1. Introduction
With rapid developments in nontraditional applied sciences such as mathematical finance [
1], image processing [
2], economics [
3], and data science [
4], there is an ever increasing demand for efficient numerical methods for computing high-dimensional integration which also becomes crucial for solving some challenging problems. Numerical methods (or quadrature rules) mostly stem from approximating the Riemann sum in the definition of integrals, hence, they are grid-based. The simplest and most natural approach for constructing numerical quadrature rules in high dimensions is to apply the same 1-d rule in each coordinate direction, this then leads to tensor-product (TP) quadrature rules. It is well known (and easy to check) that the number of integration points (and function evaluations) grows
exponentially in the dimension
d, such a phenomenon is known as
the curse of dimensionality (CoD). Mitigating or circumventing the CoD has been the primary goal when it comes to constructing efficient high-dimensional numerical quadrature rules. A lot of progress has been made in this direction in the past fifty years, this includes sparse grid (SG) methods [
1,
5,
6,
7], Monte Carlo (MC) methods [
8,
9], Quasi-Monte Carlo (QMC) methods [
10,
11,
12,
13,
14], deep neural network (DNN) methods [
15,
16,
17,
18,
19]. To some certain extent, those methods are effective for computing integrals in low and medium dimensions (i.e.,
), but it is still a challenge for them to compute integrals in very high dimensions (i.e.,
).
This is the second installment in a sequel [
20] which aims at developing fast numerical algorithms for high-dimensional numerical integration. As mentioned above, the straightforward implementation of the TP method will evidently run into the CoD dilemma. To circumvent the difficulty, we proposed in [
20] a multilevel dimension iteration algorithm (called MDI-TP) for accelerating the TP method. The ideas of the MDI-TP algorithm are to reuse the computation of function evaluation as much as possible in the tensor product method by clustering computations, which allows an efficient and fast function evaluations at integration points together, and to do clustering by a simple dimension iteration/reduction strategy, which is possible because of the lattice structure of the TP integration points. Since the idea of the MDI strategy essentially applied to any numerical integration rule whose integration points have a lattice-like structure, this indeed motivates the work of this paper by applying the MDI idea to accelerate the sparse grid method. We recall that, unlike the existing numerical integration methods, the MDI algorithm is not aiming to providing a new numerical integration method per se, instead, it is an acceleration algorithm for an efficient implementation of any tensor-product-like existing method. Thus, the MDI is not a “discretization" method but a “solver" (borrowing the numerical PDE terminologies). A well suited analogy would be high order polynomial evaluations, that is, to compute
for a given real number input
. It is well known that such a high order polynomial evaluation on a computer is notoriously unstable, inaccurate (due to roundoff errors) and expensive, however, those difficulties can be easily overcome by a simple nested iteration (or Horner’s algorithm. cf. [
21]), namely, set
and for
, set
. From the cost saving and efficiency point view, the reason for the nested iteration to be efficient and fast is that it reuses many multiplications involving
compared to the direct evaluations of each term in
. Conceptually, this is exactly the main idea of the MDI algorithm, i.e., to reuse computations of the function evaluations in an existing method as much as possible to save the computation cost and hence to make it efficient and fast.
The sparse grid (SG) method, which was first proposed by Smolyak in [
22], only uses a (small) subset of the TP integration points while still maintains a comparable accuracy of the TP method. As mentioned earlier, the SG method was one of few successful numerical methods which can mitigate the CoD in high-dimensional computation, including computing high-dimensional integration and solving high-dimensional PDEs [
1,
5,
6,
7]. The basic idea in application to high-dimensional numerical integration stems from Smolyak’s general method for multivariate extensions of univariate operators. Based on this construction, the midpoint rule [
23], the rectangle rule [
24], the trapezoidal rule [
25], the Clenshaw-Curtis rule [
26,
27], and the Gaussian-Legendre rule [
28,
29] have been used as a one-dimensional numerical integration method. A multivariate quadrature rule is then constructed by forming the TP method of each of these one-dimensional rules on the underlying sparse grid. Like TP method, the SG method is quite general and easy to implement. But unlike the TP method, its computational cost is much lower because the number of its required function evaluations grows exponentially with a smaller base. A key observation is that the function evaluations of the SG method involve a lot of computation in each coordinate direction which can be shared because each coordinate
of every integration point
is shared by many other integration points due to their tensor product structure. This observation motivates us to compute the required function evaluations in cluster and iteratively in each coordinate direction instead of computing them at the integration points independently.
The goal of this paper is to apply the MDI strategy to the SG method, the resulting algorithm, which is called the MDI-SG algorithm, provides a fast algorithm for an efficient implementation of the SG method. The MDI-SG method incorporates the MDI nested iteration idea into the sparse grid method which allows the reuse of the computation in the function evaluations at the integration points as much as possible. This saving significantly reduces the overall computational cost for implementing the sparse grid method from an exponential growth to a low-order polynomial growth.
The rest of this paper is organized as follows. In
Section 2, we first briefly recall the formulation of the sparse grid method and some known facts. In
Section 3, we introduce our MDI-SG algorithm, first in two and three dimensions to explain the main ideas of the algorithm, and then generalize it to arbitrary dimensions. In
Section 4 and
Section 5, we present various numerical experiments to test the performance of the proposed MDI-SG algorithm and compare its performances to the standard SG method and the classical MC method. These numerical tests show that the MDI-SG algorithm is much faster in low and medium dimensions (i.e.
) and in very high dimensions (i.e.
). It still works even when the MC method fails to compute. We also provide numerical tests to measure the influence of parameters in the proposed MDI-SG algorithm, including dependencies on the choice of underlying 1-d quadrature rule, and the choice of iteration step size. In
Section 6 we numerically estimate the computational complexity of the MDI-SG algorithm. This is done by using standard regression technique to discover the relationship between CPU time and dimension. It is showed that the CPU time grows at most in a polynomial order
, where
d and
N stand for respectively the dimension of integration domain and the maximum number of integration points in each coordinate direction. As a result, the proposed MDI-SG method effectively circumvents the curse of dimensionality. Finally, the paper is concluded with some concluding remarks given in
Section 7.
2. Preliminaries
In this paper,
denote a given continuous function on
for
, then
has pointwise values at every
Without loss of the generality, we set
and consider the basic and fundamental problem of computing the integral
As mentioned in
Section 1, the goal of this paper is to develop a fast algorithm for computing above integral based on the sparse grid methodology. To that end, below we briefly recall the necessary elements of sparse grid methods.
2.1. The sparse grid method
We now recall the formulation of the sparse grid method for approximating (
1) and its tensor product reformulation formula which will be crucially used later in the formulation of our fast MDI-SG algorithm.
For each positive integer index
, let
be a positive integer which denotes the number of grid points at level
l and
denote a sequence of the level
l grid points in
. The grid sets
is said to be nested provided that
. The best known example of the nested grids is the following dyadic grids:
For a given positive integer
q, the tensor product
then yields a standard tensor product grid on
. Notice that here the
qth level is used in each coordinate direction. To reduce the number of grid points in
, the sparse grid idea is to restrict the total level to be
q in the sense that
, where
is the level used in the
ith coordinate direction, its corresponding tensor product grid is
. Obviously, the decomposition
is not unique, so all such decomposition must be considered. The union
then yields the famous Smolyak sparse grid (imbedded) on
(cf. [
6]). We remark that the underlying idea of going from
to
is exactly same as going from
to
, where
denotes the set of polynomials whose degrees in all coordinate directions not exceeding
q and
denotes the set of polynomials whose total degrees not exceeding
q.
After having introduced the concept of sparse grids, we then can define the sparse grid quadrature rule. For a univariate function
g on
, we consider
d one-dimensional quadrature formula
where
and
denote respectively the integration points/nodes and weights of the quadrature rule, and
denotes the number of integration points in the
ith coordinate direction in
. Define
For example
.
Then, the sparse grid quadrature rule with accuracy level
for d-dimensional integration (
1) on
is defined as (cf. [
1])
where
We note that each term
in (
7) is the tensor product quadrature rule which uses
integration points in the
ith coordinate direction. To write
more compactly, we set
which denotes the total number of integration points in
. Let
, denote the corresponding weights and define the bijective mapping
Then, the sparse grid quadrature rule
can be rewritten as
We also note that some weights
may become negative even though the one-dimensional weights
are positive. Therefore, it is no longer possible to interpret
as a discrete probability measure. Moreover, the existence of negative weights in (
8) may cause numerical cancellation, hence, loss of significant digits. To circumvent such a potential cancellation, it is recommended in [
7] that the summation is carried out by coordinates, this then leads to the following tensor product reformulation of
:
where the upper limits are defined recursively as
In the nested mesh case (i.e.,
is nested), nested integration points are selected to form the sparse grid. We remark that different 1-d quadrature rules in (
5) will lead to different sparse grid methods in (
6). We also note that the tensor product reformulation (
9) will play a crucial role later in the construction of our MD-SG algorithm.
