1. Introduction
Mechanosensitive PIEZO ion channels are evolutionarily conserved membrane proteins whose function is critical for normal physiology in living cells and organisms (Coste, Xiao et al. 2012, Ranade, Qiu et al. 2014, Kefauver, Ward et al. 2020, Martinac 2022) ranging from single-celled ciliated protozoans to multicellular organisms, including plants, insects, worms and humans (Coste, Xiao et al. 2012, Ranade, Qiu et al. 2014, Douguet and Honoré 2019, Kefauver, Ward et al. 2020, Martinac 2022). In humans, they play a key role in sensing touch, tactile pain, breathing and blood pressure. However, they differ in their expression patterns and functions. The PIEZO1 channel is present in non-sensory tissues, with particularly high expression in the lung, bladder, and skin. By contrast, the PIEZO2 channel is predominantly present in sensory tissues, such as dorsal root ganglia (DRG) sensory neurons and Merkel cells (Gottlieb 2017, Douguet and Honoré 2019). While PIEZO1 channels are activated by the force-from-lipids indicating that they are inherently mechanosensitive (Cox, Bae et al. 2016, Syeda, Florendo et al. 2016) the inherent mechanosensitivity has not yet been demonstrated for PIEZO2 (Szczot, Nickolls et al. 2021). In addition to mechanical stimuli, PIEZO channels are also powerfully modulated by voltage (Moroni, Servin-Vences et al. 2018). Voltage modulation may be explained by the presence of an inactivation gate in the pore. Mutations that cause human diseases, such as Xerocytosis (Zarychanski, Schulz et al. 2012, Bae, Gnanasambandam et al. 2013), affect the channel inactivation and profoundly shift voltage sensitivity of the PIEZO1 channels towards the resting membrane potential, and strongly promote voltage gating (Moroni, Servin-Vences et al. 2018).
Inactivation is a general property of most types of ion channels enabling filtering out repetitive or prolonged stimuli by blocking the flow of ions via a mechanism other than the closing of the channel (Bahring and Covarrubias 2011, Bahring, Barghaan et al. 2012, Bae, Gnanasambandam et al. 2013, Vero Li, C et al. 2021, Britt, Moller et al. 2023). Structural studies reveal diverse mechanisms for inactivation, while most of the inactivation processes involve conformational changes in one or multiple inactivation gates located within the pore region. This mostly, but not necessarily happens only upon opening of the channels and generally limits the size of the permeation path for permeating ions. The inactivation state usually slowly recovers by transitioning to a closed channel state, which allows the channels to go through the open-inactive-closed cycle again before a next stimulus. Importantly, inactivation can be modified by intrinsic or extrinsic factors, with the latter ones including changes in pH, voltage, or temperature, surrounding membrane lipids and cellular components binding to the channels (Bae 2015, Douguet and Honoré 2019, Vasileva and Chubinskiy-Nadezhdin 2023).
The focus of this article is on PIEZO1 and PIEZO2 mechanosensitive channels which play a key role in sensing touch, tactile pain, breathing or blood pressure (Douguet and Honoré 2019). However, the two channels differ in their expression patterns and functions. In addition to their inherent mechanosensitivity (Cox, Bae et al. 2016, Syeda, Florendo et al. 2016) they can be modulated by the force-from-tethers transmitted to the channels by cytoskeletal or extracellular matrix molecules (Poole, Herget et al. 2014, Gaub and Muller 2017, Cox, Bavi et al. 2019, Vasileva and Chubinskiy-Nadezhdin 2023). Cryo-EM characterization of both channels reveals their very similar triskelion-like structure (Guo and MacKinnon 2017, Saotome, Murthy et al. 2018, Wang, Zhou et al. 2019, Yang 2022). A functional PIEZO channel is composed of three homomeric subunits with 38 transmembrane domains each, acting as an extensive blade-like element to transduce force to the pore. The C-terminus of the protein is composed of an extracellular cap domain, the pore-forming inner transmembrane helix (IH) and outer transmembrane helix (OH), and an intracellular C-terminal domain. Both PIEZO1 and PIEZO2 possess voltage-dependent inactivation (Lewis, Cui et al. 2017, Wu, Young et al. 2017, Zheng and Sachs 2017, Taberner, Prato et al. 2019, Zheng, Gracheva et al. 2019, Lewis and Grandl 2020, Glogowska, Arhatte et al. 2021, Sánchez Carranza 2022). Whole cell patch clamp with indentation shows a more rapid inactivation for PIEZO2, which has an inactivation time constant of around 5 ms compared to PIEZO1 of around 15 ms at a holding potential of -80 mV (Coste, Mathur et al. 2010, Glogowska, Arhatte et al. 2021) . Interestingly, only Piezo1 but not Piezo2 shows a fast inactivation inward current in the cell-attached mode (Murthy 2023). Previous studies have shown that the C terminus of the PIEZO channels, bears critical amino acids that are essential for their inactivation (Lewis and Grandl 2020). However, the precise mechanism underlying inactivation of the channels, remains elusive. In the present article, we summarize the current knowledge about the PIEZO channel inactivation with a structural insight. We also explain how the interacting partners, including lipids and proteins, exert their effects on this PIEZO channel gating property. Furthermore, we highlight our recent studies on a novel family of cellular proteins that tightly interact and regulate inactivation of these channels (Table 1). Finally, we discuss how inactivation of both ion channels is linked to human diseases and give some thoughts to what the future studies of the PIEZO channel inactivation may unveil.
