1. Introduction
Congenital anomalies (CA) are the main cause of newborn and infant deaths [
1]. Approximately 5% of newborns are found to have congenital malformations, with about 10-15% of congenital malformations being diagnosed as genetic/chromosomal abnormalities and approximately 50% classified as unknown causes [
2]. Therefore, efforts to identify environmental and genetic risk factors for causal determination have become crucial [
3]. Sufficient research resources, including in-depth data on patients and parents, as well as human-derived materials, are essential to elucidate risk factors and genetic factors associated with CA.
Rare diseases are characterized by a population of less than 20,000 individuals or conditions with unknown prevalence due to difficulties in diagnosis. More than 80% of rare diseases are genetic or congenital disorders that manifest in early childhood. Most of these diseases are serious or disabling, with a significant economic burden, as they often lack effective treatments or are associated with high-cost therapies. The National Institutes of Health (NIH) Undiagnosed Diseases Program (UDP), which has expanded nationwide as the Undiagnosed Diseases Network (UDN), is a program led by the National Institutes of Health (NIH) aimed at providing assistance in diagnosis and treatment decisions for patients. Patients provide medical information such as photographs, imaging data, and biopsy samples, and collaborate with NIH clinical centers to facilitate further diagnostic and treatment decisions. Approximately 40% of patients in the program are pediatric patients, primarily composed of CA and neurological disorders [
4]. In South Korea, the Rare Disease Management Act, Implementation Decree, and Enforcement Rule were implemented in 2016. Through the Rare Disease Registration and Statistics Project and Genetic Diagnosis Support Project, programs for the diagnosis of undiagnosed rare diseases have been developed and implemented [
5].
With the recent advancements in Next Generation Sequencing (NGS), personalized medical care tailored to individual patients has become possible for conditions that were previously difficult to diagnose and analyze molecular causes, enabling more effective treatment [
6]. The objective of this study was to establish a diagnostic research foundation for congenital malformation through clinical, epidemiological and genomic information collection protocols.
2. Methods/Design
2.1. Patient Registry Establishment and Selection of Eligible Patients
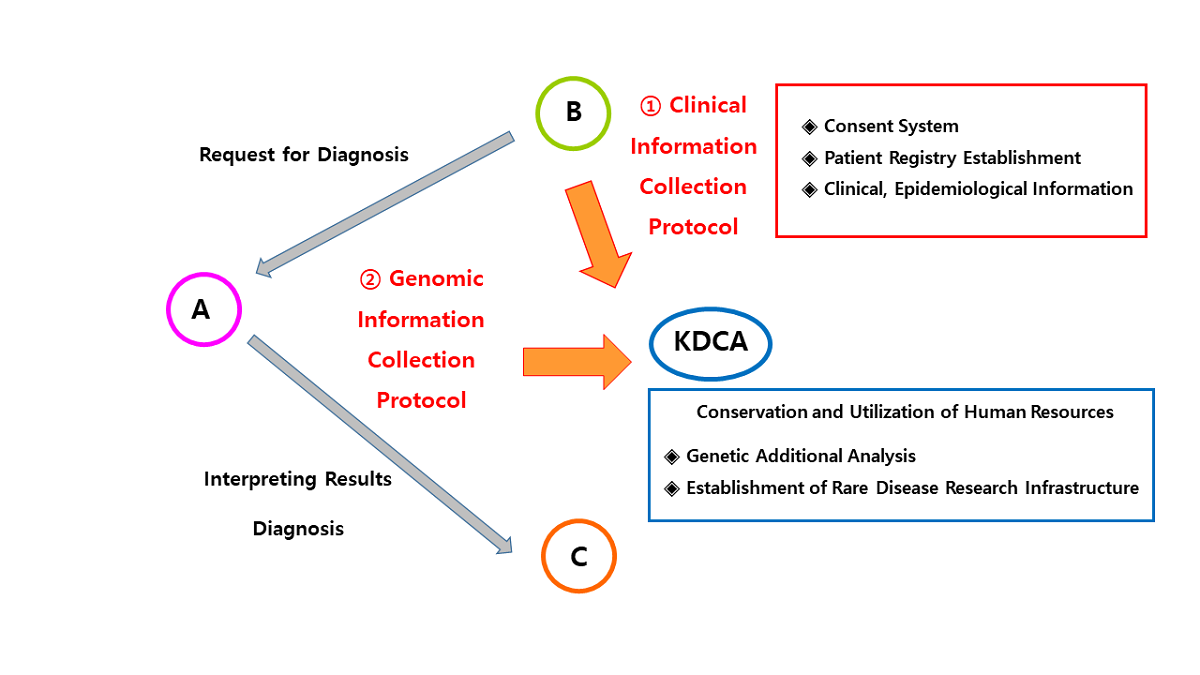
This study was developed through two academic research and development projects conducted by the Korea Centers for Disease Control and Prevention (KCDC) from April 1, 2021, to December 31, 2022 (Project No. 2021ER070600, April 1, 2021 - March 31, 2022, CM Project 2021; Project No. 2022ER060300, March 19, 2022 - December 31, 2022, CM Project 2022). Overview of the study is shown in
Figure 1.