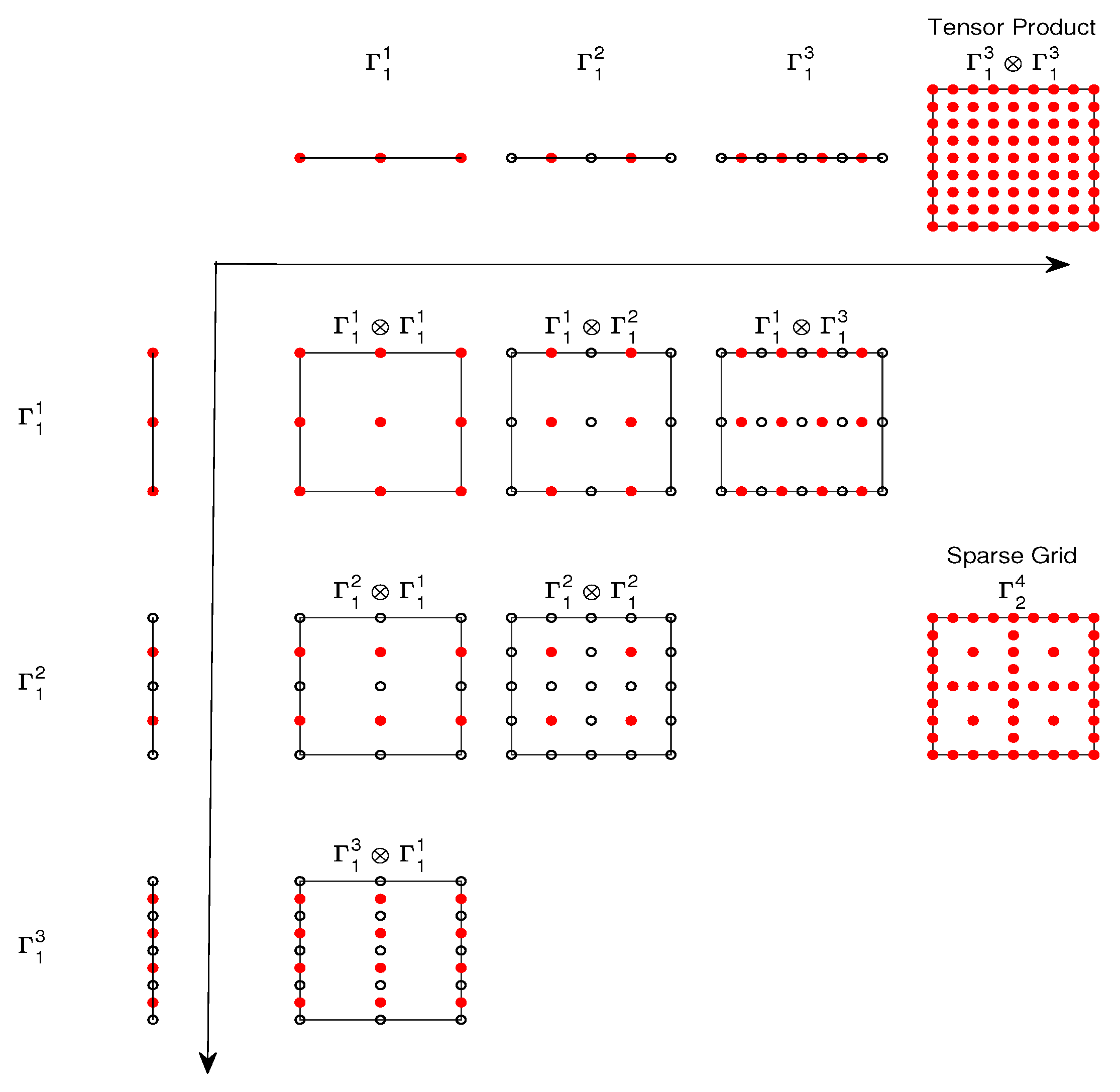
Figure 1 demonstrates the construction of a sparse grid according to the Smolyak rule when
and
. The meshes are nested, namely,
. The 1-d integration-point sequence
(
, and
) and
(
, and
) are shown at the top and left of the figure, and the tensor product points
are shown in the upper right corner. From (
6) we see that the sparse grid rule is a combination of the low-order tensor product rule on
with
. The point sets of these products and the resulting sparse grid
are shown in the lower half of the figure. We notice that some points in the sparse grid
are repeatedly used in
with
. Consequently, we would avoid the repeated points (i.e., only using the red points in
Figure 1) and use the reformulation (
9) which does not involve repetition in the summation.
2.2. Examples of sparse grid methods
As mentioned earlier, different 1-d quadrature rules in (
5) lead to different sparse grid quadrature rules in (
9). Below we introduce four widely used sparse grid quadrature rules which will be the focus of this paper.
Example 1: The classical trapezoidal rule. The 1-d trapezoidal rule is defined by (cf. [
25])
with
and
. Where the
indicates that the first and last terms in the sum are halved. The following theorem gives the error estimate, its proof can be found in [
7, pages 3-5].
Theorem 1.
Suppose , then there holds
Example 2: The classical Clenshaw-Curtis rule. This quadrature rule reads as follows (cf. [
26,
27]):
with
and the weights
for
. Where
indicates that the last term in the summation is halved.
The following error estimate holds and see [
26, Theorem 1] for its proof.
Theorem 2.
Suppose , then there holds
Example 3: The Gauss-Patterson rule. This quadrature rule is defined by (cf. [
28,
29])
with
, and
being the union of the zeroes of the polynomial
and
, where
is the
n-th order Legendre polynomial and
is the
-th order Stieltjes polynomial and the
is orthogonal to all polynomials of degree less than
with respect to the weight function
.
are defined similarly to the Gauss-Legendre case, see [
29] for details. Gauss-Patterson rules are a sequence of nested quadrature formulas with the maximal order of exactness. Its error estimate is given by the following theorem, see [
27,
28, Theorem 1].
Theorem 3.
Suppose , then there holds
Example 4: The classical Gauss-Legendre rule. This classical quadrature rule is defined by
with
for
.
are the zeroes of the
th order Legendre polynomial
, and
are the corresponding weights.
The following theorem gives the error estimate, its proof can be found in [
7, pages 3-5].
Theorem 4.
Suppose , then there holds
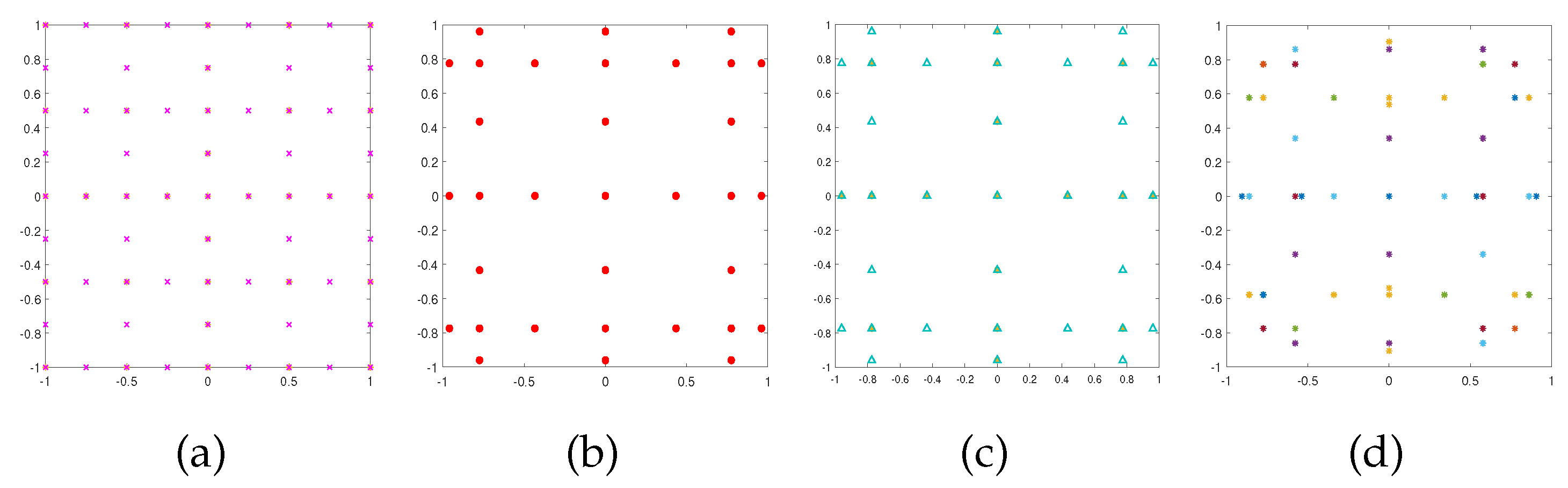
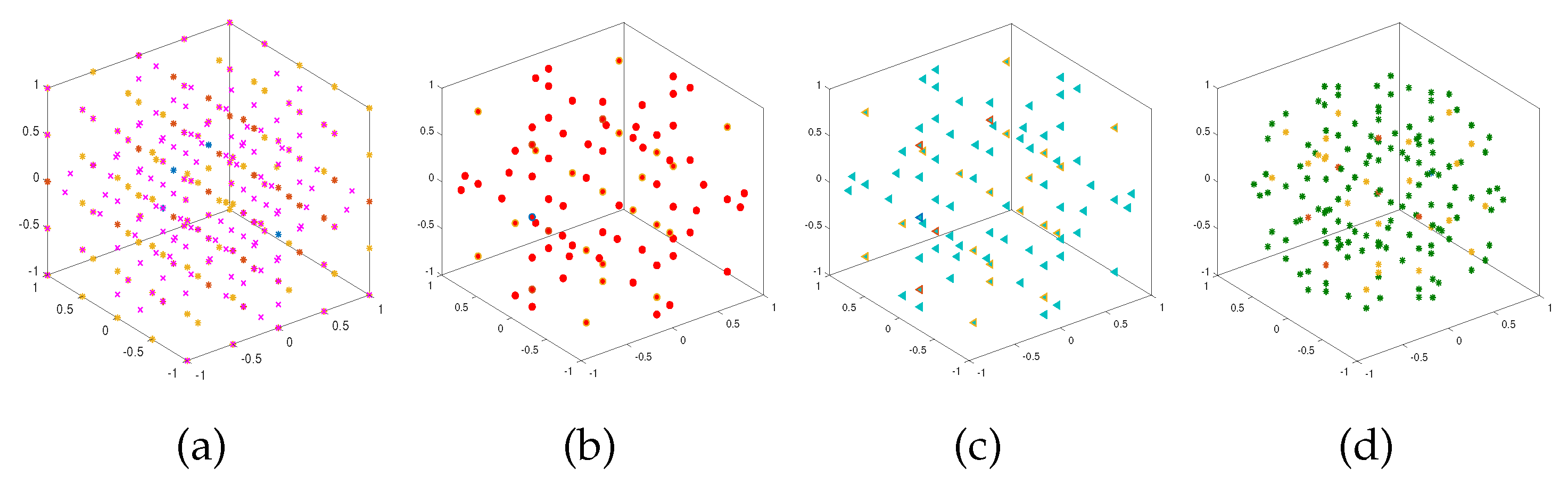
Figure 2 and
Figure 3 show the resulting sparse grids of the above four examples in both 2-d and 3-d cases with
and
respectively. We note that these four sparse grids have different structures.
We conclude this section by remarking that the error estimates of the above quadrature rules can be easily translate to error estimates for the sparse grid method (
9). For example, in the case of the Clenshaw-Curtis, Gauss-Patterson, and Gauss-Legendre quadrature rule, there holds (cf. [
26, Corollary 1])
where
We note that the above estimate indicates that the error of the sparse grid method still deteriorates exponentially in the dimension d, but with a smaller base .
3. The MDI-SG algorithm
The goal of this section is to present an efficient and fast implementation algorithm (or solver), called the MDI-SG algorithm, for evaluating the sparse grid quadrature rule (
6) via its reformulation (
9) in order to circumvent the curse of dimensionality which hampers the usage of the sparse grid method (
6) in high dimensions. To better understand the main idea of the MDI-SG method, we first consider the simple two and three dimensional cases and then to formulate the algorithm in arbitrary dimensions.
Recall that denote a continuous function on , we assume that f has a known expression.
3.1. Formulation of the MDI-SG algorithm in two dimensions
Let
,
, and
. By Fubini’s Theorem we have
Then, the two-dimensional SG quadrature rule (
9) takes the form
where
. Motivated by (and mimicking) the Fubini’s formula (
19), we rewrite (
20) as
where
We note that the evaluation of
is amount to applying the 1-d formula (
5) to approximate the integral
However, the values of
will not be computed by the 1-d quadrature rule in our MDI-SG algorithm, instead,
is formed as a symbolic function, so the 1-d quadrature rule can be called on
. Therefore, we still use the SG method to select the integration points, and then use our MDI-SG algorithm to perform function evaluations at the integration points collectively to save computation, which is the main idea of the MDI-SG method.