2. Intrinsic mechanisms underlying inactivation of the PIEZO channels
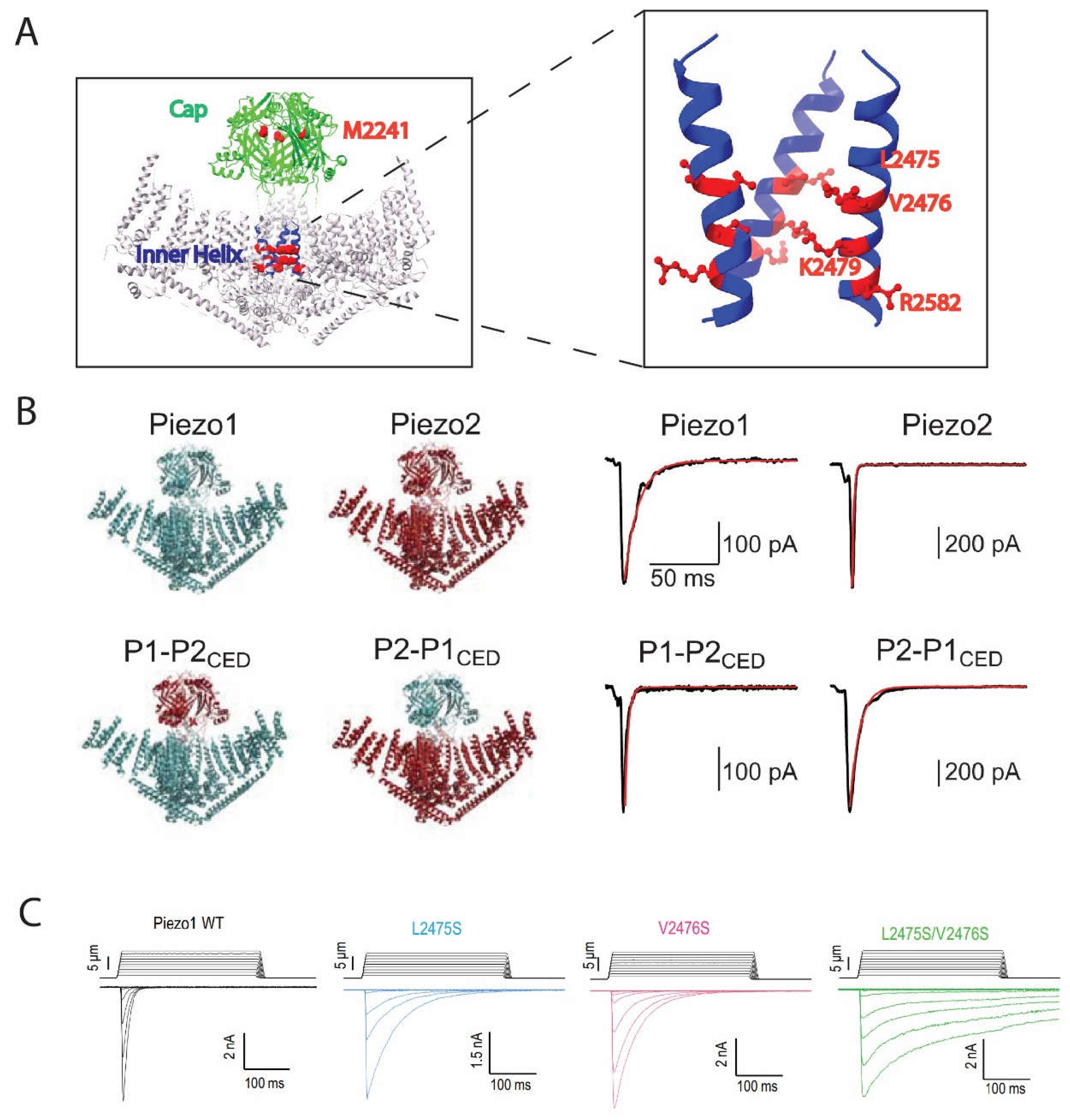
PIEZO1 and PIEZO2, exhibited fast inactivation in HEK293T or N2A cells during hyperpolarizing membrane potentials in outside-out and whole-cell patch clamping mode, while both channels inactivated slower at depolarizing membrane potentials (Gottlieb and Sachs 2012, Gottlieb 2017). PIEZO2 was further found to contribute to the rapid-inactivating, endogenous MS current in the mouse DRG neurons (Woo, Lukacs et al. 2015). Since then, several disease-causing, gain-of-function (GOF) mutants have provided critical insights into the potential mechanisms of the PIEZO1 and PIEZO2 inactivation. For PIEZO1, mutations that lead to a single amino acid substitution at human PIEZO1 M2225(M2225R) or R2456(R2456H), or mouse PIEZO1 M2241 or R2482, slow down PIEZO1 inactivation and are linked to xerocytosis
(Figure 1A
) (Zarychanski, Schulz et al. 2012, Bae, Gnanasambandam et al. 2013). A more comprehensive genetic screening identified other mutants, including R1358P, A2020T, T2127M and E2496ELE, with all of them reducing PIEZO1 inactivation.
Further studies on how the two amino acids M2225 and R2456 affect PIEZO1 inactivation reveal that these two are working independently but also cooperatively, while mutating both amino acids almost completely removed inactivation of the channel (Figure 1A). For PIEZO2, mutations at human PIEZO2 E2727(E2727del) or I802(I802F) exert similar effects as they alter PIEZO2 inactivation kinetics and cause distal arthrogryposis in humans which can be recapitulated in gene-modified mice (Coste, Houge et al. 2013). This information provided critical insights into the potential mechanisms of the PIEZO channel inactivation, while falling short of elucidating the mechanisms fully without having access to a 3D structure of the channels back then.
