Calcium-dependent lipid-binding (CaLB domain) family protein |
| A08 | Arahy.IBM9RL | 36,178,370~36,181,596 | Polycomb group protein FERTILIZATION-INDEPENDENT ENDOSPERM-like [Glycine max] |
| A08 | Arahy.W18Y25 | 36,166,807~36,175,891 | Pentatricopeptide repeat (PPR) superfamily protein |
| A08 | Arahy.C7DQ5B | 36,181,753~36,189,459 | Uncharacterized protein LOC100782617 isoform X9 [Glycine max] |
| A08 | Arahy.CPLC2W | 36,196,903~36,197,724 | Pentatricopeptide repeat (PPR-like) superfamily protein |
| A08 | Arahy.C2FWCT | 36,238,299~36,245,344 | Breast carcinoma amplified sequence 3 protein |
| A08 | Arahy.XZZ787 | 36,255,496~36,260,840 | Probable galacturonosyltransferase 12-like [Glycine max] |
| A08 | Arahy.7ZYB2E | 36,272,312~36,273,769 | Thioredoxin 2 |
| A08 | Arahy.IUT8LB | 36,278,268~36,280,173 | Oxygen-evolving enhancer protein |
| A08 | Arahy.1IX743 | 36,281,314~36,282,631 | Papain family cysteine protease |
| A08 | Arahy.14EF4H | 36,285,790~36,286,678 | Sugar transporter 11 |
| A08 | Arahy.CY6UV3 | 36,314,963~36,316,298 | Papain family cysteine protease n=3 Tax=Leptospira RepID=M6CXX2_9LEPT |
| A08 | Arahy.5Z666J | 36,306,402~36,308,262 | Unknown protein |
| A08 | Arahy.9TI2ID | 36,317,379~36,323,108 | Papain family cysteine protease n=2 Tax=Leptospira RepID=N1U715_9LEPT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