Let
W and
X denote the weight and node vectors of the 1-d quadrature rule on
,
represents the number of integration points in the
direction and we use a parameter
to indicate one of the four quadrature rule. The following algorithm implements the sparse grid quadrature formula (
21).
We note that the first do-loop forms the symbolic function which encodes all computations involving the -components at all integration points. The second do-loop evaluates the 1-d quadrature rule for the function . As mentioned above, in this paper we only focus on the four well-known 1-d quadrature rules: (i) trapezoidal rule; (ii) Clenshaw-Curtis rule; (iii) Gauss-Patterson rule; (iv) Gauss-Legendre rule. They will be represented respectively by .
Below we use a simple 2-d example to explain the mechanism of above 2d-MDI-SG algorithm. It is clear that to directly compute the SD sum
it is necessary to compute the function values of
at
points, which are often done independently. On the other hand, the 2d-MDI-SD algorithm is based on rewriting the sum as
where
denotes the symbolic function obtained in the first do-loop. Hence, the algorithm performs two separate do-loops. In the first d-loop, symbolic computations are performed to obtain the symbolic function
which is saved. In the second do-loop, the single sum
is done. When computing the symbolic function
, a lot of computations have been reused for computing the coefficients in
, and those coefficients are constants in the second do-loop. Efficiently generating the symbolic function and using it to compute the SG sum are the main reasons of saving computation and computer memory.
Take
as a concrete example. The direct computation the SG sum in (
22) requires to compute the function value
at each node
, this in turn requires three multiplications and two additions. With a total of
nodes, then computing the sum requires
a total of multiplications and additions. On the other hand, when using the 2d-MDI-SD algorithm to compute the same sum, in the first do-loop, we compute the symbolic function
which requires
“symbolic multiplications" of
(no real multiplication is needed because of its linear dependence on
) and
multiplications of
, as well as
additions. In the second do-loop, computing the
requires
multiplications of
and
multiplications of
, as well as
additions. Thus, the 2d-MDI-SD algorithm requires
a total of multiplications and additions. Therefore, the 2d-MDI-SG algorithm computes the SD sum much cheaper than the standard implementation, and this advantage will become more significant in high dimensions. See
Section 3.2 and
Section 3.3 for details.
3.2. Formulation of the MDI-SG algorithm in three dimensions
In the subsection we extend the formulation of the above 2d-MDI-SGI algorithm to the 3-d case by highlighting its main steps, in particular, how the above 2-d algorithm is utilized. First, recall that Fubini’s Theorem is given by
where
.
Second, notice that the SG quadrature rule (
9) in 3-d takes the form
where
. Mimicking the above Fubini’s formula, we rewrite (
25) as
where
We note that
is formed as a symbolic function in our MDI-SG algorithm and the right-hand side of (
26) is viewed as a 2-d sparse grid quadrature formula for
, it can be computed either directly or recursively by using
Algorithm 1. The following algorithm implements the SG quadrature formula (
26).
|
Algorithm 1 2d-MDI-SG(f, ) |
Initialize , .
for
do
for do
. end for
end for
for
do
for do
. end for
end for
returnQ. |
Where denotes the orthogonal projection (or natural embedding): , W and X stand for the weight and node vectors of the underlying 1-d quadrature rule.
From Algorithm 2 we can see the mechanism of the MDI-SG algorithm. It is based on two main ideas: (i) to use the sparse grid approach to select integration points; (ii) to use the discrete Fubini formula to efficiently compute the total sparse grid sum by reducing it to calculation of a low-dimensional (i.e., 2-d) sparse grid sum, which allows us to recursively call the low-dimensional MDI-SG algorithm.
|
Algorithm 2 3d-MDI-SG(f, ) |
Initialize .
for
do
for do
. end for
end for
.
2d-MDI-SG.
returnQ. |
3.3. Formulation of the MDI-SG algorithm in arbitrary d-dimensions
The goal of this subsection is to extend the 2-d and 3-d MDI-SG algorithms to arbitrary d-dimensions. We again start with recalling the d-dimensional Fubini’s Theorem
where
,
and
in which
and
denote respectively the orthogonal projections (or natural embeddings):
and
. The integer
m denotes the dimension reduction step length in our algorithm.
Mimicking the above Fubini’s Theorem, we rewrite the d-dimensional SG quadrature rule (
9) as follows:
where
We note that in our MDI-SG algorithm
defined by (
30) is a symbolic function and the right-hand side of (
29) is a
-order multi-summation, which itself can be evaluated by employing the dimension reduction strategy. Dimensionality can be reduced by iterating
times until
. To implement this process, we introduce the following conventions.
If
, set MDI-SG
, which is computed by using the one-dimensional quadrature rule (
5).
If , set MDI-SG 2d-MDI-SG.
If , set MDI-SG 3d-MDI-SG.
We note that when
, the parameter
m becomes a dummy variable and can be given any value. Recall that
denotes the natural embedding from
to
by deleting the first
m components of vectors in
. The following algorithm implements the sparse grid quadrature via (
29).
We remark that Algorithm 3 recursively generates a sequence of symbolic functions
, each function has
m fewer arguments than its predecessor. As mentioned earlier, our MDI-SG algorithm does not perform the function evaluations at all integration points independently, but rather iteratively along
m-coordinate directions, hence, the function evaluation at any integration point is not completed until the last step of the algorithm is executed. As a result, many computations are reused in each iteration, which is the main reason for the computation saving and to achieve a faster algorithm.
|
Algorithm 3 MDI-SG
|
.
for (the index is decreased by m at each iteration) do
. (Construct symbolic function by ( 31) below).
end for
.
for (the index is decreased by s at each iteration) do
. (Construct symbolic function by ( 31) below).
end for
return Q. |
4. Numerical performance tests
In this section, we present extensive numerical tests to guage the performance of the proposed MDI-SG algorithm and to compare it with the standard sparse grid (SG) and classical Monte Carlo (MC) methods. All numerical tests show that MDI-SG algorithm outperforms both SG and MC methods in low and medium high dimensions (i.e. ), and can compute very high-dimensional (i.e. ) integrals while others fail.
All our numerical experiments are done in Matlab 9.4.0.813654(R2018a) on a desktop PC with Intel(R) Xeon(R) Gold 6226R CPU 2.90GHz and 32GB RAM.
4.1. Two and three-dimensional tests
We first test our MDI-SG algorithm on simple 2-d and 3-d examples and compare its performance (in terms of CPU time) with SG methods.
Test 1. Let
and consider the following 2-d integrands:
Let
q denote the accuracy level of the sparse grid. The larger
q is, the more integration points we need for the 1-d quadrature rule, and the higher the accuracy of the MDI-SG quadrature. The base 1-d quadrature rule is chosen to be the Gauss-Patterson rule, hence, parameter
in the algorithm.
Table 1 and
Table 2 present the computational results (errors and CPU times) of the MDI-SG and SG methods for approximating
and
, respectively.
From
Table 1 and
Table 2, we observe that these two methods use very little CPU time, although the SG method in both tests outperforms, but the difference is almost negligible, so both methods do well in 2-d case.
Test 2. Let
and we consider the following 3-d integrands:
We compute the integral of these two functions over using the MDI-SG and SG methods. Likewise, let q denote the accuracy level of the sparse grid, choose parameters and in the algorithm.
The test results are given in
Table 3 and
Table 4. We again observe that both methods use very little CPU time although the SG method again slightly outperforms in both tests. However, as
q increases, the number of integration points increases, and the CPU times used by these two methods get closer. We would like to point out that both methods are very efficient and their difference is negligible in the 3-d case.
4.2. High-dimensional tests
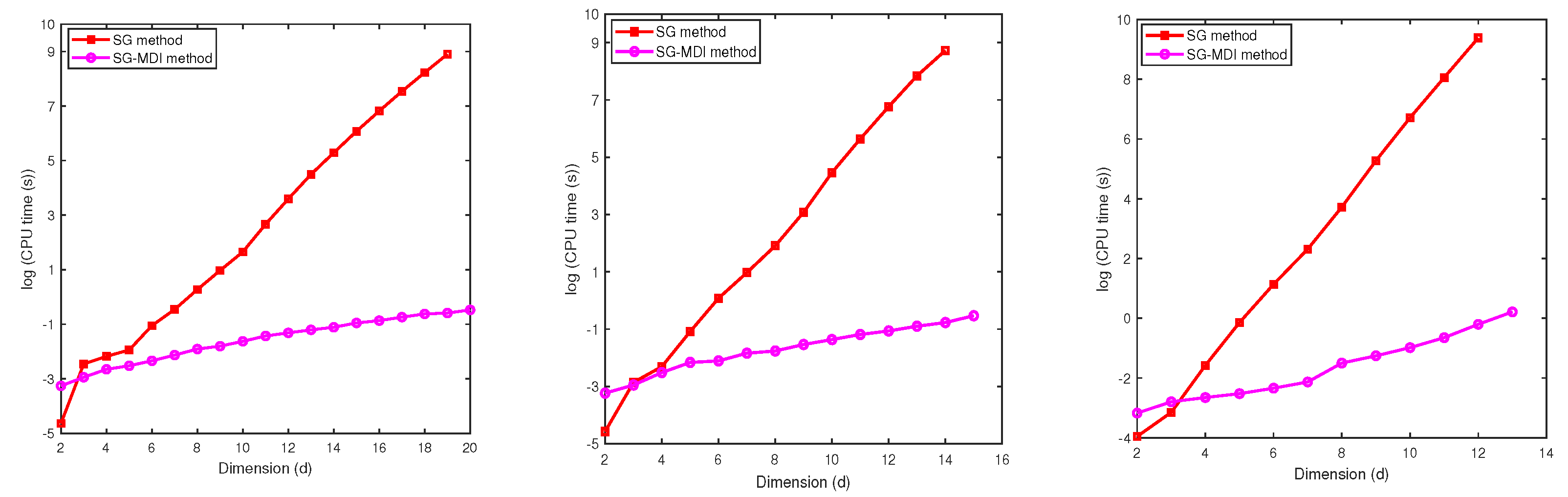
In this section we evaluate the performance of the MDI-SG method for . First, we test and compare the performance of the MDI-SG and SG methods in computing Gaussian integrals for dimensions because is the highest dimension that the SG method is able to compute a result on our computer. We then provide a performance comparison (in terms of CPU time) of the MDI-SG and classical Monte Carlo (MC) methods in computing high-dimensional integrals.
Test 3. Let
for
and consider the following Gaussian integrand:
where
stands for the Euclidean norm of the vector
.
We compute the integral by using the MDI-SG and SG methods, as done in Test 1-2. Both methods are based on the same 1-d Gauss-Patterson rule (i.e., the parameter ). We also set in the MDI-SG method and use two accuracy levels , respectively.