Thanks to the development of the high-resolution cryo-electron microscopy (cryo-EM) techniques within the last ten years, the structures of PIEZO1 and PIEZO2 have been solved over last couple of years at a resolution of ≥ 3.7 Å (Guo and MacKinnon 2017, Saotome, Murthy et al. 2018, Zhao Q 2018, Wang, Zhou et al. 2019) (Figure 1B). Since then, inactivation associated mutants were mapped back to the structures and the underlying mechanisms could be explained in more detail. For example, the M2225 and R2456 residues were found to belong to the C-terminal extracellular (cap) and IH regions, respectively (Lewis and Grandl 2020). It was also found that the cap and IH regions largely contribute to inactivation of both PIEZO1 and PIEZO2 (Zheng, Gracheva et al. 2019). Furthermore, exchanging the cap regions of PIEZO1 and PIEZO2 revealed that the cap region accounts for the difference between the gating kinetics of the two channels therefore the cap region must be important for inactivation (Wu, Young et al. 2017). In support of this idea, pulling amino acids within the cap region with magnetic nanoparticles largely reduced PIEZO1 inactivation (Wu, Goyal et al. 2016). A more precise dissection of the cap region confirmed that several subdomains within the cap of PIEZO2 were sufficient to confer the rapid inactivation of PIEZO2 to PIEZO1 (Lewis and Grandl 2020). Unlike the cap region, for which our understanding of how inactivation is affected remains unclear, our understanding of the inactivation mechanism within the inner helix has been clarified at a single amino acid level (Wu, Goyal et al. 2016). Two positively charged residues, which are K2453 and R2456 in human PIEZO1 or K2479 and R2482 in mouse PIEZO1, were found to be essential for the PIEZO1 voltage-dependent inactivation. Mutating R2482 largely removes inactivation while the remaining inactivation still exhibits voltage dependency. Size rather than charge of the residue seems to yield the effect, as R2482K or R2482Q both slow down inactivation significantly (Voltage gating of mechanosensitive PIEZO channels, 2018). In addition, the effect of R2482 is conserved in PIEZO2 as mutating the homologous mouse PIEZO2 R2756 to histidine (R2756H) or lysine (R2756K) slows down inactivation of a PIEZO1/PIEZO2 chimera (Sánchez Carranza 2022). On the other hand, neutralizing the positive charge on K2479 by mutating it to glutamine (K2479Q) or reverting the charge by mutating it to glutamic acid (K2479E) abolished voltage dependency of PIEZO1 inactivation. Importantly, they exerted opposite effects on inactivation, as K2479Q enhanced while K2479E removed inactivation, indicating the charge of this residue is essential to voltage dependency. Moreover, which L2475 and V2476 residues residing close to K2479 and R2582 in mouse PIEZO1, or L2749 and V2750 in the mouse PIEZO2, regulate inactivation of PIEZO1 and PIEZO2 in a conserved way (Figure 1C). Mutating both sites to serine (L2475S/V2476S or L2750S/V2751S) starkly removed inactivation in both channels. Structure of PIEZO1 indicates a 10 Å radius of pore size at L2475 and V2476. Thus, it is hypothesized that these two amino acids switch angles and face towards the pore, consequently narrowing down the pore size to <6 Å radius and forming a hydrophobic barrier that leads to inactivation (Zheng, Gracheva et al. 2019). Other structural motifs of PIEZO also contribute to inactivation. For example, the proximal intercellular C terminal domain (CTD) was shown to bear two very narrow constrictions at mouse PIEZO1 M2493/F2494 and E2537/P2536, with the MF constriction mildly contributing to PIEZO1 inactivation but not PIEZO2. Our recent data also indicate the involvement of the anchor domain, for which adding two glycine residues at anchor domain-outer helix linking human PIEZO1G2163 residue largely removed inactivation of PIEZO1 (Vero Li, C et al. 2021). It is worth mentioning that motifs outside the C terminus can influence channel inactivation as well (Wu, Young et al. 2017). First, a number of the gain-of-function mutants that reduce PIEZO channel inactivation, are in the peripheral transmembrane domains. However, it remains unclear what is the underlying mechanism. Second, tissue-specific alternative splicing which removes the PIEZO2 peripheral intercellular region alters PIEZO2 inactivation kinetics (Szczot, Pogorzala et al. 2017). We reason that inactivation of PIEZO channels may be affected by peripheral regions due to the force sensing or transmission, while the main inactivation gate is most likely located within the ECD cap domain and the inner helix pore-forming transmembrane domain.
3. Extrinsic factors modifying PIEZO inactivation
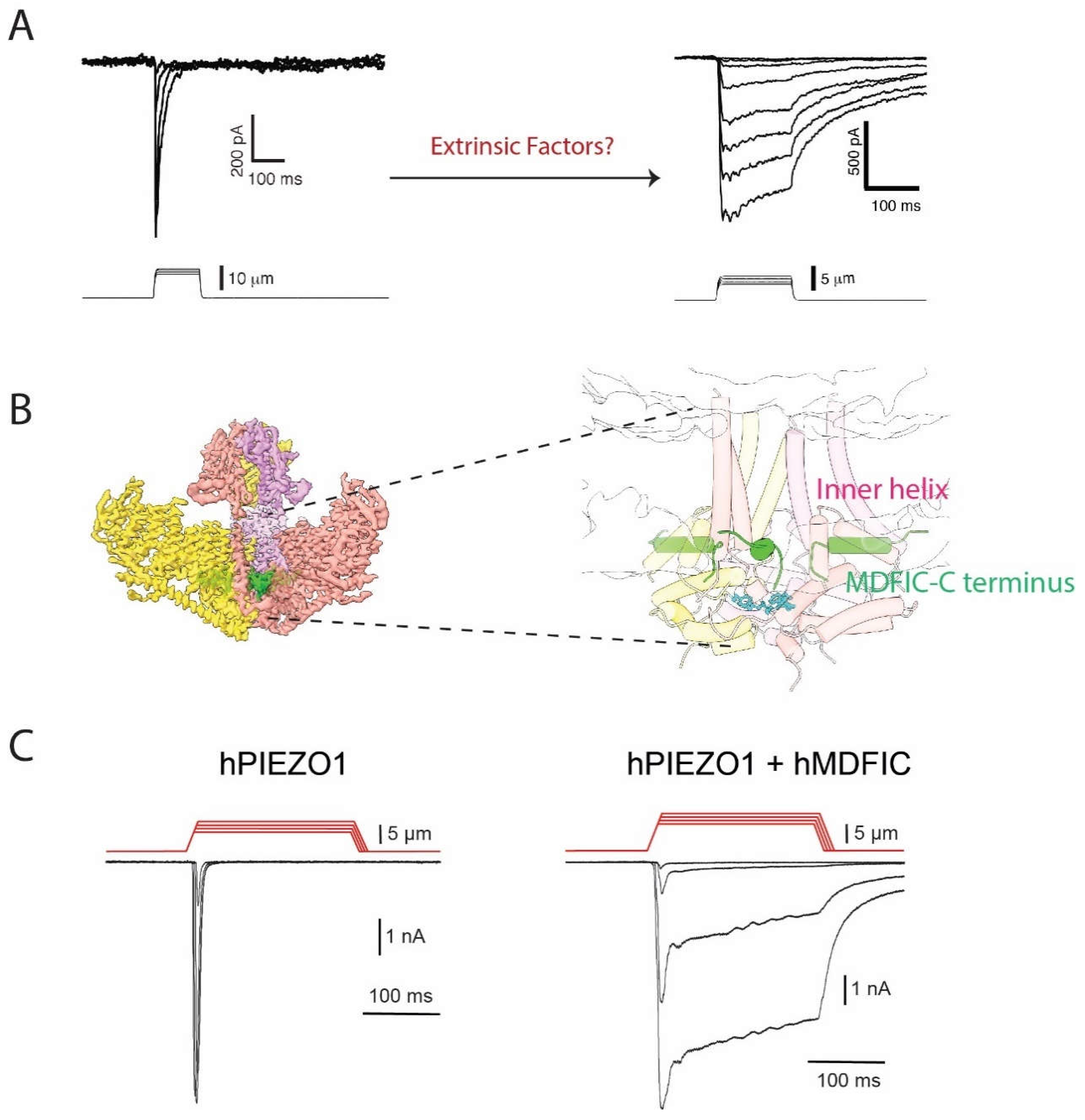
Most of our understanding of the intrinsic mechanisms of the PIEZO channel inactivation, as described above, is based on overexpressing PIEZO channels in a heterologous cell system. However, given that the activity of the channels can be modulated by their surrounding microenvironment various extrinsic factors can affect the channel kinetics, which is consistent with the fact that in some native cell lines PIEZO1 does not inactivate rapidly (Del Marmol, Touhara et al. 2018)
(Figure 2A
). In the following section, we focus on what is currently known about modulation of the PIEZO channel inactivation by extrinsic factors, including voltage, temperature, membrane lipids or intracellular proteins, which is summarized in the
Table 1.