The study targeted newborns who were negative for all items based on existing test results but had major multiple CA. The existing tests included basic complete blood count, comprehensive metabolic panel, blood gas analysis, urinalysis, newborn screening for congenital metabolic disorders, chromosomal analysis, and microarray analysis. Even if the results were negative for all items or showed positive findings that couldn't medically explain the newborn's phenotypes of multiple CA, they were invited to participate in this study. In recent rapidly advancing medical environments, there has been an increasing trend of performing targeted single gene testing or gene panel testing based on the phenotype expressed by the newborn, when there is clinical suspicion of involvement of specific genetic regions. Therefore, participation in this study was limited to cases where the results of single gene testing or gene panel testing were negative or inconclusive in explaining the newborn's phenotypes of multiple CA from a medical perspective.
A total of 45 families (135 individuals) consisting of newborns with CA and their parent trios were selected to participate in the study. In cases where genetic testing of relatives other than parents was medically necessary within the family, their inclusion in the study was determined through expert meetings on a selective basis.
2.2. Establishment of Consent System
A consent system for the collection and utilization (third-party provision) of human biological materials and related information (clinical, epidemiological and genomic information) was established, only when both parents of the patient have consented to participate in the research. The study protocols were approved by the Institutional Review Boards (IRBs) of the Samsung Medical Center (IRB no. 2021-04-189, 2022-04-054). Immediately after the start of the study, through research team meetings and extensive discussions, the items and scope of clinical/epidemiological information and genomic information to be collected in this study were pre-determined, and the appropriateness of each item was submitted to the IRB for ethics review. The ethical review process of the IRB was strictly followed, and the recommendations were adequately reflected in the consent form. In addition, the researchers completed education on the collection/production/donation of human resources conducted by the National Central Human Resources Bank/Biobank Division. The key elements of the consent form include voluntary participation, purpose/methods/procedures of the study, anticipated risks and discomfort, anticipated benefits, and personal information protection. Both the consent form for human biological material research and the donation consent form (Korea National Institute of Health, National Central Human Resources Bank) are required.
2.3. Development of Clinical Epidemiological Information Collection Protocol (Case Record Form)
For the collection of information on environmental factors related to the occurrence of CA, a questionnaire and case record form were developed to assess maternal and paternal exposure during the pregnancy period (
Figure 1). Key items included occupational history, residential area-related exposure to hazardous substances, smoking, alcohol consumption, radiation exposure, and increased body temperature and cell phone use [
7]. Regarding exposure to fine particulate matter, modeling was used to assess exposure levels if the address was available. Therefore, additional items were included such as the address of the residence for at least the previous 2 years, measures taken during periods of high fine particulate matter concentration, and the use of air purifiers. The questionnaire was supplemented with a focus on indoor environments, considering the characteristics of a prolonged period of indoor residence during pregnancy. The finalized questionnaire items, approved through expert consultation, were used to develop an electronic case report form (eCRF), which was entered and managed in the iCreaT system.
2.4. Collection of Human Biospecimens and DNA Generation
Blood samples from the study participants and their parents were collected (EDTA-treated blood). Additionally, urine samples from the parents were collected. These samples were processed to create research resources, including plasma, genomic DNA, and urine, which were stored in a -80°C freezer for preservation. In total, we obtained 138 human biological resources including plasma, genomic DNA, and urine samples, as well as 138 sets of whole genome sequencing data.
2.5. Generation of Genomic Information
Whole genome sequencing (WGS) was performed using the blood samples of the target infants and their parents (sequencing platform: NovaSeq6000). The sequencing depth was at least 30X (mean coverage), with an average read length of 151 bases. The human reference genome (hg 19) coverage was above 95% (minimum 10X threshold), and the data met the criteria of having a quality score of Q30 or higher for at least 85% of the data. The prepared data for donation includes raw data (FASTQ), processed data using the reference genome (BAM), variant calling data (VCF), and a protocol electronic document that enables reproducibility of the data analysis, including detailed instructions and commands for third-party analysis.