Table 5 gives the relative error and CPU time for approximating
using MDI-SG and SG methods with accuracy level
, and
Table 6 gives the corresponding results for
. We observe that the errors should be the same for both methods (since they use the same integration points), but their CPU times are quite different. The SG method is more efficient for
when
and for
when
, but the MDI-SG method excels for
and the winning margin becomes significant as
d and
q increase (also see
Figure 4). For example, when
and
, the CPU time required by the SG method is about 6167 seconds, which is about 2 hours, but the CPU time of the MDI-SG method is less than 1 second! Also, when
and
, the SG method fails to compute the integral due to running out of computer memory because too large number of integration points must be saved and function evaluations must be performed, but the MDI-SG method only needs about 2 seconds to complete the computation!
The classical (and quasi) Monte Carlo (MC) method is often the preferred/default method for computing high-dimensional integrals. However, due to its low order of convergence, to achieve the accuracy, a large number of function evaluations are required at randomly sampled integration points and the number grows rapidly as dimension
d increases (due to the rapid growth of variance). Below we compare the performance of the MDI-SG (with parameters
) and classical MC method. In the test, when
, we use the iteration step length
to iterate faster until
to reach the stage 2 of the iteration. We refer the reader to
Section 5.2 for a detailed analysis.
Test 4. Let
and choose the following integrands:
We use relative error as a criterion for comparison, that is, we determine a required (minimum) number of random sampling points for the MC method so that it produces a relative error comparable to that of the MDI-SG method. The computed results for
and
are respectively given in
Table 7 and
Table 8.
From
Table 7 and
Table 8, we clearly see that there is a significant difference in the CPU time of these two methods for computing
and
. When
, the classical MC method fails to produce a computed result with a relative error of order
. As explained in [
20], the MC method requires more than
randomly sampled integration points and then needs independently to compute their function values, which is a tall order to do on a regular workstation.
Next, we come to address a natural question that asks how high the dimension d can be handled by the MDI-SG method. Obviously, the answer must be computer-dependent, and that given below is obtained using the workstation at our disposal.
Test 5. Let
and consider the following integrands:
We then compute
and
using the MDI-SG algorithm for an increasing sequence of
d up to 1000 with parameters
. The computed results are shown in
Table 9. We stop the computation at
since it is already quite high and use
to minimize the computation in each iteration. This test demonstrates the promise and capability of the MDI-SG algorithm for efficiently computing high-dimensional integrals. When computing the integral of a 1000-dimensional function by the SG method, the SG sum contains
function evaluations, but the MDI-SG algorithm does not compute them one by one. Instead, the symbolic functions are efficiently degenerated through dimension iterations by reusing/sharing many calculations. It should be noted that the MDI-SG algorithm does use of a lot of computer memory when computing high-dimensional integrals.
5. Influence of parameters
There are four parameters in the d-dimensional MDI-SG algorithm, they are respectively , and q. Where represents the choice of one-dimensional numerical quadrature rule, namely, the (composite) trapezoidal rule (), Clenshaw-Curtis rule (), Gauss-Patterson rule (), and Gauss-Legendre rule (). The parameter m stands for the multi-dimensional iteration step size in the first stage of the algorithm so the dimension of the integration domain is reduced by m in each iteration. In practice, , and it is preferred to be close to q. In this section we shall evaluate the performance of the MDI-SG algorithm when . It should be noted that after iterations, the algorithm enters to its second stage and the iteration step size is changed to s. Since after the first stage, the remaining dimension small, so . It should be noted that after iterations, the residual dimension satisfies . Then in case or 3, one has two options to complete the algorithm. One either just continues the dimension reduction by calling 3d-MDI-SG or 2d-MDI-SG as explained in the definition of Algorithm 3 or compute the remaining 2- or 3-dimensional integral directly. The effect of these two choices will be tested in this section. Finally, the parameter q represents the precision level of the sparse grid method. Obviously, the larger q is, the higher accuracy of the computed results, the trade-off is more integration points must be used, hence, adds more cost. In this section we shall also test the impact of q value to the MDI-SG algorithm.
5.1. Influence of parameter r
We first examine the impact of one-dimensional quadrature rules, which are indicated by , in the MDI-SG algorithm.
Test 6: Let
and choose the integrand
f as
Below we compare the performance of the MDI-SG algorithm with different
r in computing
and
with the accuracy level
and step size
.
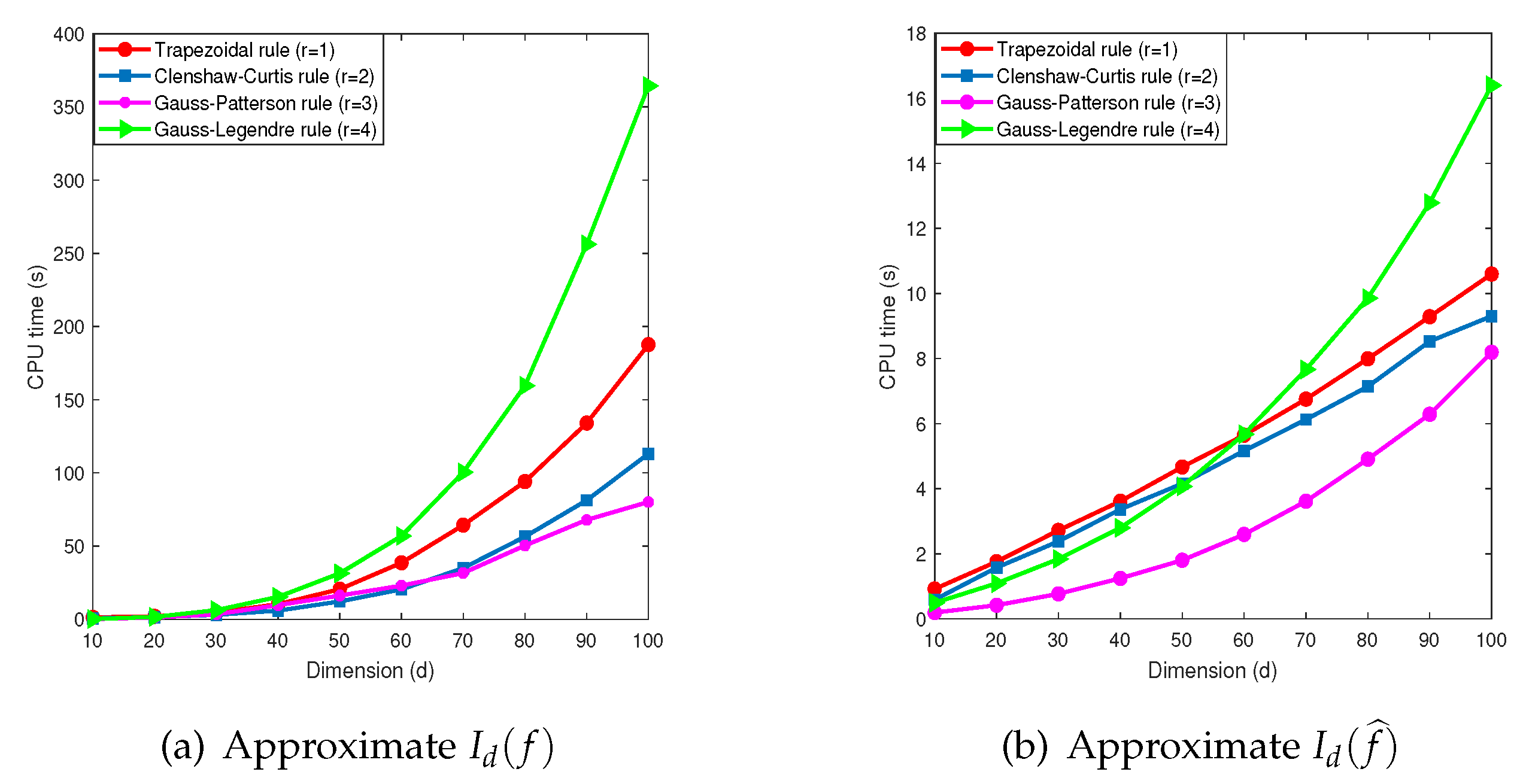
Figure 5 (a) shows the computed results of
by the MDI-SG algorithm. We observe that the choice of one-dimensional quadrature rules has a significant impact on the accuracy and efficiency of the MDI-SG algorithm. The trapezoidal rule (
) has the lowest precision and uses the most integration points, the Clenshaw-Curtis rule (
) is the second lowest, and the Gauss-Patterson (
) and Gauss-Legendre rule (
) have the highest precision. Both the Clenshaw-Curtis and Gauss-Patterson rule use the nested grids, that is, the integration points of the
th level contain those of the
qth level. Although they use the same number of integration points, the Gauss-Patterson rule is more efficient than the Clenshaw-Curtis rule. Moreover, the Gauss-Patterson rule is more efficient than the Gauss-Legendre rule (
) which uses the most CPU time and produces the most accurate solution.
This comparison suggests that the Gauss-Patterson rule is a winner among these four rules when they are used as the building blocks in the MDI-SG algorithm for high-dimensional integration.
Figure 5 (b) shows the computational results of
by the MDI-SG algorithm. Similarly, the choice of one-dimensional quadrature rules has a significant impact on the accuracy and efficiency of the MDI-SG algorithm. Because the integrand
is simple, the MDI-SG algorithm with all four 1-d quadrature rules computes this integral very fast. Again, the trapezoidal rule is least accurate and the other three rules all perform very well, but a closer look shows that the Gauss-Patterson rule is again the best performer.
Figure 5.
Performance comparison of the MDI-SG algorithm with , and
Figure 5.