The PIEZO channel inactivation exhibits strong voltage-dependency, which seems to be associated with the charged residues aligned within the pore forming inner helix (Moroni, Servin-Vences et al. 2018). As we have addressed it in the previous section, although extrinsic, membrane potential is closely linked to the intrinsic channel structure by affecting movement of the electrical charges associated with the channel conformational changes. Another remarkable extrinsic factor is pH, for which it has been found that protonation stabilizes inactivation of PIEZO1 (Bae 2015). This finding is based on the observation that PIEZO1 currents upon multiple pressure pulses decrease with decreasing pH. Change in pH does not influence the gain-of-function mutant R2456H or double mutant M2225R/R2456K, which lack inactivation already, further supporting the idea that low pH is promoting desensitization through altering inactivation of the channel. In addition, temperature is regulating inactivation of both channels across different species, as lowering temperature slows down inactivation for PIEZO1 and PIEZO2 overexpressed in heterologous system or endogenous mechanosensitive currents in the mouse DRG or duck TG cells. Mechanisms for temperature affecting inactivation is not yet clear, however stiffness of the lipid membrane, which is generally affected by temperature, seems not involved (Zheng, Nikolaev et al. 2019).
Lipids as integral component of the plasma membrane or signal molecules are also actively regulating the PIEZO channel inactivation (Vasileva and Chubinskiy-Nadezhdin 2023). Margaric acid inhibits both PIEZO1 and PIEZO2 channel activity but not the inactivation kinetics. On the other hand, linoleic acid (LA) 18:2 slows down PIEZO1 and PIEZO2 inactivation while potentiating both channels (Romero, Massey et al. 2019, Romero, Caires et al. 2023). The effect has been established by incubating cells that endogenously express PIEZO1 or PIEZO2 such as HMVEC or MCC13 cells, or N2A cells overexpressing PIEZO channels with LA followed by whole-cell patch clamping. This can be partially explained by LA increasing membrane disorder and therefore, altering membrane physical properties, as LA decreases lipid melting temperature, which also sensitizes MscL overexpressed in the N2A cells. Alteration in membrane properties by polyunsaturated fatty acids (PUFAs) on PIEZO1 and PIEZO2 channels determine the time course of the channel inactivation, as arachidonic acid (AA) 20:4 and eicosapentaenoic acid (EPA) 20:5 enhances while docosahexaenoic acid (DHA) 22:6 reduces inactivation of PIEZO1 (Romero, Massey et al. 2019). In contrast, EA or DHA do not influence PIEZO2 channel inactivation. This suggests a specific role of PUFAs on regulating PIEZO1 activity by possibly forming a lipid binding pocket within the channel. Nevertheless, LA and EPA have been shown to have potential for treating PIEZO related LOF or GOF diseases in the mouse model (Ma, Dubin et al. 2023, Romero, Caires et al. 2023). Ceramide is also implied to regulate PIEZO1 inactivation in freshly isolated second-order mesenteric artery endothelial cells (MAECs) (Shi, Hyman et al. 2020). Endogenous PIEZO1 current does not inactivate in the MAECs. By inhibiting SMPD3, which is a neutral sphingomyelinase that catalyzes the transition of sphingomyelin into ceramide, the native PIEZO1 current gain rapid inactivation. This can be rescued by incubating the cells with ceramide which restores the non-inactivating character of the PIEZO1 currents. Ceramide was also shown to be essential for MAECs’ response to flow, as the channels stay open for longer periods under continuous flow stimulation. As it is essential for the physical properties of endothelium, ceramide fails to regulate PIEZO1 overexpressed in a heterologous system. This suggests that ceramide may be required but it is not sufficient to expand the inactivation gate for which other components in the native endothelial cell are cooperatively functioning to regulate inactivation. In addition, our previous studies revealed a role of cholesterol and PIP2 in regulating PIEZO1 inactivation (Buyan, Cox et al. 2020). Cholesterol fostered PIEZO1 clustering when it is overexpressed in the HEK cells (Ridone, Pandzic et al. 2020). Removing cholesterol with methyl-β-cyclodextrin (MBCD) largely removed PIEZO1’s inactivation and lowered down PIEZO1 gating threshold to pressure. MD simulations identified multiple possible binding regions to cholesterol in PIEZO1 and provided an insight into a possible mechanism for cholesterol regulating PIEZO1 through specific binding to the channel. Interestingly, PIP2 is also suggested to bind to PIEZO1. A conserved motif in PIEZO1 K2166-K2169 is a potential binding site for PIP2 based on our simulations, while deleting these four lysine residues removed PIEZO1 inactivation (Buyan, Cox et al. 2020).