3. Discussion
Through the two projects, a standardized CA patient registry was established, and protocols necessary for collecting clinical research resources for human biospecimen banking were developed. An important aspect of developing the research consent form is to include essential information regarding consent for secondary use of resources within the preservation period after human biospecimen banking and the inclusion of personally identifiable information. The target patients in this project were newborns with major multiple CA, all of whom had negative findings in all items and was given consent by both parents and family members to participate in the research. Even if the results of previous single gene tests or genetic panel tests were positive, it would be meaningful to include infants with multiple CA that cannot be explained medically based on the phenotype for participation in the study. It is necessary to accurately collect various information such as the occupation of the mother and father, work environment, residential and living environment, exposure to teratogenic substances, medication intake, smoking history, alcohol consumption, caffeine, radiation exposure, heat increase, and dietary habits in the environmental questionnaire.
When recording and collecting major phenotypes, symptoms, and significant examination findings according to the disease, it is proposed to use HPO terms as much as possible for the diagnosis and phenotype information. Furthermore, it is crucial to select and continuously observe items that require continuous confirmation for natural course and prognosis and to continuously update clinical symptoms and newly developed genetic information that may be newly added or changed over time. In addition to the existing items, adding new items such as growth and development tests and conducting long-term follow-up on body measurements (weight, height, head circumference) according to corrected age, K-DST, Bailey's developmental test (in case of neurological abnormalities), visual acuity, hearing, readmissions, surgical procedures, and mortality would be highly meaningful [
8].
Recently, many studies on birth defects and genetic mutations in newborns have been conducted by NGS. The most important challenge is to explore new unidentified or potential neonatal mutations that can explain the corresponding phenotypes. To identify new mutations not inherited from the father or mother, parent-child trio WGS analysis can be a powerful approach, although bias interferes with identification [
6,
9].
Based on the protocols developed through these two research and development projects and the secured human resources, a foundation for establishing the basic process of early genetic diagnosis for CA after birth and a practical clinical research platform for elucidating genetic and environmental risk factors as causes of CA have been established. Specifically, WGS analysis of CA newborn-parents trio, obtaining essential consent forms based on the registry, developing a clinical information collection protocol using HPO terms as an objective tool for phenotypic quantification, introducing the basic concept of continuously updating clinical symptoms that may be newly added or changed over time, and developing a protocol that enables long-term tracking by adding new items that reflect the important characteristics of newborns such as growth and development tests have significant and innovative implications in this research.
4. Conclusion
Based on this clinical, epidemiological and genomic information and protocols of collected CA family trios, valuable information can be directly applied for early genetic diagnosis, identification of causes (genetic and environmental risk factors), and personalized tailored treatment related to each patient's CA in the future. As a follow-up study using the registration method (including consent) and protocol developed in this study, we propose a plan to fully utilize and expand the results of this study on a large scale at the national level.
References
- Verma, R.P. Evaluation and Risk Assessment of Congenital Anomalies in Neonates. Children 2021, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Vanassi, B.M.; Parma, G.C.; Magalhaes, V.S.; dos Santos, A.C.C.; Iser, B.P.M. Congenital anomalies in Santa Catarina: case distribution and trends in 2010–2018. Rev. Paul. de Pediatr. 2022, 40. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.B.; Foo, S.Y.R.; Chen, C.K. The Role of Epigenetics in Congenital Heart Disease. Genes 2021, 12, 390. [Google Scholar] [CrossRef] [PubMed]
- Stevens, S.; Miller, N.; Rashbass, J. Development and progress of the National Congenital Anomaly and Rare Disease Registration Service. Arch. Dis. Child. 2017, 103, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Lim, B.C.; Lee, J.S.; Kim, W.J.; Kim, H.; Ko, J.M.; Kim, K.J.; Choi, S.A.; Kim, H.; Hwang, H.; et al. The Korean undiagnosed diseases program: lessons from a one-year pilot project. Orphanet J. Rare Dis. 2019, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lantos, J.D. The Future of Newborn Genomic Testing. Children 2023, 10, 1140. [Google Scholar] [CrossRef]
- Baldacci, S.; Gorini, F.; Santoro, M.; Pierini, A.; Minichilli, F.; Bianchi, F. Environmental and individual exposure and the risk of congenital anomalies: a review of recent epidemiological evidence. Epidemiol Prev 2018, 42, 1–34. [Google Scholar]
- Lee, J.H.; Youn, Y.; Chang, Y.S.; Network, K.N. Short- and long-term outcomes of very low birth weight infants in Korea: Korean Neonatal Network update in 2019. Clin. Exp. Pediatr. 2020, 63, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhao, Z.; Chen, D.; He, S.; Sun, N.; Li, Z.; Liu, J.; Zhang, D.; Zhang, J.; Li, J.; et al. Detection of Clinically Relevant Genetic Variants in Chinese Patients With Nanophthalmos by Trio-Based Whole-Genome Sequencing Study. Investig. Opthalmology Vis. Sci. 2019, 60, 2904–2913. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).