Performance comparison of the MDI-SG algorithm with , and
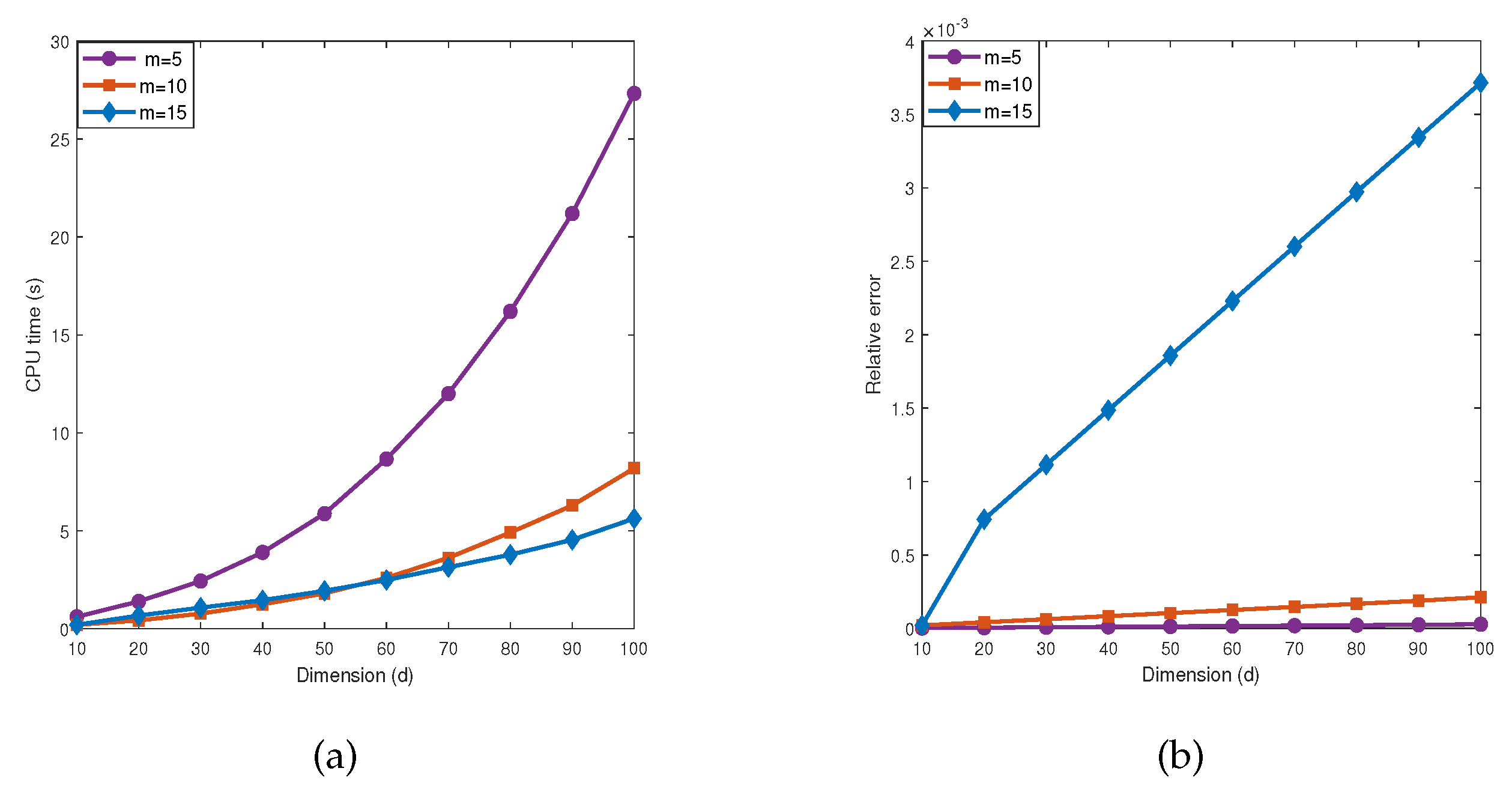
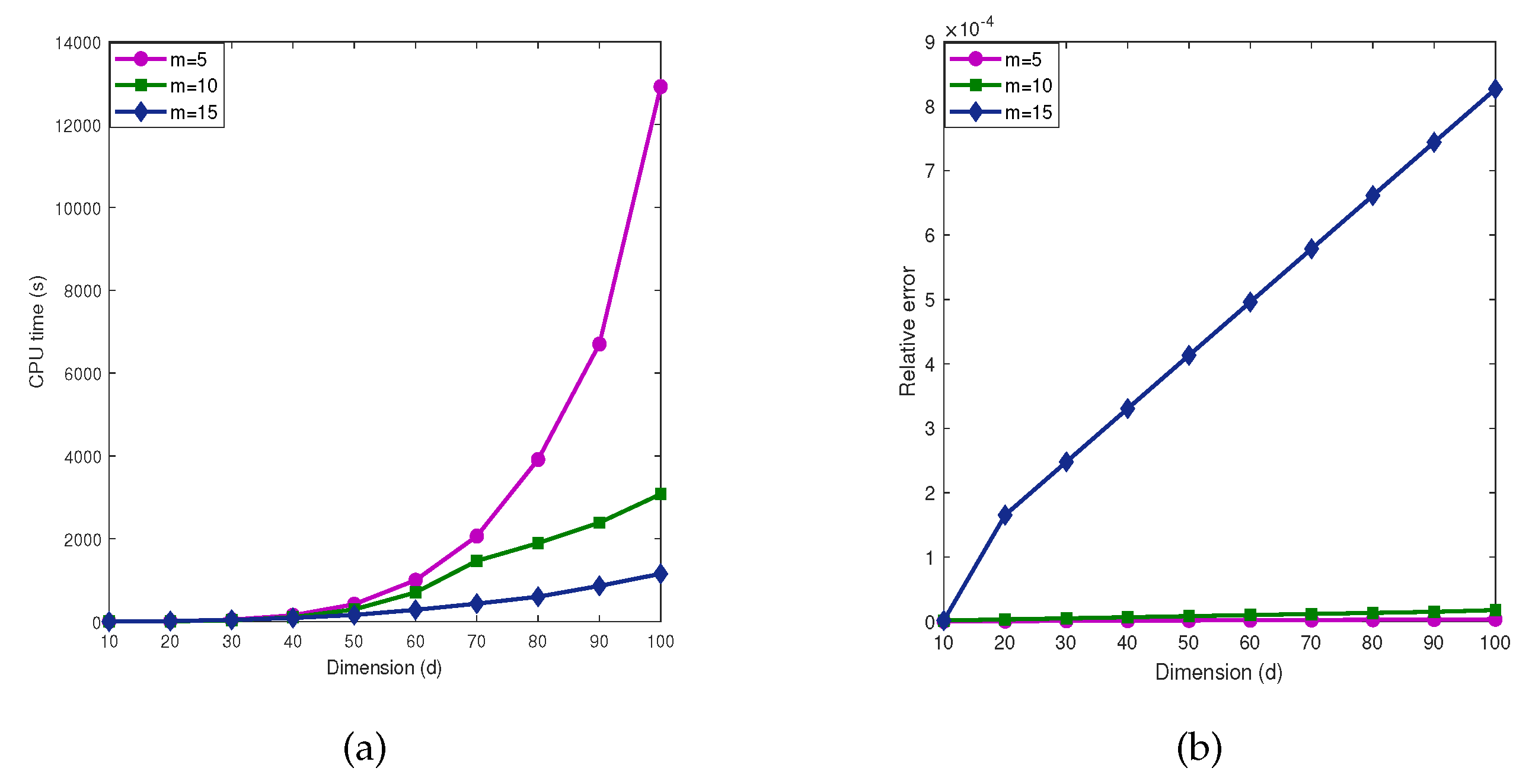
5.2. Influence of parameter m
From the
Table 5 and
Table 6 and
Figure 6 and
Figure 7, we observe that when
is fixed, as the dimension
d increases, the number of iterations by the MDI-SG algorithm also increases, and the computational efficiency decreases rapidly. In practice, the step size
m of the MDI-SG algorithm in the first stage iteration should not be too large or too small. One strategy is to use variable step sizes. After selecting an appropriate initial step size
m, it can be decreased during the dimension iteration. The next test presents a performance comparison of the MDI-SG algorithm for
.
Test 7. Let
,
f and
be the same as in (
35).
We compute these integrals using the MDI-SG algorithm with (Gauss-Patterson rule) and .
Figure 6 presents the computed results of integral
in
Test 7 by the MDI-SG algorithm. It is easy to see that the accuracy and efficiency of the MDI-SG algorithm with different
m are different, this is because the step size
m affects the number of iterations in the first stage. It shows that the larger step size
m, the smaller the number of iterations and the number of symbolic functions that need to be saved, and the fewer CPU time are used, but at the expense of decreasing accuracy and higher truncation error. On the other hand, the smaller step size
m, the more accurate of the computed results, but at the expense of more CPU time. To explain this observation, we notice that when the step size
m is large, each iteration reduces more dimensions, and the number of symbolic functions that need to be saved is less, but the truncation error generated by the system is larger, although the used CPU time is small. When the step size is small, however, more symbolic functions need to be saved. This is because each iteration reduces a small number of dimensions, the error of computed result is smaller, but the used CPU time is larger. We also observed that the MDI-SG algorithm with step size
achieves a good balance between CPU time and accuracy.
Figure 7 shows the computed results of
in
Test 7 the MDI-SG algorithm. As expected, choosing different parameters
m has a significant impact on the accuracy and efficiency of the MDI-SG algorithm. Since function evaluation of the integrand
is more complicated, the influence of different parameters
m on the MDI-SG algorithm is dramatic. Again, the MDI-SG algorithm with step size
strikes a balance between CPU time and accuracy.
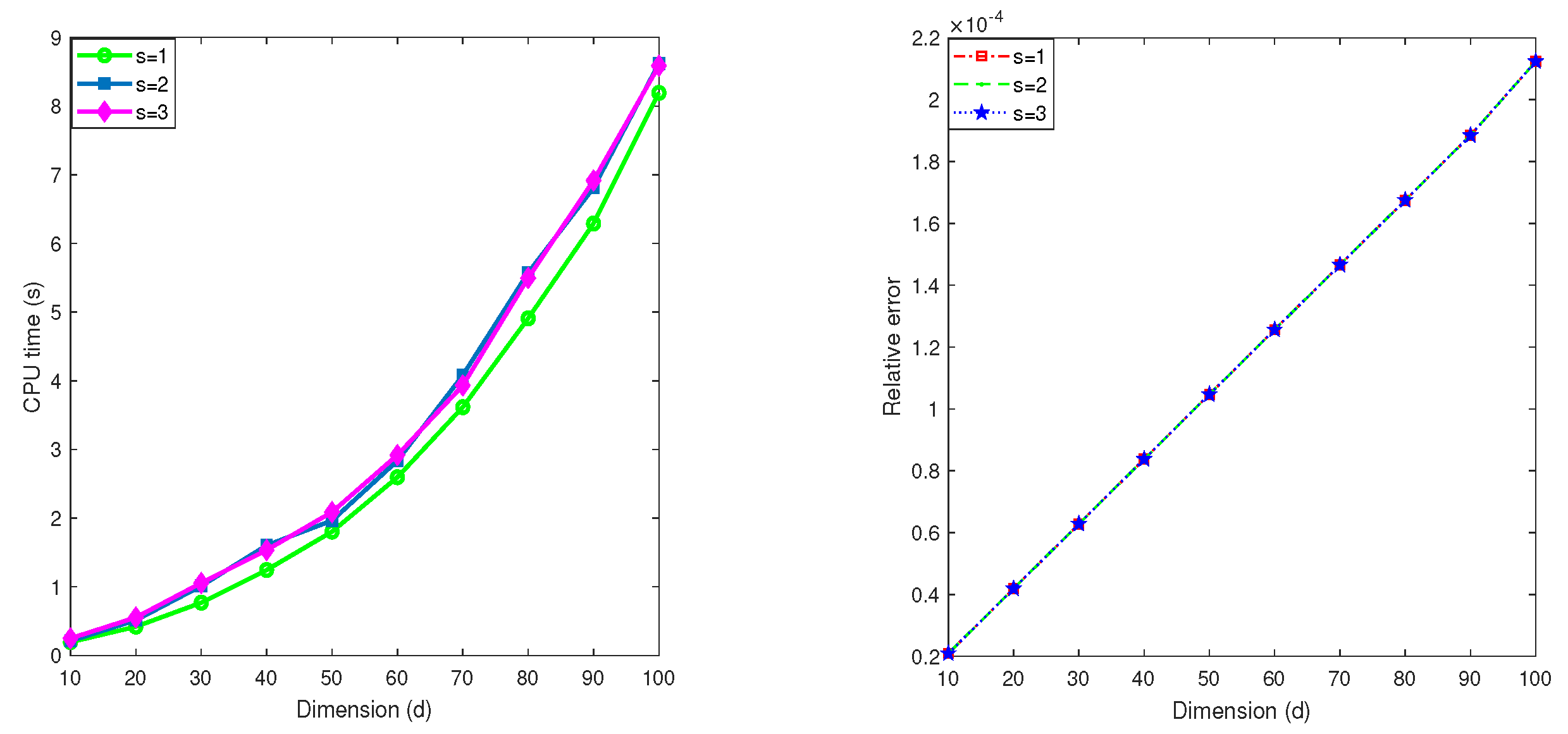
5.3. Influence of the parameter s
In this subsection, we test the impact of the step size s used in the second stage of the MDI-SG algorithm. Recall that the step size works well in the first stage. After iterations, the dimension of the integration domain is reduced to which is relatively small, hence, s should be small. In practice, we have . The goal of the next test is to provide a performance comparison of the MDI-SG algorithm with .