Importantly, PIEZO1 and PIEZO2 have been reported to interact physically and functionally with other cellular proteins. Consequently, these interacting partners potentiate or inhibit PIEZO channels activity through different mechanisms. Examples include SERCA2 binding to and reducing PIEZO1 and PIEZO2 peak current; E-cadherin binding to and potentiating PIEZO1 and PIEZO2 channel activity; STOML3 sensitizing PIEZO1 and PIEZO2 to mechanical stimuli; MTMR2 inhibiting PIEZO2 activity through PIP2; Polycyctin-2 (PKD2) interacting with PIEZO1 and reducing its function; PECAM-1 interacting with PIEZO1 at cell junctions and suppressing PIEZO1’s activity; TRPM4 interacting with PIEZO1 and amplifying PIEZO1 dependent calcium signals in cardiomyocytes, and TMEM150C interacting with PIEZO2 and positively regulating PIEZO1, PIEZO2 and TREK-1 channel activity (Qi, Andolfi et al. 2015, Anderson, Schneider et al. 2018, Wang 2022, Yu, Gong et al. 2022, Vasileva and Chubinskiy-Nadezhdin 2023) (Narayanan, Hutte et al. 2018) (Peyronnet, Martins et al. 2013). Those proteins which exert influence on PIEZO channels but not through a direct interaction with the channel, are not discussed here. It is worth noting that, along with potentiating or reducing the mechano-activated (MA) currents of PIEZO1 and PIEZO2 some of the binding partners, such as E-cadherin or PKD2, also regulate inactivation kinetics of the PIEZO channels; however, compared to the influence on the peak current their effect on inactivation seems not to be the primary cause but rather a consequence of altered channel activity. One binding partner that more specifically regulates PIEZO inactivation is TMEM150C, which was first thought to be an independent mechanosensitive channel that confers a relatively slow inactivating MA current. It was later shown that the slow inactivated MA current was the PIEZO1 current, as TEME150C overexpression did not produce MA currents in PIEZO1 knockout cells (Anderson, Schneider et al. 2018). The stark change in PIEZO1 inactivation kinetics with co-expression of TMEM150C led to further studies, which revealed an inactivation-removing effect of TMEM150C on PIEZO1, PIEZO2 and TREK-1 across multiple species, indicating that TMEM150 is a general modulator of the mechanosensitive channels (Anderson, Schneider et al. 2018). It is hypothesized that TMEM150C produces such influence on the MS channels by regulating compositions and physical properties of the membrane.
We recently identified MDFI and MDFIC that belong to the MyoD family of inhibitors as a novel family of proteins that physically interact with PIEZO1 and PIEZO2 by using affinity purification with endogenous fibroblast cell lines (Zhou 2023). We were able to resolve a high-resolution structure of PIEZO1-MDFIC complex by cryo-EM that clearly showed how a cysteine-rich, palmitoylated C terminus of MDFIC is inserted into the pore region of PIEZO1 (Figure 2B). MDFIC and MDFI seem to strongly regulate PIEZO1 and PIEZO2 inactivation kinetics since the co-expression of MDFIC or MDFI completely removed inactivation of PIEZO channels in a heterologous system (Figure 2C), while knocking down or mutating MDFIC restored a fast-inactivating endogenous PIEZO1 current in fibroblasts. We further found that the C-terminal palmitoylation is essential for the function of MDFIC interaction with both PIEZO channels. Furthermore, molecular dynamics simulations indicated that the palmitoylation chains may physically interfere with the amino acids located at the putative inactivation gate within the pore-forming helix of PIEZO1. To our knowledge, this is the first structure showing PIEZO channel complex with a binding partner. At the same time, our finding provides an important information towards better understanding of the PIEZO channel inactivation process.
4. Altered Inactivation kinetics of PIEZO channels is related to human diseases
Given their ubiquitous expression and unique function of PIEZO channels their mutations can be expected to cause severe consequences for mechanosensory transduction in a living organism. A large number of molecular mechanisms in disease-causing PIEZO channel mutations can be attributed to abnormal inactivation kinetics. Malfunction in PIEZO1 inactivation leads to hereditary Xerocytosis (or dehydrated hereditary stomatocytosis, DHS) (Zarychanski, Schulz et al. 2012, Bae, Gnanasambandam et al. 2013). DHS is characterized by fragile, dehydrated red blood cells, which results in hemolysis and severe anemia. Possible mechanism for PIEZO1 involvement in DHS is the lack of inactivation in the gain-of-function PIEZO1 mutants causing calcium overload in red blood cells (RBCs). The abundantly expressed Ca2+-dependent KCa3.1 channels are subsequently activated leading to excessive outflow of potassium, which further leads to dysregulation of ion concentration and osmolarity in the RBCs (Cahalan, Lukacs et al. 2015). On the other hand, genetic screening has located two mutations in PIEZO2 that cause Distal Arthrogryposis Type 5 (DA5) in humans (McMillin, Beck et al. 2014). Typical clinical symptom of DA5 can be described by generalized autosomal dominant contractures. PIEZO2 mutants associated with DA5 have slower inactivation kinetics, or faster recovery from inactivation. Deleting the mouse PIEZO2 E2799 residue, which mimics the human PIEZO2 mutant E2727del, is sufficient to cause DA5-like syndromes in mice. The mechanism for PIEZO2 E2727del is thought to be the hyperactivity of PIEZO2 in the proprioceptive sensory neurons, which can be partially rescued by neural transmission blocker botulinum toxin or EPA that restores PIEZO2 inactivation (Ma, Dubin et al. 2023).
In addition, a proper function of PIEZO1 was shown to be essential for lymphatic valve formation and development of the lymphatic system, given that lymphatic endothelial cell (LEC) specific PIEZO1 knockout mice exhibit postnatal lethality due to abnormal development of the lymphatic valve (Li, Hou et al. 2014). In humans, loss-of-function mutants of PIEZO1 cause general lymphatic dysplasia (GLD) that is characterized by severe lymphoedema affecting the whole body (Fotiou, Martin-Almedina et al. 2015, Lukacs, Mathur et al. 2015). The loss-of-function in these PIEZO1 mutants causing the disease has been attributed to aberrant protein trafficking and stability, or lack of the channel activity. However, most of the
PIEZO LOF mutants have not been studied in detail by the patch clamp. Unlike the removal of inactivation in the GOF mutants, possibilities of an enhanced inactivation in PIEZO channels and their disease-causing roles are less concerning partially because of the already rapid inactivation of the channels when they are overexpressed. In contrast, many native cells including the endothelia cells exhibit a non-inactivating PIEZO1 current. Our findings have demonstrated that MDFIC and MDFI contribute to a slow inactivating PIEZO1 in the native cells. Strikingly, knocking out MDFIC causes early postnatal death for the mice resulting from abnormal development of lymphatic valves. The coincidence of almost identical phenotypes in MDFIC and LEC-specific PIEZO1 knockout reminds us of a plausible mechanism for GLD, which is the insufficient removal of inactivation of PIEZO1 due to lack of MDFIC/MDFI expression or binding. Those LOF mutants within the interacting interface of PIEZO1 and MDFIC, such as PIEZO1 V2171F, are therefore of high interest to be investigated in the future.
5. Conclusions and expectations
The inactivation of PIEZO1 and PIEZO2 ion channels has emerged as a pivotal area of research with far-reaching implications for our understanding of mechanotransduction in various physiological and pathological processes. Both ion channels play essential roles in converting mechanical forces into electrical and biochemical signals, thereby influencing numerous cellular and tissue functions.