Test 8. Let
, and choose the integrand
f as
We compute this integral
using the MDI-SG algorithm with
(Gauss-Patterson rule) and
.
Figure 8 displays the computed results. We observe that the same accuracy is achieved in all cases
, which is expected. Moreover, the choice of
s has little effect on the efficiency of the algorithm. An explanation for this observation is that because
becomes small after the first stage iterations, so the number of the second stage iterations is small, the special way of performing function evaluations in the MDI-SG algorithm is not sensitive to the small variations in the choice of
.
5.4. Influence of the parameter q or N
Finally, we examine the impact of the accuracy level q on the MDI-SG algorithm. Recall that the parameter q in the SG method is related to the number of integration points N in one coordinate direction on the boundary. It is easy to check that for the trapezoidal () and Clenshaw Curtis () quadrature rules, correspond to , for the Gauss-Patterson quadrature rule (), correspond to , and for the Gauss-Legendre quadrature rule (), correspond to . Therefore, we only need to examine the impact of the parameter N on the MDI-SG algorithm. To the end, we consider the case , and (Gauss-Legendre rule) in the next test.
Test 8. Let
and choose the following integrands:
Table 10,
Table 11, and
Table 12 present respectively the computed results for
and
. We observe that the quality of approximations also depends on the behavior of the integrand. For very oscillatory and rapidly growing functions, more integration points must be used to achieve good accuracy.
6. Computational complexity
6.1. The relationship between the CPU time and N
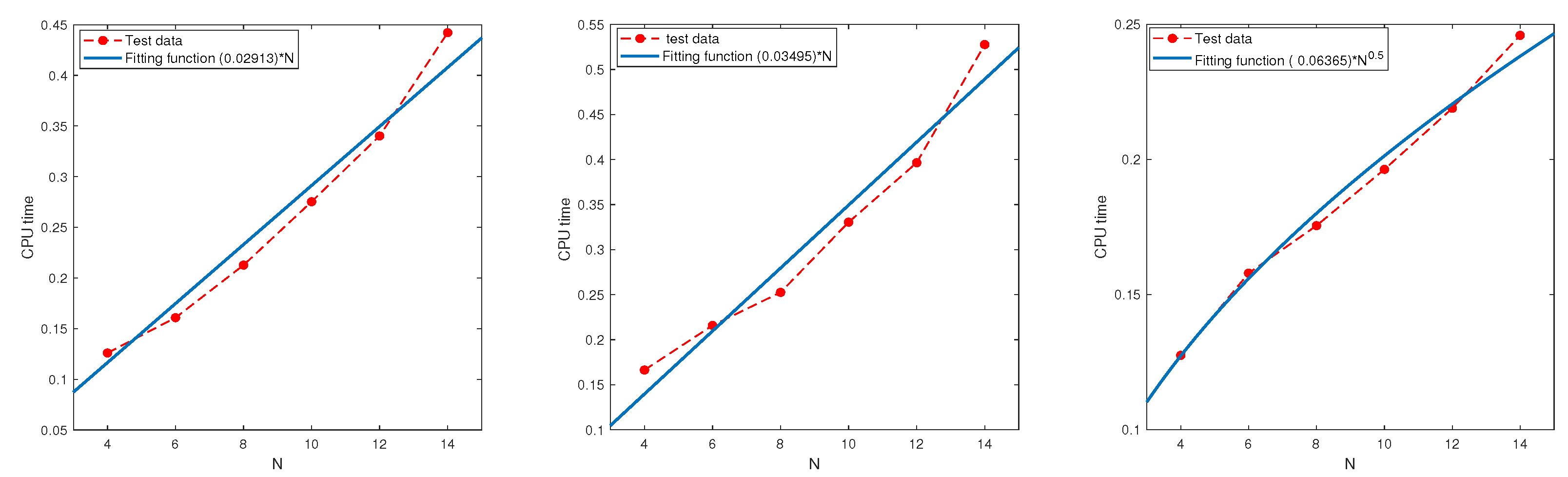
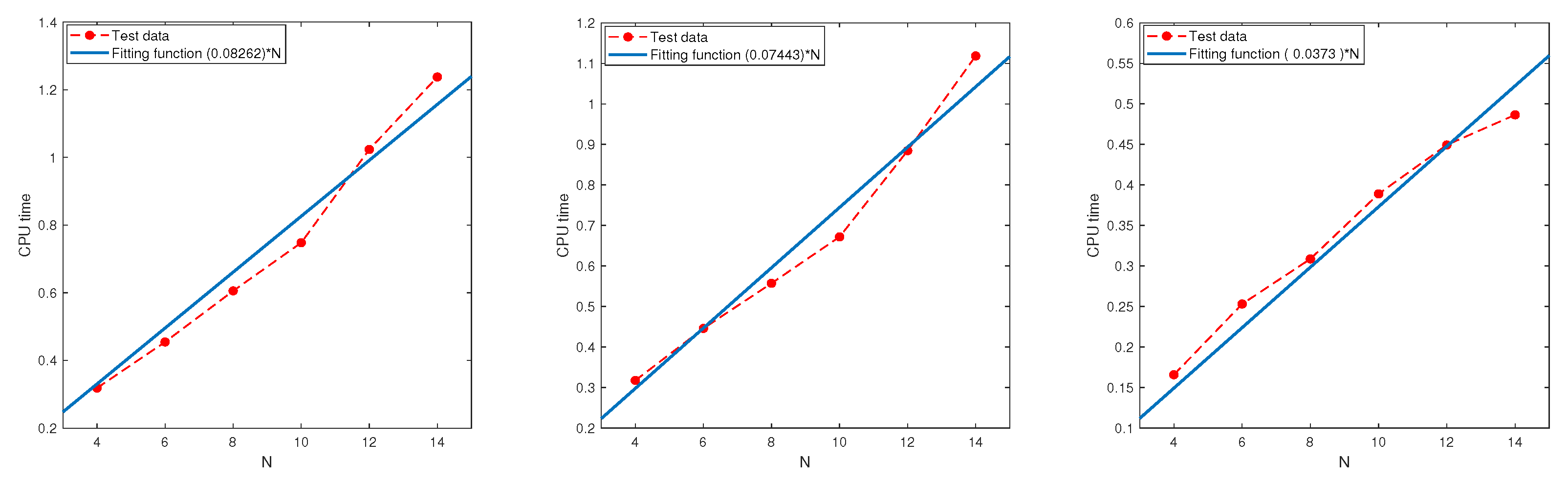
In this subsection, we examine the relationship between the CPU time and parameter N using the regression technique based on the test data.
Test 9. Let , and be the same as in Test 8.
Figure 9 and
Figure 10 show the CPU time as a function of
N obtained by the least square method and the fitting functions are given in
Table 13. All the results indicate that the CPU time grows at most linearly in
N.
6.2. The relationship between the CPU time and the dimension d
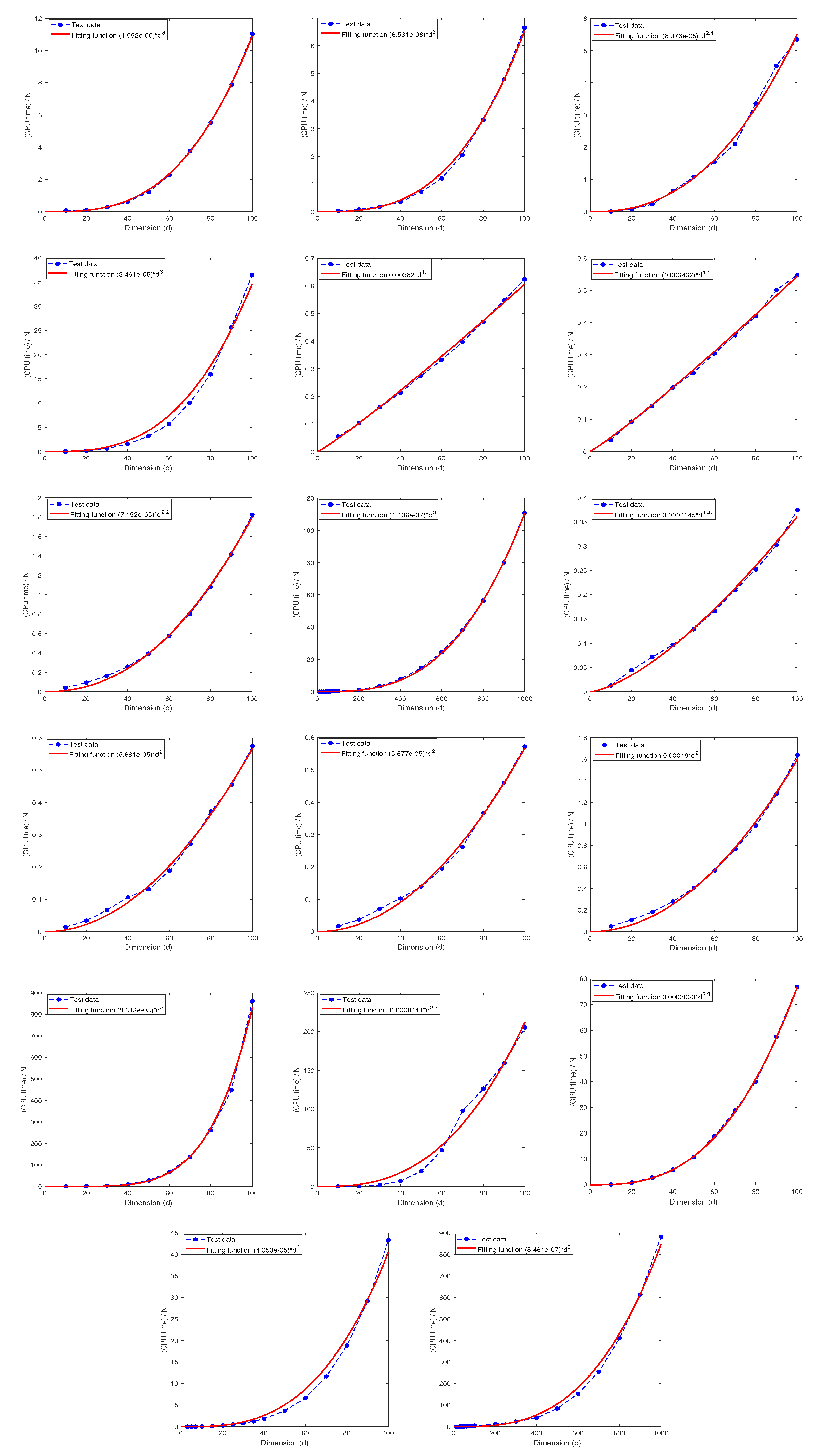
Recall that the computational complexity of the sparse grid method is of the order for computing , which grows exponentially in d with base . The numerical tests presented above overwhelmingly and consistently indicate that the MDI-SG algorithm has hidden capability to overcome the curse of dimensionality which hampers the sparse grid method. The goal of this subsection is to find out the computational complexity of the MDI-SG algorithm (in terms of CPU time as a function of d) using the regression technique based on numerical test data.
Test 10. Let
and consider the following five integrands:
Figure 11 displays the CPU time as functions of
d obtained by the least square regression method whose analytical expressions are given in
Table 14. We note that the parameters of the MDI-SG algorithm only affect the coefficients of the fitting functions, but not their orders in
d.
We also quantitatively characterize the performance of the fitted curve by the
R-square in Matlab, which is defined as
R-
. Where
represents a test data output,
refers to the predicted value, and
indicates the mean value of
.
Table 14 also shows that the
R-square of all fitting functions is very close to 1, which indicates that the fitting functions are quite accurate. These results suggest that the CPU time grows at most cubically in
d. Combining the results of
Test 8 in
Section 5.4 we conclude that the CPU time required by the proposed MDI-SG algorithm grows at most in the polynomial order
.
7. Conclusions
This paper presented an efficient and fast implementation algorithm (or solver), called the MDI-SG algorithm, for high-dimensional numerical integration using the sparse grid method. It is based on combining the idea of dimension iteration/reduction combined with the idea of computing the function evaluations at all integration points in cluster so many computations can be reused. It was showed numerically that the computational complexity (in terms of the CPU time) of the MDI-SG algorithm grows at most cubically in the dimension d, and overall in the order , where N denotes the maximum number of integration points in each coordinate direction. This shows that the MDI-SG algorithm could effectively circumvent the curse of the dimensionality in high-dimensional numerical integration, hence, makes sparse grid methods not only become competitive but also can excel for the job. Extensive numerical tests were provided to examine the performance of the MDI-SG algorithm and to carry out performance comparisons with the standard sparse grid method and with the Monte Carlo (MC) method. It was showed that the MDI-SG algorithm (regardless the choice of the 1-d base sparse grid quadrature rules) is faster than the MC method in low and medium dimensions (i.e., ), much faster in very high dimensions (i.e., ), and succeeds even when the MC method fails. An immediate application of the proposed MDI-SG algorithm is to solve high-dimensional linear PDEs based on the integral equation formulation, which will be reported in a forthcoming work.
Author Contributions
Conceptualization, X.F.; methodology, H.Z. and X.F.; code and simulation. H.Z.; writing—original draft preparation, H.Z..; writing—revision and editing, X.F. All authors have read and agreed to the published version of the manuscript.
Funding
The work of X.F. was partially supported by the NSF grants: DMS-2012414 and DMS-230962.
Data Availability Statement
All datasets generated during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
-
M. Griebel and M. Holtz, Dimension-wise integration of high-dimensional functions with applications to finance, J. Complexity, 26:455–489, 2010.
-
S. M. LaValle, K. J. Moroney and S. A. Hutchinson, Methods for numerical integration of high-dimensional posterior densities with application to statistical image models, IEEE transactions on image processing, 6(12): 1659–1672, 1997.
-
J. Barraquand and D. Martineau, Numerical valuation of high dimensional multivariate American securities, Journal of financial and quantitative analysis, 30(3): 383–405, 1995.
-
J . Quackenbush, Extracting biology from high-dimensional biological data, Journal of Experimental Biology, 210(9):1507–1517, 2007.
-
H.-J. Bungartz and M. Griebel, Sparse grids, Acta Numer., 13:147–269, 2014.
-
J. Dos Santos Azevedo and S. Pomponet Oliveira, A numerical comparison between quasi-Monte Carlo and sparse grid stochastic collocation methods, Commun. Comput. Phys., 12:1051–1069, 2012.
-
T. Gerstner and M. Griebel, Numerical integration using sparse grids, Numerical algorithms, 18(3): 209–232, 1998.
-
R. E. Caflisch, Monte Carlo and quasi-Monte Carlo methods, Acta Numer., 7:1–49, 1998.
-
Y. Ogata, A Monte Carlo method for high dimensional integration, Numer. Math., 55:137–157, 1989.
-
J. Dick, F. Y. Kuo, and I. H. Sloan, High-dimensional integration: the quasi-Monte Carlo way, Acta Numer., 22:133–288, 2013.
-
F. J. Hickernell, T. Müller-Gronbach, B. Niu, and K. Ritter, Multi-level Monte Carlo algorithms for infinite-dimensional integration on RN, J. Complexity 26:229–254, 2010.
-
F. Y. Kuo, C. Schwab, and I. H. Sloan, Quasi-Monte Carlo methods for high-dimensional integration: the standard (weighted Hilbert space) setting and beyond, ANZIAM J., 53:1–37, 2011.
-
J. Lu and L. Darmofal, Higher-dimensional integration with Gaussian weight for applications in probabilistic design, SIAM J. Sci. Comput., 26:613–624, 2004.
-
A. Wipf, High-Dimensional Integrals, in Statistical Approach to Quantum Field Theory, Springer Lecture Notes in Physics, 100:25–46, 2013.
-
W. E and B. Yu, The deep Ritz method: A deep learning-based numerical algorithm for solving variational problems, Commun. Math. and Stat., 6:1–12, 2018.
-
J. Han, A. Jentzen, and W. E, Solving high-dimensional partial differential equations using deep learning, PNAS, 115:8505–8510, 2018.
-
L. Lu, X. Meng, Z. Mao, and G. E. Karniadakis, DeepXDE: A deep learning library for solving differential equations, SIAM Rev., 63:208–228, 2021.
-
J. Sirignano and K. Spiliopoulos, DGM: A deep learning algorithm for solving partial differential equations, J. Comput. Phys., 375:1339–1364, 2018.
-
J. Xu, Finite neuron method and convergence analysis, Commun. Comput. Phys, 28:1707–1745, 2020.
-
X. Feng and H. Zhong, A fast multilevel dimension iteration algorithm for high dimensional numerical integration. preprint.
-
R. L. Burden and J. D. Faires, Numerical Analysis, 10th edition, Cengage Learning, 2015.
-
S. A. Smolyak, Quadrature and interpolation formulas for tensor products of certain classes of functions, Russian Academy of Sciences., 148(5): 1042–1045, 1963.
-
G. Baszenki and F. -J. Delvos, Multivariate Boolean midpoint rules, Numerical Integration IV, Birkhäuser, Basel, 1–11, 1993.
-
S. H. Paskov, Average case complexity of multivariate integration for smooth functions, Journal of Complexity, 9(2): 291–312, 1993.
-
T. Bonk, A new algorithm for multi-dimensional adaptive numerical quadrature,Adaptive Methods—Algorithms, Theory and Applications, Vieweg+ Teubner Verlag, Wiesbaden, 54–68, 1994.
-
E. Novak and K. Ritter, High dimensional integration of smooth functions over cubes, Numerische Mathematik, 75(1):79–97, 1996.
-
E. Novak and K. Ritter, The curse of dimension and a universal method for numerical integration,Multivariate approximation and splines. Birkhäuser, Basel, 177–187, 1997.
-
E. Novak and K. Ritter, Simple cubature formulas for d-dimensional integrals with high polynomial exactness and small error, Report, Institut für Mathematik, Universität Erlangen–Nürnberg, 1997.
-
T. N. L. PattersonThe optimum addition of points to quadrature formulae, Math. Comp., 22(104): 847–856, 1968.
Figure 1.
Construction of a nested sparse grid in two dimensions.
Figure 1.
Construction of a nested sparse grid in two dimensions.
Figure 2.
Sparse grids corresponding to the trapezoidal rule (a), Clenshaw-Curtis rule (b), Gauss-Patterson rule (c), and Gauss-Legendre rule (d) when .
Figure 2.
Sparse grids corresponding to the trapezoidal rule (a), Clenshaw-Curtis rule (b), Gauss-Patterson rule (c), and Gauss-Legendre rule (d) when .
Figure 3.
Sparse grids corresponding to the trapezoidal rule (a), Clenshaw-Curtis rule (b), Gauss-Patterson rule (c), and Gauss-Legendre rule (d) when .
Figure 3.
Sparse grids corresponding to the trapezoidal rule (a), Clenshaw-Curtis rule (b), Gauss-Patterson rule (c), and Gauss-Legendre rule (d) when .
Figure 4.
CPU time comparison of MDI-SG and SG tests: (left), (middle), (right).
Figure 4.
CPU time comparison of MDI-SG and SG tests: (left), (middle), (right).
Figure 6.
Efficiency and accuracy comparison of the MDI-SG algorithm with , and for approximating .
Figure 6.
Efficiency and accuracy comparison of the MDI-SG algorithm with , and for approximating .
Figure 7.
Efficiency and accuracy comparison of the MDI-SG algorithm with , and for approximating .
Figure 7.
Efficiency and accuracy comparison of the MDI-SG algorithm with , and for approximating .
Figure 8.
Efficiency comparison of the MDI-SG algorithm with and .
Figure 8.
Efficiency comparison of the MDI-SG algorithm with and .
Figure 9.
The relationship between the CPU time and parameter N when for computing (left), (middle), (right).
Figure 9.
The relationship between the CPU time and parameter N when for computing (left), (middle), (right).
Figure 10.
The relationship between the CPU time and parameter N when for computing (left), (middle), (right).
Figure 10.
The relationship between the CPU time and parameter N when for computing (left), (middle), (right).
Figure 11.
The relationship between the CPU time and dimension d.
Figure 11.
The relationship between the CPU time and dimension d.
Table 1.
Relative errors and CPU times of MDI-SG and SG tests with for approximating .
Table 1.
Relative errors and CPU times of MDI-SG and SG tests with for approximating .
|
|
MDI-SG |
SG |
Accuracy level
|
Total
nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
6 |
33 |
|
0.0512817 |
|
0.0078077 |
| 7 |
65 |
|
0.0623538 |
|
0.0084645 |
| 9 |
97 |
|
0.0644339 |
|
0.0095105 |
| 10 |
161 |
|
0.0724491 |
|
0.0106986 |
| 13 |
257 |
|
0.0913161 |
|
0.0135131 |
| 14 |
321 |
|
0.1072016 |
|
0.0155733 |
Table 2.