As crucial players in various sensory systems PIEZO1 and PIEZO2 channels have provided valuable insights into the mechanisms of touch and mechanical sensation, highlighting the importance of these channels in our ability to perceive and respond to the external environment. Dysfunctional PIEZO channels have been implicated in several pathological conditions, such as familial dehydrated stomatocytosis, congenital joint contractures, chronic pain syndromes and cardiovascular disorders. Understanding their inactivation mechanisms could pave the way for targeted therapeutic interventions to alleviate these conditions by modulating mechanosensory responses and may also shed light on tissue morphogenesis and regeneration processes. Consequently, detailed understanding of PIEZO channel inactivation mechanisms may inspire the design of novel drugs that can mimic or interfere with these processes.
In conclusion, the study of PIEZO1 and PIEZO2 channel inactivation and associated discoveries hold great promise for advancing our understanding of mechanosensation and its impact on various (patho)physiological processes. The implications of these discoveries extend to therapeutic interventions for diverse mechanopathologies. By delving deeper into the mechanisms of inactivation, researchers will be able to unlock new avenues for drug development and gene editing technologies, and thus potentially revolutionize the field of mechanobiology.
Acknowledgments
We thank Dr Charles Cox for critical reading of the manuscript.
References
- Anderson, E.O.; Schneider, E.R.; Matson, J.D.; Gracheva, E.O.; Bagriantsev, S.N. TMEM150C/Tentonin3 Is a Regulator of Mechano-gated Ion Channels. Cell Rep. 2018, 23, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Bae, C.; Gnanasambandam, R.; Nicolai, C.; Sachs, F.; Gottlieb, P.A. Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc. Natl. Acad. Sci. 2013, 110, E1162–E1168. [Google Scholar] [CrossRef] [PubMed]
- Bae, C.; Sachs, F.; Gottlieb, P.A. Protonation of the Human PIEZO1 Ion Channel Stabilizes Inactivation. J. Biol. Chem. 2015, 290, 5167–5173. [Google Scholar] [CrossRef] [PubMed]
- Bähring, R.; Barghaan, J.; Westermeier, R.; Wollberg, J. Voltage Sensor Inactivation in Potassium Channels. Front. Pharmacol. 2012, 3, 100. [Google Scholar] [CrossRef] [PubMed]
- Bähring, R.; Covarrubias, M. Mechanisms of closed-state inactivation in voltage-gated ion channels. Perspect. Surg. 2011, 589, 461–479. [Google Scholar] [CrossRef]
- Britt, M., E. Moller, J. Maramba, A. Anishkin and S. Sukharev (2023). "MscS inactivation and recovery are slow voltage-dependent processes sensitive to interactions with lipids." bioRxiv.
- Buyan, A.; Cox, C.D.; Barnoud, J.; Li, J.; Chan, H.S.; Martinac, B.; Marrink, S.J.; Corry, B. Piezo1 Forms Specific, Functionally Important Interactions with Phosphoinositides and Cholesterol. Biophys. J. 2020, 119, 1683–1697. [Google Scholar] [CrossRef]
- Cahalan, S.M.; Lukacs, V.; Ranade, S.S.; Chien, S.; Bandell, M.; Patapoutian, A. Piezo1 links mechanical forces to red blood cell volume. eLife 2015, 4. [Google Scholar] [CrossRef]
- Coste, B.; Houge, G.; Murray, M.F.; Stitziel, N.; Bandell, M.; Giovanni, M.A.; Philippakis, A.; Hoischen, A.; Riemer, G.; Steen, U.; et al. Gain-of-function mutations in the mechanically activated ion channel PIEZO2 cause a subtype of Distal Arthrogryposis. Proc. Natl. Acad. Sci. 2013, 110, 4667–4672. [Google Scholar] [CrossRef]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 Are Essential Components of Distinct Mechanically Activated Cation Channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef]
- Coste, B.; Xiao, B.; Santos, J.S.; Syeda, R.; Grandl, J.; Spencer, K.S.; Kim, S.E.; Schmidt, M.; Mathur, J.; Dubin, A.E.; et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 2012, 483, 176–181. [Google Scholar] [CrossRef]
- Cox, C.D.; Bae, C.; Ziegler, L.; Hartley, S.; Nikolova-Krstevski, V.; Rohde, P.R.; Ng, C.-A.; Sachs, F.; Gottlieb, P.A.; Martinac, B. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 2016, 7, 10366–10366. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D.; Bae, C.; Ziegler, L.; Hartley, S.; Nikolova-Krstevski, V.; Rohde, P.R.; Ng, C.-A.; Sachs, F.; Gottlieb, P.A.; Martinac, B. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 2016, 7, 10366–10366. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D.; Bavi, N.; Martinac, B. Biophysical Principles of Ion-Channel-Mediated Mechanosensory Transduction. Cell Rep. 2019, 29, 1–12. [Google Scholar] [CrossRef]
- Del Marmol, J. I., K. K. Touhara, G. Croft and R. MacKinnon (2018). "Piezo1 forms a slowly-inactivating mechanosensory channel in mouse embryonic stem cells." Elife 7.
- Douguet, D.; Honoré, E. Mammalian Mechanoelectrical Transduction: Structure and Function of Force-Gated Ion Channels. Cell 2019, 179, 340–354. [Google Scholar] [CrossRef]
- Fotiou, E.; Martin-Almedina, S.; Simpson, M.A.; Lin, S.; Gordon, K.; Brice, G.; Atton, G.; Jeffery, I.; Rees, D.C.; Mignot, C.; et al. Novel mutations in PIEZO1 cause an autosomal recessive generalized lymphatic dysplasia with non-immune hydrops fetalis. Nat. Commun. 2015, 6, 8085. [Google Scholar] [CrossRef] [PubMed]
- Gaub, B.M.; Müller, D.J. Mechanical Stimulation of Piezo1 Receptors Depends on Extracellular Matrix Proteins and Directionality of Force. Nano Lett. 2017, 17, 2064–2072. [Google Scholar] [CrossRef] [PubMed]
- Glogowska, E.; Arhatte, M.; Chatelain, F.C.; Lesage, F.; Xu, A.; Grashoff, C.; Discher, D.E.; Patel, A.; Honoré, E. Piezo1 and Piezo2 foster mechanical gating of K2P channels. Cell Rep. 2021, 37, 110070. [Google Scholar] [CrossRef]
- Gottlieb, P. A. (2017). "A Tour de Force: The Discovery, Properties, and Function of Piezo Channels." Curr Top Membr 79: 1-36.