Relative errors and CPU times of MDI-SG and SG tests with for approximating ).
Table 2.
Relative errors and CPU times of MDI-SG and SG tests with for approximating ).
| |
|
MDI-SG |
SG |
Accuracy level
|
Total
nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
9 |
97 |
|
0.0767906 |
|
0.0098862 |
| 10 |
161 |
|
0.0901238 |
|
0.0102700 |
| 13 |
257 |
|
0.1025934 |
|
0.0152676 |
| 14 |
321 |
|
0.1186194 |
|
0.0144737 |
| 16 |
449 |
|
0.1353691 |
|
0.0177445 |
| 20 |
705 |
|
0.1880289 |
|
0.0355606 |
Table 3.
Relative errors and CPU times of MDI-SG and SG tests with for approximating .
Table 3.
Relative errors and CPU times of MDI-SG and SG tests with for approximating .
| |
|
MDI-SG |
SG |
Accuracy level
|
Total
nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
9 |
495 |
|
0.0669318 |
|
0.0235407 |
| 10 |
751 |
|
0.0886774 |
|
0.0411750 |
| 11 |
1135 |
|
0.0902602 |
|
0.0672375 |
| 13 |
1759 |
|
0.1088353 |
|
0.0589584 |
| 14 |
2335 |
|
0.1381728 |
|
0.0704032 |
| 15 |
2527 |
|
0.1484829 |
|
0.0902680 |
| 16 |
3679 |
|
0.1525743 |
|
0.1143728 |
Table 4.
Relative errors and CPU times of MDI-SG and SG tests with for approximating .
Table 4.
Relative errors and CPU times of MDI-SG and SG tests with for approximating .
| |
|
MDI-SG |
SG |
Accuracy level
|
Total
nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
12 |
1135 |
|
0.0921728 |
|
0.0495310 |
| 13 |
1759 |
|
0.1031632 |
|
0.0644124 |
| 15 |
2527 |
|
0.1771094 |
|
0.0891040 |
| 16 |
3679 |
|
0.1957219 |
|
0.1159222 |
| 17 |
4447 |
|
0.2053174 |
|
0.1443184 |
| 19 |
6495 |
|
0.4801467 |
|
0.2259950 |
| 20 |
8031 |
|
0.6777698 |
|
0.2632516 |
Table 5.
Relative errors and CPU times of MDI-SG and SG tests with , and for approximating .
Table 5.
Relative errors and CPU times of MDI-SG and SG tests with , and for approximating .
| |
|
MDI-SG |
SG |
Dimension
(d) |
Total
nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
2 |
161 |
|
0.0393572 |
|
0.0103062 |
| 4 |
2881 |
|
0.0807326 |
|
0.0993984 |
| 8 |
206465 |
|
0.1713308 |
|
6.7454179 |
| 10 |
1041185 |
|
0.2553576 |
|
86.816883 |
| 12 |
4286913 |
|
0.3452745 |
|
866.1886366 |
| 14 |
5036449 |
|
0.4625503 |
|
6167.3838002 |
| 15 |
12533167 |
|
0.5867914 |
failed |
failed |
Table 6.
Relative errors and CPU times of MDI-SG and SG tests with , and for approximating .
Table 6.
Relative errors and CPU times of MDI-SG and SG tests with , and for approximating .
| |
|
MDI-SG |
SG |
Dimension
(d) |
Total
nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
2 |
161 |
|
0.0418615 |
|
0.0191817 |
| 4 |
6465 |
|
0.0704915 |
|
0.2067346 |
| 6 |
93665 |
|
0.0963325 |
|
3.1216913 |
| 8 |
791169 |
|
0.2233707 |
|
41.3632962 |
| 10 |
5020449 |
|
0.3740873 |
|
821.6461622 |
| 12 |
25549761 |
|
0.8169479 |
|
11887.797686 |
| 13 |
29344150 |
|
1.2380811 |
failed |
failed |
Table 7.
CPU times of the MDI-SG and MC tests with comparable relative errors for approximating .
Table 7.
CPU times of the MDI-SG and MC tests with comparable relative errors for approximating .
| |
|
MC |
MDI-SG |
Dimension
(d) |
Relative
error |
CPU time(s) |
Relative
error |
CPU time(s) |
|
5 |
|
62.1586394 |
|
0.0938295 |
| 10 |
|
514.1493073 |
|
0.1945813 |
| 20 |
|
1851.0461899 |
|
0.4204564 |
| 30 |
|
15346.222011 |
|
0.7692118 |
| 35 |
|
failed |
|
0.9785784 |
| 40 |
|
|
|
1.2452577 |
| 60 |
|
|
|
2.5959174 |
| 80 |
|
|
|
4.9092032 |
| 100 |
|
|
|
8.1920274 |
Table 8.
CPU times of the MDI-SG and MC tests with comparable relative errors for approximating .
Table 8.
CPU times of the MDI-SG and MC tests with comparable relative errors for approximating .
| |
|
MC |
MDI-SG |
Dimension
(d) |
Relative
error |
CPU time(s) |
Relative
error |
CPU time(s) |
|
5 |
|
85.2726354 |
|
0.0811157 |
| 10 |
|
978.1462121 |
|
0.295855 |
| 20 |
|
2038.138555 |
|
6.3939110 |
| 30 |
|
16872.143255 |
|
29.5098187 |
| 35 |
|
failed |
|
62.0270714 |
| 40 |
|
|
|
106.1616486 |
| 80 |
|
|
|
1893.8402620 |
| 100 |
|
|
|
3077.1890005 |
Table 9.
Computed results for and by the MDI-SG algorithm.
Table 9.
Computed results for and by the MDI-SG algorithm.
| |
|
|
|
Dimension
(d) |
Approximate
Total nodes |
Relative
error |
CPU
time(s) |
Relative
error |
CPU
time(s) |
|
10 |
|
|
0.1541 |
|
0.1945 |
| 100 |
|
|
80.1522 |
|
8.1920 |
| 300 |
|
|
348.6000 |
|
52.0221 |
| 500 |
|
|
1257.3354 |
|
219.8689 |
| 700 |
|
|
3827.5210 |
|
574.9161 |
| 900 |
|
|
9209.119 |
|
1201.65 |
| 1000 |
|
|
13225.14 |
|
1660.84 |
Table 10.
Performance comparison of the MDI-SG algorithm with for computing .
Table 10.
Performance comparison of the MDI-SG algorithm with for computing .
|
|
|
Approximate
total nodes |
Relative
error |
CPU
time(s) |
Approximate
total nodes |
Relative
error |
CPU
time(s) |
| 4(4) |
241 |
|
0.1260 |
1581 |
|
0.3185 |
| 6(6) |
2203 |
|
0.1608 |
40405 |
|
0.4546 |
| 8(8) |
13073 |
|
0.2127 |
581385 |
|
0.6056 |
| 10(10) |
58923 |
|
0.2753 |
5778965 |
|
0.7479 |
| 12(12) |
218193 |
|
0.3402 |
44097173 |
|
1.0236 |
| 14(14) |
695083 |
|
0.4421 |
112613833 |
|
1.2377 |
Table 11.
Performance comparison of the MDI-SG algorithm with for computing .
Table 11.
Performance comparison of the MDI-SG algorithm with for computing .
|
|
|
Approximate
total nodes |
Relative
error |
CPU
time(s) |
Approximate
total nodes |
Relative
error |
CPU
time(s) |
| 4(4) |
241 |
|
0.1664 |
1581 |
|
0.3174 |
| 6(6) |
2203 |
|
0.2159 |
40405 |
|
0.4457 |
| 8(8) |
13073 |
|
0.2526 |
581385 |
|
0.5571 |
| 10(10) |
58923 |
|
0.3305 |
5778965 |
|
0.6717 |
| 12(12) |
218193 |
|
0.3965 |
44097173 |
|
0.8843 |
| 14(14) |
695083 |
|
0.5277 |
112613833 |
|
1.1182 |
Table 12.
Performance comparison of the MDI-SG algorithm with for computing .
Table 12.
Performance comparison of the MDI-SG algorithm with for computing .
|
|
|
Approximate
total nodes |
Relative
error |
CPU
time(s) |
Approximate
total nodes |
Relative
error |
CPU
time(s) |
| 4(4) |
241 |
|
0.1275 |
1581 |
|
0.1657 |
| 6(6) |
2203 |
|
0.1579 |
40405 |
|
0.2530 |
| 8(8) |
13073 |
|
0.1755 |
581385 |
|
0.3086 |
| 10(10) |
58923 |
|
0.1963 |
5778965 |
|
0.3889 |
| 12(12) |
218193 |
|
0.2189 |
44097173 |
|
0.4493 |
| 14(14) |
695083 |
|
0.2459 |
112613833 |
|
0.4864 |
Table 13.
The relationship between the CPU time and parameter N.
Table 13.
The relationship between the CPU time and parameter N.
Integrand
|
r |
m |
d |
Fitting function |
R-square
|
|
3 |
1 |
5 |
|
0.9683 |
|
3 |
1 |
5 |
|
0.9564 |
|
3 |
1 |
5 |
|
0.9877 |
|
3 |
1 |
10 |
|
0.9692 |
|
3 |
1 |
10 |
|
0.9700 |
|
3 |
1 |
10 |
|
0.9630 |
Table 14.
The relationship between CPU time as a function of the dimension d.
Table 14.
The relationship between CPU time as a function of the dimension d.
Integrand
|
r |
m |
s |
|
Fitting function |
R-square
|
|
1 |
10 |
1 |
10(33) |
|
0.9995 |
| |
2 |
10 |
1 |
10(33) |
|
0.9977 |
| |
3 |
10 |
1 |
10(15) |
|
0.9946 |
| |
4 |
10 |
1 |
10(10) |
|
0.9892 |
|
1 |
10 |
1 |
10(33) |
|
0.9985 |
| |
2 |
10 |
1 |
10(33) |
|
0.9986 |
| |
3 |
5 |
1 |
10(15) |
|
0.9983 |
| |
3 |
10 |
1 |
10(15) |
|
0.9998 |
| |
3 |
15 |
1 |
10(15) |
|
0.9955 |
| |
3 |
15 |
2 |
10(15) |
|
0.9961 |
| |
3 |
15 |
3 |
10(15) |
|
0.9962 |
| |
4 |
10 |
1 |
10(10) |
|
0.9965 |
|
3 |
5 |
1 |
10(15) |
|
0.9977 |
| |
3 |
10 |
1 |
10(15) |
|
0.9844 |
| |
3 |
15 |
1 |
10(15) |
|
0.9997 |
|
3 |
10 |
1 |
10(15) |
|
0.9903 |
|
3 |
10 |
1 |
10(15) |
|
0.9958 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).