- Gottlieb, P. A. and F. Sachs (2012). "Piezo1: properties of a cation selective mechanical channel." Channels (Austin) 6(4): 214-219.
- Guo, Y. R. and R. MacKinnon (2017). "Structure-based membrane dome mechanism for Piezo mechanosensitivity." Elife 6.
- Kefauver, J.M.; Ward, A.B.; Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nature 2020, 587, 567–576. [Google Scholar] [CrossRef]
- Lewis, A.H.; Cui, A.F.; McDonald, M.F.; Grandl, J. Transduction of Repetitive Mechanical Stimuli by Piezo1 and Piezo2 Ion Channels. Cell Rep. 2017, 19, 2572–2585. [Google Scholar] [CrossRef]
- Lewis, A.H.; Grandl, J. Inactivation Kinetics and Mechanical Gating of Piezo1 Ion Channels Depend on Subdomains within the Cap. Cell Rep. 2020, 30, 870–880. [Google Scholar] [CrossRef]
- Li, J.; Hou, B.; Tumova, S.; Muraki, K.; Bruns, A.; Ludlow, M.J.; Sedo, A.; Hyman, A.J.; McKeown, L.; Young, R.S.; et al. Piezo1 integration of vascular architecture with physiological force. Nature 2014, 515, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, V.; Mathur, J.; Mao, R.; Bayrak-Toydemir, P.; Procter, M.; Cahalan, S.M.; Kim, H.J.; Bandell, M.; Longo, N.; Day, R.W.; et al. Impaired PIEZO1 function in patients with a novel autosomal recessive congenital lymphatic dysplasia. Nat. Commun. 2015, 6, 8329. [Google Scholar] [CrossRef]
- Ma, S.; Dubin, A.E.; Romero, L.O.; Loud, M.; Salazar, A.; Chu, S.; Klier, N.; Masri, S.; Zhang, Y.; Wang, Y.; et al. Excessive mechanotransduction in sensory neurons causes joint contractures. Science 2023, 379, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Martinac, B. 2021 Nobel Prize for mechanosensory transduction. Biophys. Rev. 2022, 14, 15–20. [Google Scholar] [CrossRef] [PubMed]
- McMillin, M. J., A. E. Beck, J. X. Chong, K. M. Shively, K. J. Buckingham, H. I. Gildersleeve, M. I. Aracena, A. S. Aylsworth, P. Bitoun, J. C. Carey, C. L. Clericuzio, Y. J. Crow, C. J. Curry, K. Devriendt, D. B. Everman, A. Fryer, K. Gibson, M. L. Giovannucci Uzielli, J. M. Graham, Jr., J. G. Hall, J. T. Hecht, R. A. Heidenreich, J. A. Hurst, S. Irani, I. P. Krapels, J. G. Leroy, D. Mowat, G. T. Plant, S. P. Robertson, E. K. Schorry, R. H. Scott, L. H. Seaver, E. Sherr, M. Splitt, H. Stewart, C. Stumpel, S. G. Temel, D. D. Weaver, M. Whiteford, M. S. Williams, H. K. Tabor, J. D. Smith, J. Shendure, D. A. Nickerson, G. University of Washington Center for Mendelian and M. J. Bamshad (2014). "Mutations in PIEZO2 cause Gordon syndrome, Marden-Walker syndrome, and distal arthrogryposis type 5." Am J Hum Genet 94(5): 734-744.
- Moroni, M.; Servin-Vences, M.R.; Fleischer, R.; Sánchez-Carranza, O.; Lewin, G.R. Voltage gating of mechanosensitive PIEZO channels. Nat. Commun. 2018, 9, 1096. [Google Scholar] [CrossRef]
- Murthy, S.E. Deciphering mechanically activated ion channels at the single-channel level in dorsal root ganglion neurons. J. Gen. Physiol. 2023, 155. [Google Scholar] [CrossRef]
- Narayanan, P., M. Hutte, G. Kudryasheva, F. J. Taberner, S. G. Lechner, F. Rehfeldt, D. Gomez-Varela and M. Schmidt (2018). "Myotubularin related protein-2 and its phospholipid substrate PIP(2) control Piezo2-mediated mechanotransduction in peripheral sensory neurons." Elife 7.
- Peyronnet, R., J. R. Martins, F. Duprat, S. Demolombe, M. Arhatte, M. Jodar, M. Tauc, C. Duranton, M. Paulais, J. Teulon, E. Honore and A. Patel (2013). "Piezo1-dependent stretch-activated channels are inhibited by Polycystin-2 in renal tubular epithelial cells." EMBO Rep 14(12): 1143-1148.
- Poole, K.; Herget, R.; Lapatsina, L.; Ngo, H.-D.; Lewin, G.R. Tuning Piezo ion channels to detect molecular-scale movements relevant for fine touch. Nat. Commun. 2014, 5, 3520. [Google Scholar] [CrossRef]
- Qi, Y.; Andolfi, L.; Frattini, F.; Mayer, F.; Lazzarino, M.; Hu, J. Membrane stiffening by STOML3 facilitates mechanosensation in sensory neurons. Nat. Commun. 2015, 6, 8512. [Google Scholar] [CrossRef]
- Ranade, S. S., Z. Qiu, S. H. Woo, S. S. Hur, S. E. Murthy, S. M. Cahalan, J. Xu, J. Mathur, M. Bandell, B. Coste, Y. S. Li, S. Chien and A. Patapoutian (2014). "Piezo1, a mechanically activated ion channel, is required for vascular development in mice." Proc Natl Acad Sci U S A 111(28): 10347-10352.
- Ridone, P.; Pandzic, E.; Vassalli, M.; Cox, C.D.; Macmillan, A.; Gottlieb, P.A.; Martinac, B. Disruption of membrane cholesterol organization impairs the activity of PIEZO1 channel clusters. J. Gen. Physiol. 2020, 152. [Google Scholar] [CrossRef]
- Romero, L.O.; Caires, R.; Victor, A.K.; Ramirez, J.; Sierra-Valdez, F.J.; Walsh, P.; Truong, V.; Lee, J.; Mayor, U.; Reiter, L.T.; et al. Linoleic acid improves PIEZO2 dysfunction in a mouse model of Angelman Syndrome. Nat. Commun. 2023, 14, 1–20. [Google Scholar] [CrossRef]
- Romero, L.O.; Massey, A.E.; Mata-Daboin, A.D.; Sierra-Valdez, F.J.; Chauhan, S.C.; Cordero-Morales, J.F.; Vásquez, V. Dietary fatty acids fine-tune Piezo1 mechanical response. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Sánchez Carranza, O. , Chakrabarti, S., Kühnemund, J., Schwaller, F., Bégay, V. and Lewin, G.R. (2022). "Piezo2 voltage-block regulates mechanical pain sensitivity." bioRxiv.
- Saotome, K.; Murthy, S.E.; Kefauver, J.M.; Whitwam, T.; Patapoutian, A.; Ward, A.B. Structure of the mechanically activated ion channel Piezo1. Nature 2017, 554, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Hyman, A.J.; De Vecchis, D.; Chong, J.; Lichtenstein, L.; Futers, T.S.; Rouahi, M.; Salvayre, A.N.; Auge, N.; Kalli, A.C.; et al. Sphingomyelinase Disables Inactivation in Endogenous PIEZO1 Channels. Cell Rep. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Syeda, R.; Florendo, M.N.; Cox, C.D.; Kefauver, J.M.; Santos, J.S.; Martinac, B.; Patapoutian, A. Piezo1 Channels Are Inherently Mechanosensitive. Cell Rep. 2016, 17, 1739–1746. [Google Scholar] [CrossRef] [PubMed]
- Szczot, M.; Nickolls, A.R.; Lam, R.M.; Chesler, A.T. The Form and Function of PIEZO2. Annu. Rev. Biochem. 2021, 90, 507–534. [Google Scholar] [CrossRef]
- Szczot, M.; Pogorzala, L.A.; Solinski, H.J.; Young, L.; Yee, P.; Le Pichon, C.E.; Chesler, A.T.; Hoon, M.A. Cell-Type-Specific Splicing of Piezo2 Regulates Mechanotransduction. Cell Rep. 2017, 21, 2760–2771. [Google Scholar] [CrossRef]
- Taberner, F.J.; Prato, V.; Schaefer, I.; Schrenk-Siemens, K.; Heppenstall, P.A.; Lechner, S.G. Structure-guided examination of the mechanogating mechanism of PIEZO2. Proc. Natl. Acad. Sci. 2019, 116, 14260–14269. [Google Scholar] [CrossRef]
- Vasileva, V.; Chubinskiy-Nadezhdin, V. Regulation of PIEZO1 channels by lipids and the structural components of extracellular matrix/cell cytoskeleton. J. Cell. Physiol. 2023, 238, 918–930. [Google Scholar] [CrossRef]
- Li, J.V.; Cox, C.D.; Martinac, B. The anchor domain is critical for Piezo1 channel mechanosensitivity. Channels 2021, 15, 438–446. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, J.; Yang, X.; Zhou, G.; Wang, L.; Xiao, B. Tethering Piezo channels to the actin cytoskeleton for mechanogating via the cadherin-β-catenin mechanotransduction complex. Cell Rep. 2022, 38, 110342. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, H.; Zhang, M.; Liu, W.; Deng, T.; Zhao, Q.; Li, Y.; Lei, J.; Li, X.; Xiao, B. Structure and mechanogating of the mammalian tactile channel PIEZO2. Nature 2019, 573, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.-H.; Lukacs, V.; de Nooij, J.C.; Zaytseva, D.; Criddle, C.R.; Francisco, A.; Jessell, T.M.; A Wilkinson, K.; Patapoutian, A. Piezo2 is the principal mechanotransduction channel for proprioception. Nat. Neurosci. 2015, 18, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Goyal, R.; Grandl, J. Localized force application reveals mechanically sensitive domains of Piezo1. Nat. Commun. 2016, 7, 12939. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Young, M.; Lewis, A.H.; Martfeld, A.N.; Kalmeta, B.; Grandl, J. Inactivation of Mechanically Activated Piezo1 Ion Channels Is Determined by the C-Terminal Extracellular Domain and the Inner Pore Helix. Cell Rep. 2017, 21, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lin, C.; Chen, X.; Li, S.; Li, X.; Xiao, B. Structure deformation and curvature sensing of PIEZO1 in lipid membranes. Nature 2022, 604, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.-Y.; Gong, H.; Kesteven, S.; Guo, Y.; Wu, J.; Li, J.V.; Cheng, D.; Zhou, Z.; Iismaa, S.E.; Kaidonis, X.; et al. Piezo1 is the cardiac mechanosensor that initiates the cardiomyocyte hypertrophic response to pressure overload in adult mice. Nat. Cardiovasc. Res. 2022, 1, 577–591. [Google Scholar] [CrossRef]
- Zarychanski, R.; Schulz, V.P.; Houston, B.L.; Maksimova, Y.; Houston, D.S.; Smith, B.; Rinehart, J.; Gallagher, P.G. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood 2012, 120, 1908–1915. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, H.; Chi, S.; Wang, Y.; Wang, J.; Geng, J.; Wu, K.; Liu, W.; Zhang, T.; Dong, M.-Q.; et al. Structure and mechanogating mechanism of the Piezo1 channel. Nature 2018, 554, 487–492. [Google Scholar] [CrossRef]
- Zheng, W., E. O. Gracheva and S. N. Bagriantsev (2019). "A hydrophobic gate in the inner pore helix is the major determinant of inactivation in mechanosensitive Piezo channels." Elife 8.
- Zheng, W.; Nikolaev, Y.A.; Gracheva, E.O.; Bagriantsev, S.N. Piezo2 integrates mechanical and thermal cues in vertebrate mechanoreceptors. Proc. Natl. Acad. Sci. 2019, 116, 17547–17555. [Google Scholar] [CrossRef]
- Zheng, W.; Sachs, F. Investigating the structural dynamics of the PIEZO1 channel activation and inactivation by coarse-grained modeling. Proteins: Struct. Funct. Bioinform. 2017, 85, 2198–2208. [Google Scholar] [CrossRef]
- Zhou, Z. , Ma, X., Lin, Y.C., Cheng, D., Bavi, N., Li, J.V., Janbandhu, V., Sutton, D.L., Scott, H.S., Yao, M., Harvey, R.P., Harvey, N., Corry, B., Zhang, Y. and Cox, C.D. (2023). "MyoD family inhibitor proteins act as auxiliary subunits of Piezo channels." Science (in press).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).